The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase (original) (raw)
Abstract
Trypanosomatids contain an unusual DNA base J (β-d-glucosylhydroxymethyluracil), which replaces a fraction of thymine in telomeric and other DNA repeats. To determine the function of base J, we have searched for enzymes that catalyze J biosynthesis. We present evidence that a protein that binds to J in DNA, the J-binding protein 1 (JBP1), may also catalyze the first step in J biosynthesis, the conversion of thymine in DNA into hydroxymethyluracil. We show that JBP1 belongs to the family of Fe2+ and 2-oxoglutarate-dependent dioxygenases and that replacement of conserved residues putatively involved in Fe2+ and 2-oxoglutarate-binding inactivates the ability of JBP1 to contribute to J synthesis without affecting its ability to bind to J-DNA. We propose that JBP1 is a thymidine hydroxylase responsible for the local amplification of J inserted by JBP2, another putative thymidine hydroxylase.
INTRODUCTION
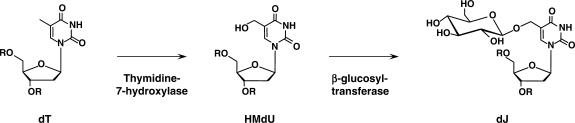
Eukaryotic unicellular kinetoplastid flagellates, such as Trypanosoma and Leishmania species, contain a unique modified base in their nuclear DNA, β-d-glucosylhydroxymethyl-uracil, or J (1–4). In Trypanosoma brucei, J replaces 0.5–1.0% of thymine (T) in nuclear DNA (5) and somewhat lower values have been found in other trypanosomatids (2) and Euglena (6). J is predominantly present in repetitive DNA sequences in T. brucei (7–9), and most prominently in the telomeric GGGTTA repeats (7). Indirect evidence indicates that J is made in two steps (Figure 1): hydroxylation of specific Ts in DNA is followed by addition of a glucose to the hydroxymethyluracil (HMU) formed (10–12). We have tried to find the thymidine hydroxylase and the glucosyl transferase that we expect to be involved in J biosynthesis in cell extracts, by complementation, and by RNAi knockdown of candidate genes, but these attempts have failed thus far. Hence, we have not been able to generate J-less parasites to unravel the function of base J.
Figure 1.
Proposed pathway for J biosynthesis. First, a thymidine hydroxylase catalyzes the formation in DNA of HMdU. Second, HMdU in DNA is converted into J by a putative glucosyl transferase.
A protein that specifically binds to J-containing duplex DNA was found in T. brucei, in Leishmania species and in the insect trypanosome Crithidia fasciculata (13–15). In Leishmania this 90-kDa J-binding protein (JBP1) is essential (16), but in T. brucei it is not. The absence of JBP1 in T. brucei has no effect on growth, repeat stability or gene expression, but results in a 20-fold decrease in J level relative to wild-type cells (17). This decrease in J level was not due to loss of protection by JBP1 against removal of J from DNA and suggested that JBP1 is somehow involved in J maintenance (17).
A large protein with substantial homology with JBP1 (34% identity; 47% similarity) in its N-terminal half turned up in a database search (18). In its C-terminal half, this J-binding protein 2 (JBP2) contains a region homologous to proteins with SWI2/SNF2-like chromatin remodeling activity. Although there is no evidence that JBP2 can bind to J-DNA, DiPaulo et al. were able to show that JBP2 does affect J biosynthesis, because it can induce J synthesis in insect form trypanosomes, which normally do not express JBP2 (18) and are devoid of J (19,20). JBP2 is neither essential in T. brucei, nor in Leishmania (unpublished data).
As both JBP1 and JBP2 are able to increase the level of J in trypanosomes, we hypothesized that both proteins actually catalyze the first, and rate-limiting step in J biosynthesis, the hydroxylation of T in DNA. The homology of the N-terminal halves of JBP1 and JBP2 would then be due to the presence of the thymidine hydroxylase function in this part of the protein. Since the hydroxylation of T resembles the hydroxylation of methylated bases by the repair enzyme AlkB (21,22), we further hypothesized that the thymidine hydroxylase, like AlkB, would be a member of the family of dioxygenases that use Fe2+ and 2-oxoglutarate as cofactors (23–25). In this article, we provide evidence supporting this hypothesis for JBP1. Our results lead to a simple (but still speculative) model for the first step of J biosynthesis. In this model, JBP2 is responsible for the de novo region-specific synthesis of J, whereas JBP1 is responsible for the local maintenance and expansion of J.
MATERIALS AND METHODS
Bioinformatics
JBP1 and JBP2 sequences were obtained by PSI-BLAST (26) searches in the nr database combined with the genome databases of T. brucei (27), T. cruzi and Leishmania major (28) prior to their incorporation into nr. Their homologous regions were aligned using MUSCLE (29). Distant homologies of this common domain with other proteins were sought using the sensitive sequence comparison and fold-recognition tools of the Meta server (30). This server conveniently collects the results of analysis by various modern sequence comparison and fold-recognition methods and applies the consensus 3D-Jury method (31) to them. The HHsearch software (32) for comparison of hidden Markov model representations of protein alignments was also used in conjunction with the domain databases Pfam (33), Smart (32), COG (34) and KOG (35). Structures were browsed in the SCOP database (36).
