A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs (original) (raw)
Abstract
Microarrays to examine the global expression levels of microRNAs (miRNAs) in a systematic in-parallel manner have become important tools to help unravel the functions of miRNAs and to understand their roles in RNA-based regulation and their implications in human diseases. We have established a novel miRNA-specific microarray platform that enables the simultaneous expression analysis of both known and predicted miRNAs obtained from human or mouse origin. Chemically modified 2′-_O_-(2-methoxyethyl)-(MOE) oligoribonucleotide probes were arrayed onto Evanescent Resonance (ER) microchips by robotic spotting. Supplementing the complementary probes against miRNAs with carefully designed mismatch controls allowed for accurate sequence-specific determination of miRNA expression profiles obtained from a panel of mouse tissues. This revealed new expression signatures of known miRNAs as well as of novel miRNAs previously predicted using bioinformatic methods. Systematic confirmation of the array data with northern blotting and, in particular, real-time PCR suggests that the described microarray platform is a powerful tool to analyze miRNA expression patterns with rapid throughput and high fidelity.
INTRODUCTION
miRNAs play important roles in a variety of physiological processes (1–6). Determination of spatial and temporal expression of miRNAs is a useful means to gain insight into the function of these small biomolecules. Northern blot analysis and molecular cloning strategies have been frequently used to determine approximate miRNA expression levels in biological samples (7–12). Recently, more quantitative methodologies such as the Invader assay (13), bead-based hybridization assays (14) and RT-PCR assays (15–19) have been developed. The number of experimentally verified miRNAs is continuously growing and it is predicted that the total number of mammalian miRNAs will be in excess of more than a thousand (3), possibly tens of thousands (20).
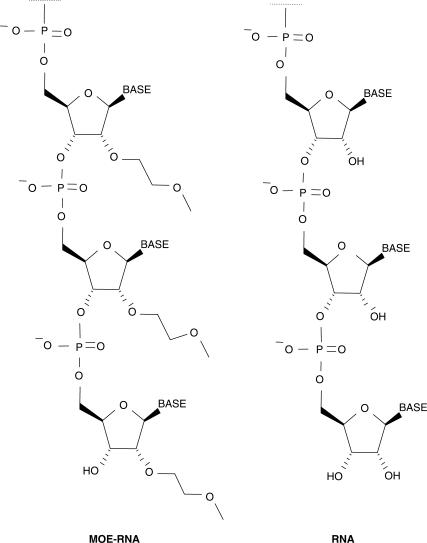
It is likely that microarrays will become the method of choice for global miRNA profiling studies since they are able to screen large numbers of molecules simultaneously and allow for flexible probe design. Due to the short length of miRNAs, the principle challenge in the generation of microarray-based miRNA assays is the optimization of hybridization conditions that offer maximal binding affinity without compromising specificity. Most miRNA array platforms that have been described to date make use of DNA-oligonucleotides as capture probes (21–28). However, several nucleic acid analogs have emerged over the last years that demonstrate more favorable hybridization characteristics as compared to standard DNA-based probes. Some of the analogs, including peptide nucleic acids (PNA) (29) and locked nucleic acids (LNA) (30), have been used on classical mRNA microarrays. Recently, Castoldi et al. have described the application of mixed LNA/DNA-modified capture probes in array-based profiling of miRNAs and have reported superior sensitivity over conventional DNA-based miRNA arrays (31). From a technical point of view, one of the most amenable oligonucleotide modifications comprises chemical manipulations at the 2′-position of the ribose. In particular, the 2′-_O_-(2-methoxyethyl)-(MOE) moiety (Figure 1), which has been the subject of extensive research in antisense-based technology, was shown to bind with high affinity and specificity to natural RNA (32,33). These properties encouraged us to apply MOE-modified oligoribonucleotides as capture probes for the microarray-based detection of miRNAs. We have recently introduced a novel generation of microchips that rely on evanescent resonator (ER) technology which have shown improved sensitivity in mRNA expression experiments when compared with conventional microarrays due to their unique optical features (34).
Figure 1.
Structure of chemically modified 2′-_O_-MOE-RNA compared with natural RNA.
In this article, we describe a miRNA microarray which combines the advantages of ER-based microarray detection with the favorable properties of MOE-oligoribonucleotide as capture probes. The MOE-probes were arranged on the ER-microarrays by robotic spotting which results in a flexible platform which can be rapidly updated in order to deal with newly identified validated or predicted miRNA candidates. Extensive validation studies were performed by comparing miRNA array data generated from human cell lines and mice tissues with conventional Northern blots as well as semi-quantitative RT-PCR. miRNA expression profiles obtained with the MOE-ER array revealed novel signatures for several known miRNAs as well as for computationally predicted miRNAs, demonstrating the suitability of this novel microarray platform for robust high-throughput analysis of miRNA expression.
MATERIALS AND METHODS
Design and synthesis of MOE oligonucleotides
MOE capture probes were designed for 200 human or mouse miRNAs including 50 miRNAs that have been predicted in silico but not experimentally confirmed yet (35). The length of these miRNAs varies from 20 to 24 nucleotides (nts). Since cloning and sequencing procedures employed to identify miRNAs often cannot precisely define the 5′ and 3′ extremities of the miRNA, the MOE capture probes were complementary to the central 19 nts of the miRNA. For 20-nt-long miRNAs, the capture probe was complementary to 19 5′-proximal nucleotides of the miRNA, for 21-nt-long and longer miRNAs, the capture probes were complementary to nucleotides 2–20 of the miRNA.
