Complex SNP-Based Haplotypes in Three Human Helicases: Implications for Cancer Association Studies (original) (raw)
Abstract
We have initiated a candidate gene approach to study variation and predisposition to cancer in the four major ethnic groups that constitute the U.S. population (African Americans, Caucasians, Hispanics, and Asians). We resequenced portions of three helicase genes (BLM, WRN, and RECQL) identifying a total of 37 noncoding single nucleotide polymorphisms (SNPs). Haplotype inference predicted 50 haplotypes in BLM, 56 in WRN, and 47 in RECQL in a sample of 600 chromosomes. Approximately 10% of the predicted haplotypes were shared among all ethnic groups. Linkage disequilibrium and recombination effects showed that each locus has taken a diverse evolutionary path. Primate DNA analysis of the same loci revealed one human haplotype per gene shared with the great apes, indicating that the observed diversity occurred since the divergence of humans from the last common ancestor. In BLM, we confirmed the presence of a founder haplotype among Ashkenazi Jews homozygous for the _blm_Ash mutation. The cosegregating haplotype was seen in all (6/6) samples of Ashkenazi descent, whereas in the general population it has a low frequency (0.02) and was not found in African Americans. In WRN, ethnic samples were studied for their haplotype content and the presence or absence of six previously described coding SNPs (cSNPs). Hispanic individuals carrying two of these cSNPs showed a 60% increase in the frequency of a common haplotype (haplotype No. 28). In the pooled sample, no association was found. Because (1) the majority of the haplotypes are population specific and (2) the patterns of linkage disequilibrium, recombination, and haplotype diversity are markedly different between gene regions, these data show the importance of either ethnically matched controls or within-family-based disease-gene association studies.
[The sequence data described in this paper have been submitted to the GenBank data library under accession no. AC006559. Online supplemental material available at http://www.genome.org]
With the completion of a human reference sequence through the Human Genome Project, the ability to define the role of genomic variation in disease risk is at hand. A starting point is to assess the type and distribution of sequence variation within and between ethnic populations. In recent years, single nucleotide polymorphisms (SNPs) have been favored as more tractable genotypic markers. SNPs, although reduced in allelic diversity, have a number of advantages over microsatellites, such as abundance (one every 750–1000 bp; Kwok et al. 1996; Wang et al. 1998), stability, and the capacity for highly automated analysis. As a consequence, SNPs are being utilized extensively as markers of choice in human genetic studies ranging from comparative population variation (Halushka et al. 1999; Goddard et al. 2000; Kidd et al. 2000; Ober et al. 2000) to disease linkage studies (Kruglyak 1997, 1999; Lai et al. 1998; Martin et al. 2000).
Several investigators (Spielman et al. 1993; Risch and Merikangas 1996; Long and Langley 1999) have suggested that association-based studies are more powerful than linkage studies in identifying genes or variants that contribute to complex trait phenotypes. This is based on the assumption that one of the markers used in the analysis will in fact be the polymorphism contributing to the disease variation or that neutral markers will be in linkage disequilibrium with the disease-causing site. These assumptions led to suggestions of genome-wide (Risch and Merikangas 1996) or candidate-gene scans (Halushka et al. 1999; Long and Langley 1999; Martin et al. 2000) using dense sets of SNPs as the means to identify sequences contributing to variation in complex traits.
In this study we set out to test the use of common variants and their associated haplotypes for detecting functional variants contributing to cancer risk. Given that numerous genetic lesions are found in tumors, along with the fact that carcinogenesis requires several somatic hits (Knudson 1996), we have begun a candidate-gene approach and concentrated our efforts on detecting variation in genes that play a role in DNA maintenance and chromosomal integrity. We have utilized primarily noncoding sequence variants. The rationale for this choice was to avoid bias that might be introduced by studying sites with functional effects. Our goal was to build haplotypes capable of marking chromosomes carrying common and functionally significant variants. However, we preferred not to assume the type of variant that might be relevant. Therefore, exclusive focus on relatively rare coding sequence changes might not allow common, functionally relevant intronic or promoter variants to be uncovered. A further benefit of such an approach is that it allows a more complete analysis of the population history of the loci under study (Bonnen et al. 2000).