Mutagenesis
The mutations were made by site-directed mutagenesis using the Quik-Change site-directed mutagenesis kit (Stratagene) following the instruction of the manufacturer. A Lt GFP-JBP1 gene fusion or a Tb GFP-JBP1 gene fusion cloned in a T. brucei or a Leishmania tarentolae expression vector were used as a template. The different oligonucleotides used for the mutagenesis reaction as well as maps of the expression vectors used can be found in Supplementary Figure 1. Mutations were verified by sequencing using an ABI Prism 3700 DNA Analyzer (Applied Biosystems). The GFP-JBP1 constructs were linearized and transfected into the JBP1 null trypanosomes or transfected as episomes in Leishmania following standard protocols (37,38). Leishmania was cultured in SDM-79 medium (39) and the bloodstream form of T. brucei in HMI9 medium (40).
Anti-J-DNA immunoblot
Genomic DNAs of the JBP1 null trypanosomes transfected with the various GFP-JBP1 DNA constructs were isolated, serially diluted in steps of two, denatured, spotted on a nitrocellulose membrane and crosslinked. J levels were determined by incubation with the J antiserum as described before (8). Briefly, the membrane was blocked in skim milk, incubated with a polyclonal J antiserum, washed, incubated with a goat anti-rabbit antibody (horse-radish peroxidase conjugated) and washed, followed by enhanced chemiluminescence detection. The membrane was later hybridized with a telomeric or tubulin DNA probe in a formamide-based hybridization buffer at 42°C overnight. Washes were done with 6× SSC (0.9 M NaCl, 0.09 M Na citrate, pH 7.0), 0.1% SDS. The signal was detected by autoradiography.
Western blotting
Protein lysates were made by spinning down the parasites followed by resuspension of the pellet in Laemmli buffer (41). Proteins were run on 8% SDS-PAGE gel, blotted according to previously described protocols (41). Membranes were blocked in skim milk and incubated with the Lt JBP1 or the Tb JBP1 antibodies as described previously (42).
Protein purification and nuclear extracts
A His tag version of the various JBP1 mutants was generated by PCR and was ligated in a pET-46 Ek/LIC or pET-16b E. coli expression vector (Novagen) following the instructions of the supplier. The constructs were transformed in the BL21(DE3)pLysS or in the Rosetta2(DE3)pLysS E. coli strains (Novagen). Protein expression was induced by addition of IPTG (isopropyl-beta-d-thiogalactopyranoside) at a final concentration of 0.5 mM in Luria-Bertani broth, for 3 h at 37°C or overnight at 15°C. Proteins were purified using His-Select Nickel Magnetic Beads (Sigma) according to the instructions given by the manufacturer and eluted with imidazole. Nuclear extracts of the Leishmania transfected with the Lt GFP-JBP1 mutants were made as described before (13).
Electromobility shift assay
The partially purified proteins were used in an electromobility shift assay following published protocols (13). Briefly, the purified proteins were incubated with radioactively labeled double-stranded J-containing DNA oligonucleotides in a buffer containing 25 mM Hepes-NaOH pH 7.9, 100 mM KCl, 5 mM MgCl2, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 0.1 μg poly-dIdC per μl and 0.5 mg bovine serum albumin (BSA) per ml. Proteins were run on a 4.5% native polyacrylamide gel for 2 h in 0.5× TBE (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA). The gel was dried and the signal detected by autoradiography. The Lt His-JBP1/J-DNA complex was supershifted using an Lt JBP1 antiserum (42). The Lt GFP-JBP1/J-DNA complex was supershifted using a GFP antiserum (Clontech).
Microscopy
GFP localization of the Lt JBP1-GFP fusions in the Tb JBP1 null trypanosomes
Trypanosomes were spun down and washed in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3) and stained with DAPI. The samples were then embedded in low melting 2% agarose in 1× PBS, spotted on a glass slide, sealed with a cover slip and placed at 4°C for a minimum of 1.5 h to ensure little movement of the trypanosomes. Images were acquired using a motorized Zeiss Axioplan 2 and a Axiocam MRm camera which were controlled by the Axiovisons 4.4 software.
GFP localization of the Tb JBP1-GFP in the Tb JBP1 null trypanosomes
Trypanosomes were fixed in 1% formaldehyde in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3). They were then spotted and dried on a glass slide and embedded in 5 μM ToPro3 in Vectashield mounting medium (Vector Laboratories, Inc.). Images were acquired using a Leica-NT confocal microscope and the Leica confocal software.