For each perfectly matched (0MM) probe, single nt mismatch (1MM) control oligomers were designed by replacing an existing base at a non-terminal position by applying one of the four base permutation rules: A → C, C → A, U → G or G → U. To avoid cross-matches of the controls, we aligned all probe sequences to all non-parental miRNA probes using the Smith–Waterman algorithm from the FASTA 3.4 suite (expect value = 20, ungapped, both strands, base scoring +5/−4. G–U base pairs were not considered as mismatches but as a partial match with positive contribution of +2 to the score). The position of the mismatch was initially central (base number 10) and the smallest cross-match distance was extracted. The replacement position was then gradually shifted out of the center towards the ends and the homology search was repeated in order to identify a position of the replacement which increases the distance of the probe from the closest non-parental miRNA sequence. Terminal positions (1,19) were not selected by the algorithm. An additional 2 nt mismatch (2MM) control oligomer was generated for each accepted 1MM control sequence by changing a second base using the same algorithm. 2′-MOE-modified oligonucleotides were prepared as described elsewhere (36).
Cell culture
Human HeLa cells were grown at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% FCS and 2 mM l-Glutamine.
Preparation of labeled miRNAs from human HeLa cells or mouse organs
According to the protocols provided by the manufacturer, approximately 1.5 × 107 cells were washed with PBS and total RNA was isolated using Trizol® Reagent (Life-TechnologiesTM, cat no: 15596-018) and genomic DNA potentially present in the RNA fraction was digested using RNase-free DNase I (Ambion, cat no: 2222). Following phenol/chloroform extraction, total RNA was ethanol precipitated, resuspended in RNase-free water and 100 µg of RNA was applied to a RNeasy® mini spin column (Qiagen, cat no: 74104) following the RNA cleanup protocol of the manufacturer. The RNA-species smaller than ∼200 nts contained in the flow through were ethanol-precipitated, resuspended in RNase-free water. The amount of recovered RNA was estimated by measuring optical density at 260 nm.
Mouse organs were collected from 0.9% perfused 18-week-old BL6 male mice. The organs were homogenized in the presence of Trizol (1 ml Trizol/100 mg tissue) using a polytron homogenizer. RNA recovery and DNA digestion was performed as described above.
MiRNAs present in the column purified RNA fraction were labeled by incorporation of a single Cy5 label at the 3′-end of RNA molecules as described by Garnier et al. (37). In brief, 9 µg RNA dissolved in 9 µl RNase-free water was oxidized to dialdehyde by adding 1 µl freshly prepared 100 mM sodium periodate followed by an 1 h incubation at room temperature in the dark. Excess of oxidant was removed by adding 1 µl of a 200 mM solution of sodium sulfite and incubation for 20 min at room temperature. After adding 12 µl of 50 mM sodium acetate buffer (pH 4), 5 µl of 20 mM ethylenediamine hydrochloride (pH 7.2) was added to the oxidized RNA. The reaction mixture was incubated for 1 h at 37°C, and the aldimine bond between the RNA and the spacer was reduced with 2 µl freshly prepared 200 mM sodium cyanoborohydride in acetonitrile. The mixture was incubated for 30 min at room temperature and precipitation was effected with 2 volumes of 2% lithium perchlorate in acetone for 1 h at 0°C. The sample was spun at 14 000 rpm for 45 min at 4°C and, after removal of the supernatant, the RNA pellet was washed twice with acetone and air dried. Conjugation was carried out by resuspending the amino-modified RNA in 5 µl DEPC-treated water and adding 5 µl of 30 mM Cy5 N-hydroxysuccinimidyl active ester in 1 M sodium phosphate buffer (pH 7.8). After incubation for 1 h at room temperature in the dark, the RNA was precipitated with 2.5 volumes of 100% ice-cold ethanol for 1 h at −20°C. The sample was spun at 14 000 rpm for 30 min at 4°C and after removal of supernatant, the Cy5-RNA pellet was washed twice with ice-cold 70% aqueous ethanol and air dried. Labeled RNA was quantified by measuring optical density at 260 nm.
Preparation of microarrays
The ER microchip transducers (dimensions 25 × 75 mm) were manufactured by Unaxis AG, Liechtenstein, according to specifications described elsewhere (38). Glass slides were nanostructured in order to create a fully corrugated surface (grating period 360 nm). Subsequently, the corrugated surface was coated with Ta2O5 as dielectric material (150 nm thickness, refractive index material n = 2.1). Chips were printed essentially as described (34,38). Briefly, MOE-oligomer probes were spotted as 10 µM solutions in 4 × SCC containing 0.001% Sarcosyl in quadruplicates on the chip surface. A MicroGrid Arrayer (Genomic Solutions, Inc.) equipped with SMP 3 pins (Telechem, Inc.) was used for the production of the microarrays. The dimensions of the printed area were approximately 2 cm2. Printing was carried out at 23°C at humidity adjusted to 55–60%. A Tecan LS scanner in scatter mode (red laser excitation, fluorescence filter removed) was used to check the presence of the spots printed as quality control.