BLM, WRN, and RECQL are genes encoding proteins with significant similarity with the Escherichia coli RecQ-type DNA helicases (Puranam and Blackshear 1994; Ellis et al. 1995; Yu et al. 1996) involved in duplex DNA unwinding (Umezu et al. 1990; Puranam and Blackshear 1994; Gray et al. 1997; Karow et al. 1997). Although to date no disease has been associated with RECQL, homozygous loss of function in BLM and WRN result in two distinct syndromes, Bloom's and Werner's, respectively (Ellis et al. 1995; Foucault et al. 1997; Moser et al. 1999). Both syndromes exhibit high tumor incidence attributed to high levels of chromosome instability and somatic mutation (Goto et al. 1981, 1996; Fukuchi et al. 1989; German 1993). Variation in these genes in the general population may play a role in predisposition to tumor formation or progression.
We carried out a pilot study of functionally neutral variation in the three helicases in four ethnic populations (Caucasians, African Americans, Hispanics, and Asians). We employed a resequencing method as the means to detect and identify SNPs in the genomic regions of human BLM, WRN, and RECQL. The aim was to develop a series of biallelic markers, 8–12 SNPs per gene, and use these to infer haplotypes that could allow chromosomal distinction. Although most efforts of defining variation in genes playing a role in cancer have concentrated on coding sequence (Helland et al. 1998; Josefsson et al. 1998; Storey et al. 1998; Li et al. 1999; Wagner et al. 1999), we focused on intronic and other noncoding sequences. A genomic sequence length of 150 kb was targeted so markers clustered within the region would potentially be in disequilibrium, facilitating the identification of functional variants of the gene through haplotype association.
The SNP data presented here allowed definition of haplotypes that mark the variation observed in the helicases in the four ethnic groups. Creation of complex haplotypes based on multiple markers can strengthen the power of association studies, and such data will be further useful in comparisons of haplotype frequencies and distributions across the human genome and the human population.
RESULTS
SNP Detection and Distribution of Variation
The available genomic sequence spanned a region of ∼154 kb, 186 kb, and 180 kb for BLM, WRN, and RECQL, respectively. Repetitive elements (40%–50% of each locus) were masked, and PCR primers were designed to generate products in the range of 350 bp to 600 bp for SNP detection. Products allowed scanning of between 15% and 20% of each locus by resequencing in five individuals.
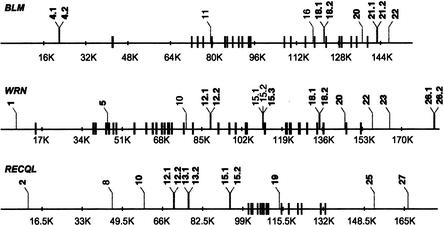
The majority of the amplicons screened for polymorphisms spanned intronic or intragenic sequence (22/26 in BLM, 21/26 in WRN, and 27/29 in RECQL). A total of 15, 10, and 12 biallelic polymorphisms were identified in WRN, BLM, and RECQL, respectively (Table 1). All were noncoding and involved a single base change, the majority being transitions (6/10 in BLM, 8/15 in WRN, and 8/12 in RECQL). Therefore, SNPs were found at an average (over the three helicases) frequency of 1 in every 1290 bp. Figure 1 shows a schematic of the gene structure and the identified SNPs in each locus.
Table 1.