RESULTS
JBP1 and JBP2 share a conserved domain with the hydroxylase superfamily
JBP1 and JBP2 share amino acid sequence identity over a region of ∼270 residues. Iterative database searches with PSI-BLAST (26) starting from this region pick out the JBP1/2 family but nothing more. Similarly, extensive iterative PSI-BLAST (26) searches starting from nucleic-acid-modifying members of the Fe2+- and 2-oxoglutarate-dependent dioxygenase superfamily, such as AlkB, (43) did not reach JBP1 or JBP2 sequences. In order to explore the possibility of a remote relationship, undetectable even by PSI-BLAST, further analyses were done. Secondary structure predictions for the 270 residue homologous region showed an all-β portion flanked by two largely α-helical parts. We worked with this all-β region, as well as with the entire JBP1/2 conserved region. Both native and derived consensus sequences were submitted for analysis to the Meta server (30). Matches to Fe2+- and 2-oxoglutarate-dependent dioxygenases were indeed present, although rarely top-scoring by individual methods. The most significant result by an individual method was the score of 3.84 (corresponding to a 90% confidence level) given to the central all-β portion of the conserved domain of T. brucei JBP1, in a hit to clavaminate synthase by FUGUE (44). Although scores were below confidence levels (45), the top hits by the 3D-Jury consensus method, when applied to the same sequence, were dominated by the dioxygenases and by proteins with the cupin fold. In the Structural Classification of Proteins Database (36), these architectures are united at the fold level as they share β-helix structure. At large evolutionary distances, it is no surprise that the two are confused by fold-recognition programs. Analysis with the hidden Markov model comparison program HHsearch (46) was also supportive of structural correspondence between JBP1/2 and Fe2+- and 2-oxoglutarate-dependent dioxygenases; dioxygenases monopolized the top hits in the domain databases tested, although the best E-value obtained was only 0.2.
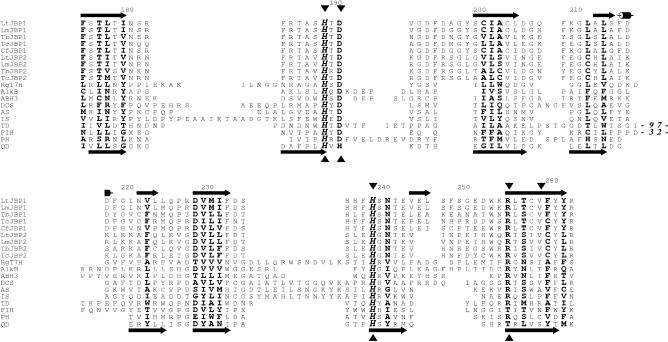
Two arguments further supported the existence of a distant homology. The first was the excellent correspondence between JBP1/2-predicted secondary structure and the actual secondary structures of the dioxygenases in the conserved all-β core (a structural fold comprising of eight β-strands found in all Fe2+/2-oxoglutarate-dependent dioxygenases) (Figure 2). The second and more telling factor was the presence in the JBP1/2 sequences of conserved residues at positions matching the key Fe2+ binding residues of the dioxygenases. Two His residues and one Asp residue are usually involved in Fe2+ binding and are invariant in the aligned JBP1/2 sequences (Figure 2). In addition, an Arg residue, also highly conserved in dioxygenases and important for binding the 2-oxoglutarate, is present in the JBP1/2 sequences (Figure 2). As Figure 2 and recent analyses (23,24) make clear, the all-β core with these four largely conserved residues is the only typical feature within the structures of the dioxygenase superfamily. The structures of family members outside this core cannot generally be superimposed (data not shown), and even within the core substantial insertions are sometimes present (Figure 2). This unusual degree of structural variability probably explains the relatively poor scores for dioxygenase folds produced for the JBP1/2 common domain.
Figure 2.
Sequence alignment of the conserved all-β core (see text) of JBP1 and JBP2 sequences with diverse members, of known structure, from the Fe2+- and 2-oxoglutarate-dependent dioxygenase superfamily. JBP1 and JBP2 sequences are given with species as follows: Tb, T. brucei; Tc, T. cruzi; Lt, L. tarentolae; Lm, L. mexicana and Cf, Crithidia fasciculata. The gi codes in the nr database (Wheeler et al., 2005) are: 62361410 for Lt JBP1, 68124616 for Lm JBP1, 6018041 for Tb JBP1, 71421637 for Tc JBP1, 6018043 for Cf JBP1, 68125217 for Lm JBP2, 71750205 for Tb JBP2 and 71422266 for Tc JBP2. The dioxygenase structures are as follows: DCS, deacetoxycephalosporin C synthase (PDB code 1dcs) (47); AS, anthocyanidin synthase (1gp6); IS, isopenicillin N synthase (1obn); TD, taurine dioxygenase (1os7); FIH, factor inhibiting HIF (1iz3); PH, proline 3-hydroxylase (1e5s); QD, quercetin 2,3-dioxygenase (1y3t). The better conserved positions within the conserved core are shown in bold font with invariant positions additionally italicized. Numbers in the alignment represent the positions of the amino acids in Lt JBP1 and mark the positions of large deletions that are not shown for clarity. The predicted secondary structure of Lt JBP1 is shown above the alignment and the actual secondary structure of quercetin 2,3-dioxygenase (PDB code 1y3t) is shown below the alignment. The long arrows represent β-sheets and the tubes α-helices. Arrowheads below the alignment indicate the four key residues, largely conserved throughout the Fe2+- and 2-oxoglutarate-dependent dioxygenase superfamily that bind the iron (His, Asp) or 2-oxoglutarate (Arg) (23,24). The residues of Lt JBP1 that were mutated to alanines: H189, D191, H239, R255, V259, are shown by the arrowheads above the alignment. Note that there are near identical pairs of sequences for both JBP1 and JBP2 in the T. cruzi genome: only one of each pair is shown in the alignment.