Hybridization and scanning
An HS 4800 Hybridization station from Tecan, Inc. was used for the hybridization of microarrays. First, a pre-wash consisting of 2 cycles was performed with washbuffer (WB) containing 20 mM Na-phosphate pH 6.5 (>99%, Merck), 50 mM NaCl (≥99.5%, Fluka), 1 mM EDTA (Invitrogen) and 0.1% (w/v) SDS (Fluka), each wash for 20 s at 75°C, followed by two washes for 20 s with WB at 50°C.
Typically, 0.15–8.0 µg size-fractionated HeLa RNA and 2 µg size-fractionated mouse organ RNA dissolved in 100 µl hybridization buffer [70% (v/v) ExpressHyb (Clontech BD, cat no: 8015-1) in formamide (Fluka, cat no: 47671) containing 100 µg/ml sonicated salmon sperm DNA (Stratagene)] was injected into the flow chambers in liquid phase at 50°C and the samples were agitated at the same temperature for one minute. The temperature was then increased to 75°C (for 10 min) to denature any hybridized strands. Subsequently, the temperature was adjusted to 52°C and hybridization was carried out for 16 h with agitation. A post-wash consisted of four 20 s washes with WB at 52°C followed by an additional wash at 23°C. Finally the slides were washed three times with WB, dried under a stream of N2, and scanned immediately with an unmodified Agilent LS fluorescence scanner (gain 80%). An external spike was used as landing mark for the grid alignment and could be optionally used as control for the hybridization experiments.
Image analysis
The fluorescence images were analyzed by means of Array Pro (Mediacybernetics, Inc. US). Net signals were calculated by subtracting the local background ring median from trimmed mean of each spot (20% trimming on both sides of the intensity histogram). Typically, the data represent a mean value of the quadruplicates printed for each sequence onto the slides. No normalization was employed. Contaminated spots were manually excluded from analysis, in such cases the mean value was based on less than four spots. Raw data were analyzed in GENESPRING software, version 7.0 (Agilent Technologies/Silicon Genetics, Redwood City, CA). The observed mean background standard deviation was typically approximately 15 counts (cts). In general, miRNAs were considered as absent if the fluorescence intensity of the perfectly matched probe was not 2-fold higher than the mean background fluorescence. A particular miRNA was considered as present if the fluorescence intensity from the perfectly matched probe was found to be greater than the corresponding 1 and 2MM control probes.
Northern blotting
Mir-141 (5′-ccatctttaccagacagtgtta-3′), mir-194 (5′-tccacat-ggagttgctgttaca-3′), mir-31 (5′-cagctatgccagcatcttgcct-3′), mir-215 (5′-gtctgtcaaatcataggtcta-3′) probes were labeled with [γ-32P]ATP (MP Biomedicals, 7000 Ci/mmole) and T4 polynucleotide kinase (New Englands Biolabs). Pir-5/mir-499 (5′-ttaaacatca-ctgcaagtcttaa-3′) and U6 snRNA (5′-gccatgctaatcttctctgta-tc-3′) probes were 5′-digoxigenin-labeled. All probes were obtained from Microsynth GmbH.
For Northern analysis, 5–30 µg of total tissue RNA was separated on denaturing 15% polyacrylamide/7M urea gels run in TBE and transferred to Hybond N + membranes (Amersham) in TBE using semi-dry conditions (Biorad). After UV-cross-linking at 120 mJ, the membranes were equilibrated for 1h at 37°C in 0.75 M NaCl, 3.75 mM Na2EDTA, 50 mM NaH2PO4 (pH 7.4), supplemented with 0.04% Ficoll Type 400, 0.04% polyvinlylpyrrolidone, 0.04% BSA, 20 µg/ml salmon sperm DNA and 4 µg/ml yeast tRNA. 32P-labeled probes were hybridized to the membranes over-night at 37°C in the above buffer. After washing of the membranes for 10 min in 2 × SSC containing 0.1% SDS, 30 min in 0.1 × SSC containing 0.1% SDS and 5 min in 2 × SSC, radioactivity was analyzed using a Storm 860 PhosphorImager (Molecular Dynamics). Northern blots hybridized with digoxigenin-labeled probes were washed, blocked with blocking solution (Roche, cat no: 1585762) and incubated with an alkaline phosphatase conjugated anti-digoxigenin antibody (Roche, cat no: 1093274) according to the manufacturer's protocol. Membranes were incubated in ready-to-use CDP-Star (Roche, cat no: 2041677) and chemiluminescense was detected using the ChemiDoc XRS (BioRad).
miRNA stem-loop Real-Time PCR analysis
Quantification of miRNAs by TaqMan Real-Time PCR was carried out as described by the manufacturer (Applied Biosystems). Briefly, 0.1–1 ng of template RNA was reverse transcribed using stem-loop primer (ABI, cat no: 4365409) high capacity cDNA Archive Kit (ABI, cat no: 4322171) and RNAase inhibitor (ABI, cat no: N8080119). The 15 µl reactions were incubated in a thermocycler (Applied Biosystems 9800) in 96-well plates for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C.