Effects of amino acid replacements in JBP1 on its ability to rescue a double inactivation of JBP1 in Leishmania
If JBP1 is an Fe2+-and 2-oxoglutarate-dependent thymidine hydroxylase catalyzing the first step of J biosynthesis (see Figure 1), mutation of the three residues essential for Fe2+-binding, or the Arg residue important for the binding of 2-oxoglutarate, should abolish the putative hydroxylase activity. To test this, we mutated the His 189, the Asp 191, the His 239 and the Arg 255 of a GFP-tagged version of the JBP1 of L. tarentolae to alanines by site-directed mutagenesis (Figure 2, Supplementary Figure 1). As JBP1 is essential in Leishmania (16), we could not assess the functionality of the mutants by transfecting the constructs in a JBP1 null Leishmania. We therefore attempted to inactivate both wild-type chromosomal JBP1 alleles in cell lines that expressed the wild-type, or a mutant Lt GFP-JBP1 from a rescue plasmid. This was possible with the wild type JBP1 construct, but not with the mutant JBP1 constructs (as seen by the generation of cell lines still expressing the endogenous wild-type protein) (Supplementary Figure 2), even though the mutant protein routed normally to the nucleus of Leishmania (data not shown).
Effects of amino acid replacements in JBP1 on its ability to support J biosynthesis in the JBP1 null trypanosomes
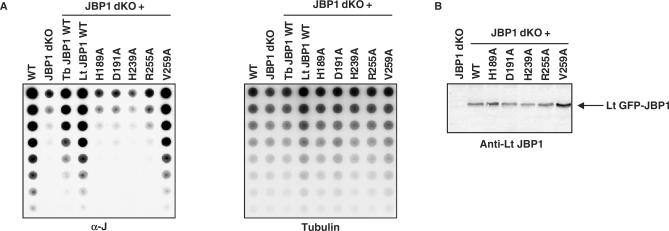
Our Leishmania experiments show that replacement of amino acids critical for the putative hydroxylase function of JBP1 abrogates the ability of JBP1 to complement JBP1 null mutants, but these experiments do not prove that JBP1 converts T into HMU. To test this putative hydroxylase function more directly we turned to T. brucei, in which the absence of JBP1 is not lethal, but results in a 20-fold reduction of J in DNA (17). We transfected wild-type and mutant Lt GFP-JBP1 gene constructs into the JBP1 null trypanosomes and determined the level of J in the genomic DNAs of the transfectants using the J antiserum (8). Wild-type Lt GFP-JBP1 was equally effective as the Tb GFP-JBP1 in raising the J levels of the JBP1 null trypanosomes approximately to wild-type levels (Figure 3A), but the H189A, the D191A, the H239A and the R255A mutants were not (Figure 3A and Table 1). As a control, we replaced Val 259. This residue is conserved in all JBPs analyzed and close to the ‘hydroxylase’ domain, but it is not conserved in members of the Fe2+- and 2-oxoglutarate-dependent dioxygenase superfamily (Figure 2). Its replacement might therefore not affect the putative hydroxylase function. Indeed, the Val 259 mutant protein did complement the JBP1 null trypanosomes and resulted in wild-type J levels (Figure 3A and Table 1).
Figure 3.
Determination of J levels in the DNA of JBP1 null trypanosomes expressing the Lt JBP1 mutants. (A) Genomic DNA of wild-type T. brucei bloodstream form 427 line (WT), JBP1 null trypanosomes (JBP1 dKO) and JBP1 null trypanosomes expressing the wild-type Tb GFP-JBP1 fusion (Tb JBP1 WT), the wild-type Lt GFP-JBP1 fusion (Lt JBP1 WT) or the mutant Lt GFP-JBP1 fusions (H189A, D191A, H239A, R255A, V259A) were serially diluted in steps of two, denatured, spotted on a nitrocellulose membrane and incubated with a polyclonal J antiserum (left panel). The membrane was hybridized with a tubulin DNA probe as a loading control (right panel). (B) Western blot on lysates of the JBP1 null trypanosomes (JBP1 dKO) and the JBP1 null trypanosomes expressing the wild-type (WT) and the mutant (H189A, D191A, H239A, R255A, V259A) Lt GFP-JBP1 fusions using an Lt JBP1 antiserum.
Table 1.