For the PCR reaction, 2 µl of RT-product, equivalent to approximately 0.13 ng total RNA, was mixed with the Universal PCR Master mix (ABI, cat no: 4324018) and the TaqMan MicroRNA Assay Human Panel (ABI, cat no: 4365409). The reactions were incubated in 96-well plates on the Applied Biosystems 7500 FAST Sequence Detection System at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. An assay specific for an Arabidopsis thaliana miRNA (ath-mir159A) and two assays specific for Caenorhabditis elegans miRNAs (Cel-miR-2 and Cel-lin-4) were used as negative controls. If not indicated otherwise, both RT- and PCR-reactions were performed in duplicate. miRNAs were considered as present when Ct-values (TaqMan threshold cycle) were lower than 34.
RESULTS AND DISCUSSION
miRNA array design
MOE oligonucleotides to 150 human and mouse miRNAs were designed to perfectly complement miRNA sequences deposited in the miRNA Registry Database (39,40). In addition, we designed oligomer probes complementary to 50 predicted putative mammalian miRNAs for which experimental validation was lacking (35), as well as four miRNA-unrelated MOE oligonucleotides as internal controls. One of the critical issues in short-oligomer microarray technology is the occurrence of false positive signals due to cross-hybridization of surface-bound probes with highly homologous miRNAs belonging to the same family and other non-target-related RNA sequences. In order to address this problem, we designed for each perfect matched probe (0MM) two additional mismatched probes containing one (1MM) and two (2MM) mismatches placed close to the center of the sequence. The purified MOE oligoribonucleotides containing no additional modifications were directly deposited onto the silanized ER slides by robotic pin printing.
miRNA expression profiling in human HeLa cells and mouse organs
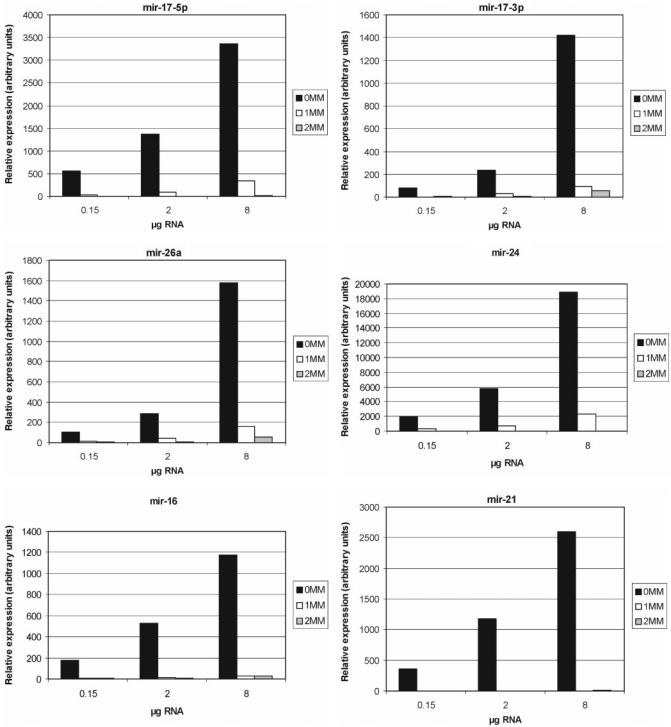
In order to label the miRNA population in a complex RNA sample, we have adapted a chemical labeling technique (37) allowing for the incorporation of Cy5 at the 3′-terminus of RNA. Since this approach requires an intact 2′,3′-diol terminus, background signals due to cross-hybridization with contaminant RNA species bearing 3′-phosphate groups or DNA can be minimized. An added benefit of this labeling method is that it does not require enzymatic or amplification steps that bear the danger of modifying the original RNA sample content. Hybridization conditions for miRNA profiling were optimized by testing the effects of hybridization conditions and probe concentration using a panel of chemically synthesized 3′-end-labeled miRNAs (data not shown). Subsequently, we investigated the performance of the MOE-ER array platform when challenged with a complex biological sample. For this, different amounts of labeled HeLa RNA were hybridized on the arrays. A dose response and mismatch discrimination of fluorescence signals was obtained for the majority of the miRNAs as exemplified for mir-17-5p, mir-17-3p, mir-26a, mir-24, mir-16 and mir-21 which represent miRNAs known to be expressed in HeLa cells (7,41). miRNA signals were considered as present if the fluorescence intensity of the perfectly matched probes was higher than that of the corresponding mismatch probes, and if the perfectly matched signal increased with increasing RNA concentration (Figure 2 and Supplementary Table S1). Importantly, in most cases no to minimal signals were obtained with the mismatch controls, demonstrating the specificity of the MOE-ER array.
Figure 2.
Sensitivity and specificity of MOE-ER-based miRNA arrays. A dose response and mismatch (MM) discrimination of fluorescence signals was obtained for mir-17-5p, mir-17-3p,mir-26a, mir-24, mir-16 and mir-21 which represent miRNAs known to be expressed in HeLa cells. MOE-ER arrays were probed with the indicated amounts of the size-fractionated (<200 nt) Cy5-labeled HeLa cell RNA. Expression values for all individual miRNA are shown in Supplementary Table S1.