Characteristics of the Lt JBP1 mutants. The position of the amino acids replaced in these mutants is indicated in Figure 2
| Lt JBP1 mutants | WT | H189A | D191A | H189A + D191A | H239A | R255A | V259A |
|---|---|---|---|---|---|---|---|
| Rescues J levels in JBP1 null T. brucei | Yes | No | No | No | No | No | Yes |
| Nuclear location in T. brucei | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Binds to J-DNA | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
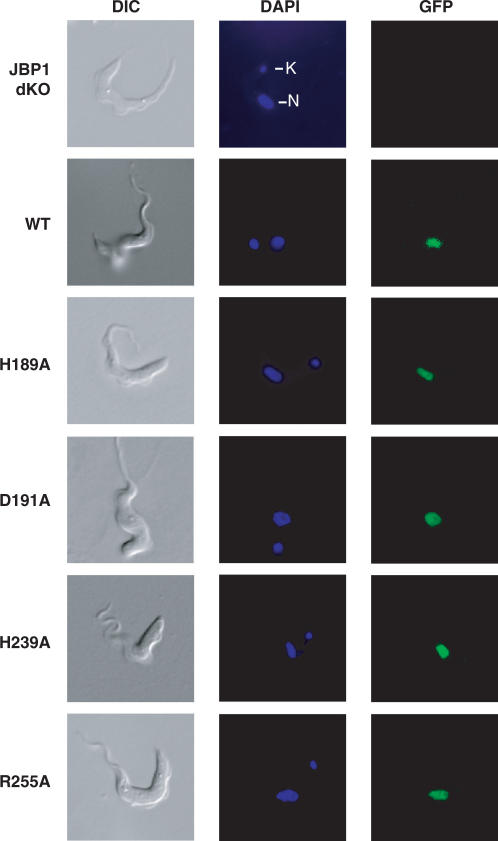
We also verified that the putative Fe2+-binding His 213 and Asp 215 of T. brucei JBP1 (corresponding to the His 189 and the Asp 191 of Lt JBP1) are essential for the putative hydroxylase function. Replacement of these residues by serines resulted in mutant proteins that failed to restore the J-level in the JBP1 null trypanosomes to wild-type levels, whereas a wild-type Tb GFP-JBP1 fusion did (Supplementary Figure 3, Supplementary Table 1). The expression levels of the wild type and mutants of the Lt and Tb GFP-JBP1 fusions were comparable, excluding the possibility that the mutants did not complement the J levels because they were poorly expressed in the parasite (Figure 3B, data not shown). All wild type and mutant GFP-JBP1 fusions of Lt JBP1 and Tb JBP1 tested were targeted to the nucleus of T. brucei ruling out the possibility that the mutants were not functional because they were not correctly routed to the nucleus (Table 1, Figure 4, Supplementary Table 1 and Supplementary Figure 4).
Figure 4.
Localization of mutant Lt GFP-JBP1 in JBP1 null trypanosomes. JBP1 null trypanosomes (JBP1 dKO) were transfected with the wild-type (WT) or the mutant (H189A, D191A, H239A, R255A) Lt GFP-JBP1 fusions, DAPI-stained and visualized live on agarose slides by microscopy. DAPI staining shows the location of the nucleus (N) and of the kinetoplast (mitochondrial DNA; K).
Effects of amino acid replacements in JBP1 on its ability to bind to J-DNA
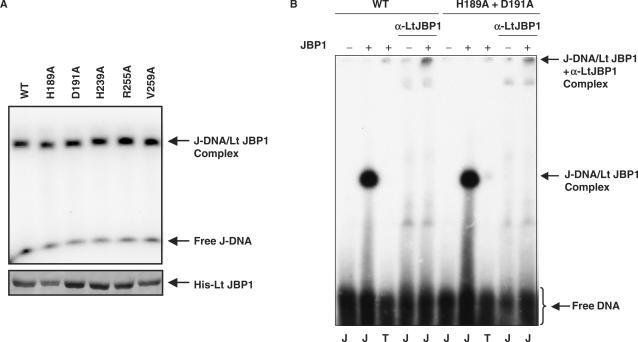
We previously found that mutant versions of JBP1 that lost their J-binding property do not restore the J levels in the JBP1 null trypanosomes (our unpublished data). The fact that the putative hydroxylase mutants did not rescue the JBP1 inactivation in Leishmania and did not restore the J levels in the JBP1 null T. brucei could therefore be due to their inability to bind to J-DNA. To exclude this possibility, we purified His-tagged recombinant wild-type and mutant Lt JBP1 from E. coli (Figure 5A) and tested these for J-binding in electromobility shift assays using radioactively labeled double-stranded J-containing DNA oligonucleotides (13,14). A J-DNA-dependent bandshift was obtained with the wild type as well as with all the mutated versions of Lt JBP1 we tested (Figure 5 and Table 1). The shifted J-DNA band, which contains a 1:1 ratio of JBP1:DNA, could be supershifted using an Lt JBP1 antiserum (42), implying that the shift was indeed due to JBP1 (Figure 5B). As reported previously (13,14), purified JBP1 does not bind to DNA without J under our assay conditions (Figure 5B). The results in Figure 5 indicate that wild-type and mutant forms of Lt JBP1 have similar affinity for J-DNA. Protein titration of the double mutant (H189A plus D191A) supports this conclusion (Supplementary Figure 5).
Figure 5.
Determination of the J-binding activity of purified Lt JBP1 mutants by electromobility shift assay. (A) Upper panel: Electromobility shift assay with the purified His-tagged wild-type (WT) and mutant (H189A, D191A, H239A, R255A, V259A) Lt JBP1 proteins. Proteins (10 fmol) were incubated with a radioactively labeled double-stranded oligonucleotide containing J (J-DNA) (10 fmol) and run on 4.5% native acrylamide gel. The gel was dried and the signal detected by autoradiography. The bands corresponding to the J-DNA/Lt JBP1 and the free J-DNA are indicated. Lower panel: Segment of a Coomassie-brilliant-blue-stained acrylamide gel with the His-tagged wild-type (WT) and mutant (H189A, D191A, H239A, R255A, V259A) Lt JBP1 proteins purified from E. coli. Only the segment of the gel with the His-tagged Lt JBP1 band is shown. (B) Example of a determination of the J-binding activity of mutant Lt JBP1 by electromobility shift assay. His-tagged wild-type (WT) and H189A + D191A mutant Lt JBP1 proteins were purified from E. coli and incubated with a radioactively labeled double-stranded oligonucleotide containing J (J) or lacking J (T) in the presence or absence of a Lt JBP1 antibody (α-Lt JBP1). Samples were run on a 4.5% native acrylamide gel. The gel was dried and the signal detected by autoradiography. Lanes without recombinant protein are indicated by ‘–’. The position of the free DNA, the J-DNA/Lt JBP1 complex and the J-DNA/Lt JBP1 + α-Lt JBP1 complex is indicated.
To exclude the possibility that the GFP-tagged versions of the mutant proteins produced in Leishmania would not bind to J-DNA, whereas the His-tagged versions produced in E. coli would (Figure 5), we made nuclear extracts of the Leishmania cell lines expressing the H189A, the D191A and the H189A + D191A Lt GFP-JBP1 mutant proteins and tested these for J-binding activity in electromobility shift assays. Mutant proteins bound to J-DNA are shown in Supplementary Figure 6. We conclude from these results that the H189A, the D191A, the H239A and the R255A Lt JBP1 mutants failed to rescue the J levels in the JBP1 null trypanosomes because they lost their putative thymidine hydroxylase activity, not because they can no longer bind to J-DNA.
The relation of JBP1 and JBP2 to other members of the dioxygenase superfamily
Given the experimental evidence for the correctness of the dioxygenase fold assignment for JBP1 and JBP2, the relationships of JBP1/2 with other branches of the dioxygenase superfamily were explored further. Do JBP1/2 bear a particular similarity with other enzymes that modify nucleic acids such as AlkB and, especially, the thymine 7-hydroxylase, cloned recently from Rhodotorula glutinis (48), which also converts T into HMU? Alternatively, are they unexpectedly more closely related to other enzymes such as deacetoxycephalosporin C synthase (DCS) which featured in the top hits of several fold-recognition programs? These questions were addressed using PSI-BLAST and HHsearch. PSI-BLAST runs to convergence when initiated with R. glutinis T7H or AlkB, but failed to reach JBP1/2 sequences, indicating that there is only a distant relationship between JBP1/2 and these nucleic-acid-modifying dioxygenases. Interestingly, R. glutinis T7H bears only a distant relationship to AlkB, being more closely related to enzymes such as isopenicillin N synthase and gibberellin 20-oxidase (data not shown). Also, JBP1/2 sequences were not retrieved in PSI-BLAST searches starting from DCS. We then chose 22 sequences to cover the full range of the dioxygenase superfamily in the Pfam database PF03171 (33) and initiated PSI-BLAST runs with each. No run retrieved JBP1/2 sequences, confirming that only a very distant evolutionary relationship exists between JBP1/2 and all other dioxygenase superfamily members characterized to date. HHsearch-based comparison of a JBP1/2 conserved domain alignment with alignments of 55 AlkB sequences, 11 DCS proteins and 8 T7H sequences told a similar tale: E-values were similar—0.01 for T7H, 0.02 for AlkB and 0.05 for DCS—and indicative of only very distant homologies. Therefore, it appears that the AlkB, T7H and JBP1/2-catalyzed thymidine hydroxylase activities, all base modifying, represent three phylogenetically distant branches within the Fe2+- and 2-oxoglutarate-dependent dioxygenase superfamily.
DISCUSSION
Our results show that JBP1 has the properties to be expected of a thymidine hydroxylase catalyzing the first step in J biosynthesis:
- JBP1 has all the sequence/structural hallmarks of an Fe2+- and 2-oxoglutarate-dependent dioxygenase/hydroxylase.
- Replacement of amino acids predicted to be essential for hydroxylase function abolishes the ability of JBP1 to stimulate J synthesis. We have mutated the His and Asp residues essential for Fe2+ binding and the Arg important for 2-oxoglutarate binding in other members of this dioxygenase family. Replacement of these residues with Ala or Ser inactivates the putative hydroxylase function, as shown by the fact that the mutated JBP1 is unable to complement the JBP1 null trypanosomes. The mutations were introduced in a GFP-JBP1 fusion gene, allowing us to show that the mutant fusion protein is made in T. brucei and Leishmania cells in similar amount as wild-type protein and that it is properly routed to the nucleus.
- Replacement of residues essential for the putative hydroxylase function does not affect the ability to recognize and bind J-DNA. In experiments with purified mutant and wild-type recombinant Lt JBP1 proteins, we have shown that the affinity of the JBP1 mutants for J-DNA is not substantially altered. Moreover, the GFP-tagged mutant proteins still bind to J-DNA in extracts of L. tarentolae as shown by bandshift of J-containing duplex oligonucleotides. The J-DNA binding by JBP1 is therefore independent of the putative hydroxylase function.