More recently, PCR-based methods have been developed to assay quantitatively the abundance of individual miRNAs (18,19,42,43). We compared the expression levels of 107 miRNAs measured with the MOE-ER array and with commercially available stem-loop RT-PCR reagents (17,44), using HeLa RNA. Taking into account that both methods have potential biases and technical limitations, the expression data determined by the MOE-ER array were in excellent agreement with the results obtained by using RT-PCR (Table 1). Applying 150 ng size-fractionated HeLa RNA, ∼75% from the miRNAs classified as present by the array were confirmed by the PCR. On the other hand, ∼75% of miRNAs classified as absent in HeLa cells by the array were also recorded negative by RT-PCR. For eight miRNAs, a clear assignment was not possible since the corresponding array signals did not match the established specificity criteria. The discrepancies between the two analysis methods may be caused by several reasons. Due to the limited possibilities for miRNA probe design, subjecting all probes to identical hybridization conditions is a compromise which may lead to some false negatives as well as false positives. Thus, the ‘absence’ of a specific signal on the array does not necessarily imply that the miRNAs are not present in the sample but may be due to the hybridization signal not passing the arbitrary threshold value at the applied experimental conditions. With two exceptions (mir-137 and mir-140) in all instances where miRNAs were clearly recorded by RT-PCR, they also became detectable by the array when the amount of input RNA was increased (Supplementary Table S1). The cases when a miRNA was scored as positive on the MOE-ER array but was not detected by RT-PCR (mir-214, mir-185 and mir-146) may represent non-specific hybridization of other cellular RNAs to the array probes. These considerations emphasize the importance of validating miRNA array data by an independent method.
Table 1.
Comparison of miRNA expression in HeLa cells recorded by MOE-ER array (150 ng size-fractionated RNA) and stem-loop RT-PCR methodology (15 pg RNA per PCR reaction). miRNAs are ranked in the order of decreasing relative abundance as determined by RT-PCR (inverse correlation with the threshold cycle (Ct) value). Ath-mir-159, cel-mir-2 and cel-lin4 miRNA RT-PCR assays were included as negative controls. Signals considered as present are highlighted in bold. Values in italics depict cases where a clear assignment was not possible as the corresponding array probe sets did not pass the established specificity criteria (n/a, not available)
| miRNA ID | PCR (Ct) | Array (counts) | miRNA ID | PCR (Ct) | Array (counts) | miRNA ID | PCR (Ct) | Array (counts) |
|---|---|---|---|---|---|---|---|---|
| mir-21 | 23.5 | 362 | mir-29b | 30.5 | 23 | mir-200a | 40.0 | 9 |
| mir-17-5p | 25.9 | 565 | let-7d | 30.7 | 547 | mir-146 | 40.0 | 66 |
| mir-19a | 26.1 | 61 | mir-151 | 30.8 | 18 | mir-154 | 40.0 | 7 |
| mir-23a | 26.3 | 2873 | mir-181a | 31.1 | 32 | mir-197 | 40.0 | 23 |
| mir-16 | 26.4 | 177 | mir-30e | 31.2 | 48 | mir-199a | 40.0 | 7 |
| mir-20 | 26.7 | 455 | mir-28 | 31.5 | 33 | mir-9 | 40.0 | 3 |
| mir-106a | 26.8 | 432 | mir-152 | 31.5 | 12 | mir-141 | 40.0 | 4 |
| mir-30b | 27.1 | 125 | mir-140 | 31.5 | 14 | mir-223 | 40.0 | 3 |
| mir-29c | 27.1 | 61 | mir-182 | 31.6 | 33 | mir-134 | 40.0 | 9 |
| mir-29a | 27.1 | 319 | mir-195 | 31.8 | 9 | mir-219 | 40.0 | 2 |
| mir-30c | 27.3 | 327 | mir-193 | 31.9 | 203 | mir-200b | 40.0 | 9 |