- Absence of JBP1 in T. brucei reduces the J level in nuclear DNA 20-fold (17). Although this could be an indirect effect, the result is fully compatible with a hydroxylase function for JBP1. This also holds for the ability of JBP1 to promote retention of J introduced into DNA by growing trypanosomes in medium containing HMdU (17). This nucleoside is randomly incorporated into DNA, both in bloodstream form and in insect-form trypanosomes (10), and in the presence of JBP1 this extra J is more sluggishly lost than in its absence (17).
Although these results support our hypothesis that JBP1 is an Fe2+-and 2-oxoglutarate-dependent dioxygenase that catalyzes the hydroxylation of T in DNA to yield HMU, our attempts to demonstrate hydroxylase activity with purified JBP1 have failed thus far (see Supplementary Data, Materials and Methods for a detailed description of these experiments). Dioxygenases belonging to this enzyme family are often able to catalyze the oxidative decarboxylation of 2-oxoglutarate in the absence of primary substrate (24), but we have been unable to detect succinate formation or oxygen consumption by JBP1 with or without J-containing oligonucleotide substrate, using a highly purified His-tagged JBP1 preparation from C. fasciculata. Negative results were also obtained in another in vitro assay in which the conversion of T into HMU by Cf JBP1 or Lt JBP1 (in the presence of Fe2+, 2-oxoglutarate and a J-DNA oligonucleotide) was analyzed with a specific DNA base excision glycosylase for HMU (11). Finally, we looked in vain for thymidine hydroxylase activity in E. coli overproducing JBP1 of L. tarentolae using the HMU-specific glycosylase.
These negative results might raise the question whether JBP1 is really a hydroxylase or only affecting thymidine hydroxylation indirectly, e.g. by recruiting a hydroxylase to the DNA. We consider this highly unlikely. First, the possibility that a domain shared by members of the Fe2+- and 2-oxoglutarate-dependent dioxygenase superfamily is present in a protein recruiting a thymidine hydroxylase is small. Second, the likelihood that replacement of conserved amino acids in this domain affects the recruitment of the hydroxylase is remote. Third, it is hard to believe that residues critical for hydroxylase function would be conserved over the long evolutionary time separating T. brucei and Crithidia, if these residues would not serve the same function as in other members of the dioxygenase family. The fact that we have not yet been able to demonstrate hydroxylase activity in our JBP1 preparations, does not mean much, like most negative results. Nothing is known yet about the nature of the DNA substrate for JBP1. It may simply require longer DNA than the oligonucleotides used thus far, DNA packaged in chromatin, or additional cofactors, etc.
The analysis of JBP2 remains to be carried out, but the available evidence supports a hydroxylase function for JBP2 as well: Its N-terminal half contains all conserved elements characteristic of Fe2+- and 2-oxoglutarate-dependent dioxygenases. Absence of JBP1 in T. brucei does not result in a complete loss of J, indicating the presence of a second hydroxylase, i.e. JBP2 (17). Expression of JBP2 in insect form T. brucei (which does not normally contain any J) results in the formation of J (18).
On the basis of our results, we propose that the de novo modification of T to HMU is exclusively catalyzed by JBP2 and that it is JBP2 that determines the region, and context specificity of J in nuclear DNA (18). The low level of HMU made by JBP2 is then converted into J and locally amplified 20-fold by JBP1 (17). Although this procedure for HMU formation looks unduly complex, the need for two enzymes could stem from the requirement to insert HMU in very specific places in repetitive DNA sequences packed in chromatin. The initial precise recognition of this sequence may be a slow process, or the number of recognition sites for JBP2 might be limited. This could explain that a second enzyme is required to finish the job and raise the J level further, making use of the HMU already inserted by JBP2 (after the conversion of HMU into J). The presence of two hydroxylases, each with a unique sequence and substrate specificity, may explain why we have never been able to find a common theme in the sequences containing J, even though these sequences are highly non-random (7).