| mir-100 | 27.5 | 492 | let-7e | 32.1 | 166 | mir-129 | 40.0 | 3859 |
| mir-15b | 27.6 | 287 | mir-132 | 32.2 | 234 | mir-127 | 40.0 | 12 |
| let-7i | 27.6 | 2055 | mir-10a | 32.7 | 14 | mir-142-5p | 40.0 | 6 |
| mir-23b | 27.6 | 2175 | mir-137 | 32.7 | 12 | mir-9* | 40.0 | 2 |
| let-7a | 27.7 | 672 | let-7b | 33.0 | 192 | mir-124a | 40.0 | 254 |
| mir-92 | 27.7 | 462 | mir-107 | 33.7 | 111 | mir-144 | 40.0 | 6 |
| mir-99a | 27.9 | 503 | mir-155 | 34.0 | 0 | mir-147 | 40.0 | 17 |
| mir-27b | 28.0 | 2091 | mir-139 | 34.1 | 4 | mir-150 | 40.0 | 8 |
| mir-26a | 28.2 | 106 | mir-181c | 34.1 | 15 | mir-154 | 40.0 | 7 |
| mir-31 | 28.2 | 646 | mir-138 | 34.1 | 31 | mir-184 | 40.0 | 0 |
| let-7g | 28.3 | 89 | mir-149 | 34.1 | 14 | mir-187 | 40.0 | 6 |
| mir-130a | 28.3 | 43 | mir-203 | 34.1 | 5 | mir-198 | 40.0 | 737 |
| mir-224 | 28.6 | 72 | mir-194 | 34.3 | 8 | mir-205 | 40.0 | 18 |
| mir-186 | 28.7 | 5 | mir-213 | 34.5 | 11 | mir-211 | 40.0 | 5 |
| mir-145 | 28.7 | 35 | mir-126 | 34.6 | 6 | mir-216 | 40.0 | 1 |
| mir-103 | 28.8 | 101 | mir-148a | 34.6 | 4 | mir-220 | 40.0 | 8 |
| mir-25 | 29.0 | 169 | mir-214 | 35.0 | 132 | ath-mir-159a | 40.0 | n/a |
| mir-26b | 29.1 | 60 | mir-133a | 35.4 | 6 | cel-mir-2 | 40.0 | n/a |
| mir-221 | 29.2 | 131 | mir-189 | 35.7 | 22 | cel-lin-4 | 40.0 | n/a |
| mir-27a | 29.3 | 2928 | mir-185 | 35.7 | 635 | |||
| mir-181b | 29.4 | 12 | mir-199a* | 40.0 | 2 | |||
| mir-130b | 29.4 | 21 | mir-199b | 40.0 | 24 | |||
| mir-125a | 29.5 | 148 | mir-218 | 40.0 | 46 | |||
| mir-30d | 29.5 | 185 | mir-215 | 40.0 | 8 | |||
| mir-191 | 29.6 | 96 | mir-96 | 40.0 | 33 | |||
| mir-222 | 29.8 | 146 | mir-122a | 40.0 | 3 | |||
| mir-98 | 29.9 | 4 | mir-135a | 40.0 | 2 | |||
| mir-30a-3p | 30.1 | 50 | mir-105 | 40.0 | 13 | |||
| mir-15a | 30.3 | 187 | mir-190 | 40.0 | 5 |
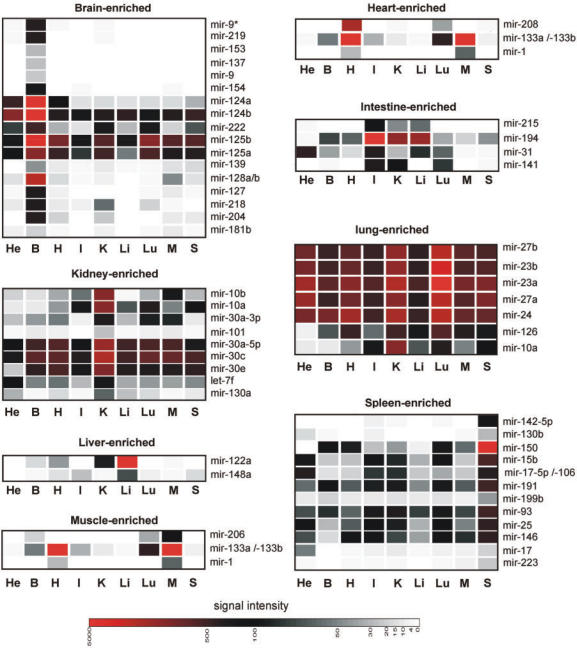
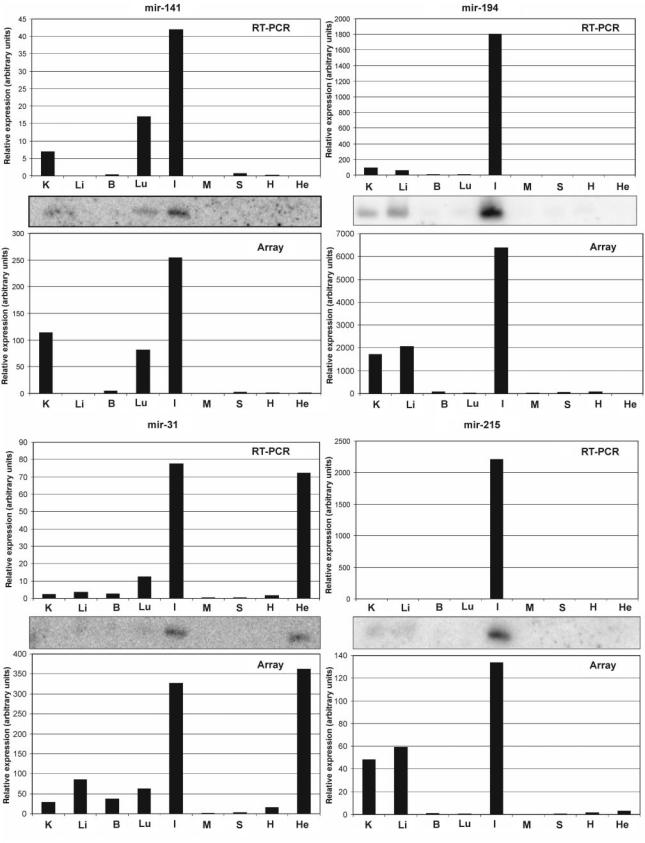
As a next step, we have used the MOE-array to define new expression profiles of known miRNAs and also to verify the existence and expression of bioinformatically predicted miRNAs (complete data set in Supplementary Table S2). RNA samples (<200 nt) originating from eight different mouse organs were labeled and hybridized individually to MOE-ER arrays. Approximately half of the miRNAs represented on the array exhibited a significant expression in at least one tissue investigated, with several miRNAs showing a strong enrichment in some tissues (Figure 3). The tissue miRNA profiling data obtained with the MOE-ER array were comparable with the results obtained by other array-based or unrelated methodologies (8,27,28,45–50) (see also Supplementary Table S3). However, the MOE-array data also revealed new information about expression profiles of known miRNAs. Specifically, a small group of known miRNAs, consisting of mir-31, mir-141, mir-194 and mir-215, was found to have elevated expression levels in the small intestine. The expression profiles of these miRNAs were also analyzed by Northern blot and RT-PCR analysis (Figure 4). Intriguingly, comparable relative expression patterns were obtained in all cases independent of the method of analysis. The array data correlates with Northern blot and RT-PCR in that the four miRNAs (mir-141, mir-194, mir-31 and mir-215) are found to be enriched in small intestine. These data illustrate the exquisite selectivity of the MOE-ER methodology. The stem-loop RT-PCR reagents used in the present study have been demonstrated to effectively discriminate between mature miRNA and corresponding primary and precursor miRNA (17). Since we could demonstrate that the MOE-ER array data correlate very well with corresponding stem-loop PCR data (Figure 4) we conclude that the array detects primarily the mature miRNA. In addition, in the majority of cases investigated here, Northern blot analysis was not able to detect signals that could be clearly assigned to the pre-miRNA molecule. This is in agreement with other reports (18,31) indicating that for most miRNAs the steady-state level of the precursor form is very low as compared to the mature miRNA. In a recent study (51), the authors demonstrate, using Northern blot analysis, that the mature form of mir-138 is predominantly expressed in brain whereas mir-138 precursor is ubiquitously expressed. Interestingly, both the MOE-ER array as well as stem-loop RT-PCR suggest that mir-138 is predominantly expressed in brain, supporting the notion that the MOE-ER array mainly detects mature miRNAs (Supplementary Figure S4). We speculate that the thermodynamically stable hairpin structure of the precursors makes them less available for array probe hybridization.
Figure 3.
The MOE-ER array profiling of miRNAs in eight different mouse tissues reveals novel miRNA expression signatures. A selection of miRNAs displaying tissue-enriched expression patterns is shown. Colors depict relative intensities of array hybridization signals: High (red), medium (black), low (gray). Quantification of expression of all individual miRNA is presented in Supplementary Table S2. He: HeLa; B: brain; H: heart; I: small intestine; K: kidney; Li: liver; Lu: lung; M: skeletal muscle; S: spleen. Data was clustered to represent tissue-enriched distribution of expression.
Figure 4.
Comparison of the relative abundance of miRNAs determined using MOE-ER array, Northern blot and stem-loop RT-PCR analyses of human HeLa cell and mouse tissue RNAs. He: HeLa; B: brain; H: heart; I: small intestine; K: kidney; Li: liver; Lu: lung; M: skeletal muscle; S: spleen.
In addition to known miRNA-probe sequences, the microarray described here also carried probe sets against a series of computationally-predicted (pir) miRNAs (35). Seven of the putative miRNAs (pirs) produced positive signals on the MOE-ER array for which Northern blot analysis failed to detect a RNA species corresponding to the expected size (data not shown). Development of more sensitive assays, such as RT-PCR, would be required to confirm the authenticity of these putative miRNAs.
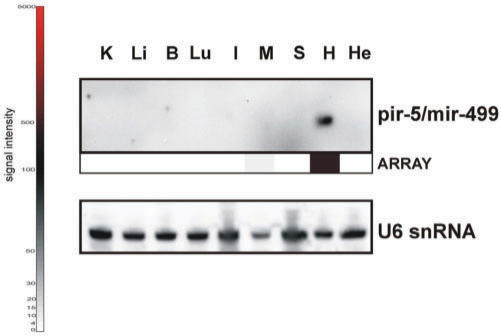
From all the pirs represented on the MOE-ER array, the most distinct expression pattern was obtained for mmu-pir-5 having a strong expression in mouse heart but no detectable expression in any other tissue tested. The enrichment of pir-5 in heart was verified by Northern blot analysis (Figure 5). During the course of this study, Mineno et al. reported the existence of pir-5 in whole mouse embryos and subsequently annotated it as mmu-mir-499 (52). It would be of great interest to identify the functional significance of the heart-specific expression of this miRNA.
Figure 5.
Verification of computational-predicted miRNA pir-5 by MOE-ER array and northern blot analyses. Colours depict relative intensities of array hybridization signals: High (red), medium (black), low (grey). Loading control was performed with U6 snRNA. He: HeLa; B: brain; H: heart; I: small intestine; K: kidney; Li: liver; Lu: lung; M: skeletal muscle; S: spleen.