Supplementary Material
[Supplementary Material]
ACKNOWLEDGEMENTS
We thank Dr Saara Vainio, Dr Fred van Leeuwen, Dr Bas van Steensel, Dr Hein te Riele and Dr Jos Jonkers for critical reading of this manuscript. This work was funded by a grant from the Netherlands Organization for Scientific Research and Chemical Sciences (NWO-CW) to P.B., NIH grant A1063523 to R.S. and NIH grant GM063584 to R.P.H. We are indebted to Torsten Ochsenreiter, Lenny Brocks and Lauran Oomen, for help with microscopy. The authors have no financial interest conflicting with this article. Funding to pay the Open Access publication charge was provided by the Netherlands Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gommers-Ampt JH, Van Leeuwen F, De Beer ALJ, Vliegenthart FG, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. β-D-Glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan. Trypanosoma brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 2.Van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. Beta-D-Glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J. 1995;9:1034–1042. doi: 10.1096/fasebj.9.11.7649402. [DOI] [PubMed] [Google Scholar]
- 4.Borst P, Van Leeuwen F. Beta-D-glucosyl-hydroxymethyluracil, a novel base in African trypanosomes and other Kinetoplastida. Mol. Biochem. Parasitol. 1997;90:1–8. doi: 10.1016/s0166-6851(97)00170-9. [DOI] [PubMed] [Google Scholar]
- 5.Van Leeuwen F, De Kort M, van der Marel GA, Van Boom JH, Borst P. The modified DNA base beta-D-glucosyl-hydroxymethyluracil confers resistance to micrococcal nuclease and is incompletely recovered by 32P-postlabeling. Anal. Biochem. 1998;258:223–229. doi: 10.1006/abio.1998.2587. [DOI] [PubMed] [Google Scholar]
- 6.Dooijes D, Chaves I, Kieft R, Dirks-Mulder A, Martin W, Borst P. Base J originally found in Kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel GA, Van Boom JH, Borst P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, Van Boom JH, Borst P. Localisation of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Leeuwen F, Kieft R, Cross M, Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by β-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;109:133–145. doi: 10.1016/s0166-6851(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 10.Van Leeuwen F, Kieft R, Cross M, Borst P. Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Cell. Biol. 1998;10:5643–5651. doi: 10.1128/mcb.18.10.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulbert S, Cross M, Boorstein RJ, Teebor GW, Borst P. Expression of the human DNA glycosylase hSMUG1 in Trypanosoma brucei causes DNA damage and interferes with J biosynthesis. Nucleic Acids Res. 2002;30:3919–3926. doi: 10.1093/nar/gkf533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulbert S, Eide L, Seeberg E, Borst P. Base J, found in nuclear DNA of Trypanosoma brucei, is not a target for DNA glycosylases. DNA Repair. 2004;3:145–154. doi: 10.1016/j.dnarep.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Cross M, Kieft R, Sabatini R, Wilm M, De Kort M, van der Marel GA, Van Boom J, Van Leeuwen F, Borst P. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J. 1999;18:6573–6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatini R, Meeuwenoord N, Van Boom JH, Borst P. Recognition of base J in duplex DNA by J-binding protein. J. Biol. Chem. 2002;277:958–966. doi: 10.1074/jbc.M109000200. [DOI] [PubMed] [Google Scholar]
- 15.Sabatini R, Meeuwenoord N, Van Boom JH, Borst P. Site-specific interactions of JBP with base and sugar moieties in duplex J-DNA. Evidence for both major and minor groove contacts. J. Biol. Chem. 2002;277:28150–28156. doi: 10.1074/jbc.M201487200. [DOI] [PubMed] [Google Scholar]
- 16.Genest PA, ter Riet B, Dumas C, Papadopoulou B, Van Luenen HG, Borst P. Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania. Nucleic Acids Res. 2005;33:1699–1709. doi: 10.1093/nar/gki304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross M, Kieft R, Sabatini R, Dirks-Mulder A, Chaves I, Borst P. J-binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol. 2002;46:37–47. doi: 10.1046/j.1365-2958.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- 18.DiPaolo C, Kieft, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol. Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Blundell PA, Van Leeuwen F, Brun R, Borst P. Changes in expression site control and DNA modification in Trypanosoma brucei during differentiation of the bloodstream form to the procyclic form. Mol. Biochem. Parasitol. 1998;93:115–130. doi: 10.1016/s0166-6851(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 20.Van Leeuwen F, Dirks-Mulder A, Dirks RW, Borst P, Gibson W. The modified DNA base beta-D-glucosyl-hydroxymethyluracil is not found in the tsetse fly stage of Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;94:127–130. doi: 10.1016/s0166-6851(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 21.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 22.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygeneases. Genome Biol. 2001;2:research 7.1–research 7.8. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 25.Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J. Inorg. Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 28.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bujnicki JM, Elofsson A, Fischer D, Rychlewski L. Structure prediction meta server. Bioinformatics. 2001;17:750–751. doi: 10.1093/bioinformatics/17.8.750. [DOI] [PubMed] [Google Scholar]
- 31.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 32.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11:3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carruthers VB, Van der Ploeg LHT, Cross GAM. DNA-mediated transformation of bloodstream form Trypanosoma brucei. Nucleic Acids Res. 1993;21:2537–2538. doi: 10.1093/nar/21.10.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brun R, Schönenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi defined medium. Acta Tropica. 1979;36:289–292. [PubMed] [Google Scholar]
- 40.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 41.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 42.Toaldo CB, Kieft R, Dirks-Mulder A, Sabatini R, Van Luenen HG, Borst P. A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1. Mol. Biochem. Parasitol. 2005;143:111–115. doi: 10.1016/j.molbiopara.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falnes PO, Rognes T. DNA repair by bacterial AlkB proteins. Res. Microbiol. 2003;154:531–538. doi: 10.1016/S0923-2508(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 44.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 45.Ginalski K, Rychlewski L. Detection of reliable and unexpected protein fold predictions using 3D-Jury. Nucleic Acids Res. 2003;31:3291–3292. doi: 10.1093/nar/gkg503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 47.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr, Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochim. Biophys. Acta. 2005;1723:256–264. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]