CONCLUSIONS
In analogy to mRNA expression profiling studies, approaches to systematically determine expression patterns of miRNAs have the potential to rapidly accumulate essential information concerning the involvement of these small RNAs in regulatory networks and human diseases (for recent reviews see (2,53–55). In light of the growing list of characterized miRNAs and claims of thousands of putative miRNAs, we have developed a novel flexible microarray that takes advantage of the highly sensitive ER microarrays, in combination with the favorable biophysical properties of chemically modified 2′-MOE oligonucleotides as capture probes for natural RNA. In accordance with data published by others (25), we found that discrimination between miRNAs differing by only one or two nucleotides at the end-proximal positions is difficult due to cross-hybridization as exemplified by the analysis of let-7 family members (Supplementary Table S5). The let-7 family represents the most challenging set of highly related sequences which to our knowledge has so far not been completely resolved by any array-based methodology. Although carefully designed mismatch probes used in the described array platform helped to minimize this limitation, our present study also suggests that interpretation of miRNA array data should be cautious until complemented by the use of other technologies such as Northern blot and RT-PCR analysis. In the present study, miRNA array profiling data has been systematically confirmed using such complementary methodologies, at the same time validating the reliability of the MOE-ER-array approach. Taken together, the array described here revealed novel expression signatures for known and novel computationally predicted miRNAs. This tool will be valuable in future studies to facilitate analysis of the biological function of miRNAs.
Supplementary Material
[Supplementary Material]
ACKNOWLEDGEMENTS
We would like to thank Dr. Heidi Lane for critical reading of the manuscript and Veronique Drephal, Sandro Gfeller, Isabelle Keller, Christian Fuhrimann, Edgar Baer, David Kirk and Werner Zürcher for excellent technical assistance in oligonucleotide synthesis and microarray We also thank Drs Maciej Drozdz and Petr Svoboda for their help during isolation of mouse organs. F.A.K. was the recipient of a long-term fellowship from the Human Frontier Science Program. The Friedrich Miescher Institute for Biomedical Research is supported by the Novartis Research Foundation.
REFERENCES
- 1.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 11.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 12.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, Takova T, Neri BP, Lyamichev VI. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10:1153–1161. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Fu HJ, Zhu J, Yang M, Zhang ZY, Tie Y, Jiang H, Sun ZX, Zheng XF. A novel method to monitor the expression of microRNAs. Mol. Biotechnol. 2006;32:197–204. doi: 10.1385/MB:32:3:197. [DOI] [PubMed] [Google Scholar]
- 16.Lao K, Xu NL, Yeung V, Chen C, Livak KJ, Straus NA. Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochem. Biophys. Res. Commun. 2006;343:85–89. doi: 10.1016/j.bbrc.2006.02.106. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 21.Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 24.Liang RQ, Li W, Li Y, Tan CY, Li JX, Jin YX, Ruan KC. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005;33:e17. doi: 10.1093/nar/gni019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiler J, Gausepohl H, Hauser N, Jensen ON, Hoheisel JD. Hybridisation based DNA screening on peptide nucleic acid (PNA) oligomer arrays. Nucleic Acids Res. 1997;25:2792–2799. doi: 10.1093/nar/25.14.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaupinnen S, Nielsen PS, Mouritzen P, Nielsen AT, Vissing H, Moller S, Ramsing NB. LNA microarrays in genomics. PharmaGenomics. 2003;3:24–34. [Google Scholar]
- 31.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin P. A new access to 2-O-alkylated ribonucleosides and properties of 2-O-alkylated oligoribonucleotides. Helvetica Chimica Acta. 1995;78:486–504. [Google Scholar]
- 33.Lind KE, Mohan V, Manoharan M, Ferguson DM. Structural characteristics of 2′-O-(2-methoxyethyl)-modified nucleic acids from molecular dynamics simulations. Nucleic Acids Res. 1998;26:3694–3799. doi: 10.1093/nar/26.16.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budach W, Neuschafer D, Wanke C, Chibout SD. Generation of transducers for fluorescence-based microarrays with enhanced sensitivity and their application for gene expression profiling. Anal. Chem. 2003;75:2571–2577. doi: 10.1021/ac026390k. [DOI] [PubMed] [Google Scholar]
- 35.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 36.Natt F, Martin P. Preparation of phosphorothioate-containing DNA using aminodithiazolthiones as sulfuration reagents. patents: EP 991–19768, US 981–68447, CAN 132:279477, AN 2000:240734
- 37.Garnier A, Husken D, Weiler J. New approaches towards fluorescence labelling of messenger RNA transcripts. Nucleosides Nucleotides Nucleic Acids. 2001;20:1181–1185. doi: 10.1081/NCN-100002515. [DOI] [PubMed] [Google Scholar]
- 38.Neuschafer D, Budach W, Wanke C, Chibout SD. Evanescent resonator chips: a universal platform with superior sensitivity for fluorescence-based microarrays. Biosens. Bioelectron. 2003;18:489–497. doi: 10.1016/s0956-5663(02)00272-5. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 43.Lu DP, Read RL, Humphreys DT, Battah FM, Martin DIK, Rasko JEJ. PCR-based expression analysis and identification of microRNAs. Journal of RNAi and Gene Silencing. 2005;1:44–49. [PMC free article] [PubMed] [Google Scholar]
- 44.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 49.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 51.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong H, Liu CM, Liu DP, Liang CC. The role of small RNAs in human diseases: potential troublemaker and therapeutic tools. Med. Res. Rev. 2005;25:361–381. doi: 10.1002/med.20023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 55.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]