Sequence Variants Identified in BLM, WRN, and RECQL.
| Locus | SNP name | Position | Variation |
|---|---|---|---|
| BLM | B4.1 (IVS1-2056lt-c) | 21812a | T→C |
| B4.2 (IVS1-20290g-a) | 22083a | G→A | |
| B11 (IVS5-302a-g) | 79526 | A→G | |
| B16 (IVS16-954a-g) | 118225 | A→G | |
| B18.1 (IVS17-425a-g) | 122762a | T→C | |
| B18.2 (IVS17-345c-g) | 122842a | C→G | |
| B20 (IVS22-2082c-a) | 136931a | G→T | |
| B21.1 (IVS22+3336c-g) | 142615a | C→G | |
| B21.2 (IVS22+3401a-c) | 142680a | A→C | |
| B22 (IVS22+9303c-t) | 148582a | G→A | |
| WRN | W1 (IVS1-8213g-a) | 6114a | G→A |
| W5 (IVS4+176a-g) | 45121a | T→C | |
| W10 (IVS17+1084c-a) | 78453 | C→A | |
| W12.1 (IVS19-3173t-c) | 88968a | T→C | |
| W12.2 (IVS19-3145t-a) | 88996a | T→A | |
| W15.1 (IVS24-210c-a) | 111587 | C→A | |
| W15.2 (IVS24-209a-g) | 111588 | A→G | |
| W15.3 (IVS24-191c-t) | 111606a | G→A | |
| W18.1 (IVS32+845c-t) | 135048a | G→A | |
| W18.2 (IVS32+859g-t) | 135062a | G→T | |
| W20 (IVS34-628t-g) | 145865a | A→C | |
| W22 (IVS35+4302t-c) | 157465a | T→C | |
| W23 (IVS35+11737g-c) | 164900a | G→C | |
| W26.1 (IVS53+30673c-t) | 183836a | G→A | |
| W26.2 (IVS35+30764c-a) | 183927a | G→T | |
| RECQL | R2 (IVS1-89964t-g) | 10998a | A→C |
| R8 (IVS1-55823a-g) | 45139 | A→G | |
| R10 (IVS1-4258lt-c) | 58381a | T→C | |
| R12.1 (IVS1-30638g-c) | 70324a | G→C | |
| R12.2 (IVS1-30329g-t) | 70633a | G→T | |
| R13.1 (IVS1-24228g-a) | 76734a | G→A | |
| R13.2 (IVS1-24159c-t) | 76803a | G→A | |
| R15.1 (IVS1-7216a-g) | 93746a | T→C | |
| R15.2 (IVS1-7166g-a) | 93796a | G→A | |
| R19 (IVS10-1078g-a) | 113771a | G→A | |
| R25 (IVS15+19546t-c) | 152798a | T→C | |
| R27 (IVS15+33444t-c) | 166696a | T→C |
Figure 1.
Schematic of SNP positions within BLM, WRN, and RECQL. Black vertical bars indicate exon positions and gray jagged lines the positions of identified SNPs in each gene. “.1” and “.2” denote SNPs found within the same PCR product. SNP numbering reflects their position in a 5′ to 3′ orientation. SNPs used in haplotype determination are in bold.
Allele Specific Oligonucleotide Hybridization (ASO) Analysis and Allele Frequencies
Eight of the 10 SNPs identified within BLM, 12 of the 15 SNPs in WRN, and 11 of the 12 SNPs in RECQL (Table 1) proved to be robust in PCR amplification and allele-specific oligonucleotide hybridization and were used in subsequent studies. Table 2 shows the frequencies of the rarer allele for each of the 31 SNPs detected in the three genes for each of the populations tested. Both alleles were observed in each ethnic group for 27 of 31 SNPs, with rare allele frequencies ranging from 0.01 to 0.5. The remaining four SNPs (W23, W26.1, W26.2, and R27) were excluded from the population analysis because they were monomorphic or contained very low frequency rare alleles.
Table 2.
Rare Allele Frequencies in BLM, WRN, and RECQL
| BLM SNPs | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | N | B4.1 | B4.2 | B18.1 | B18.2 | B20 | B21.1 | B21.2 | B22 | ||||
| T/C | G/A | T/C | C/G | G/T | C/G | A/C | G/A | ||||||
| AfAm | 148 | C.27 | A.02 | T.40 | G.18 | T.19 | G.36 | C.31 | A.18 | ||||
| Asian | 78 | T.49 | A.28 | C.26 | G.18 | T.15 | G.21 | C.19 | A.19 | ||||
| Cauc | 154 | C.28 | A.12 | C.29 | G.20 | T.30 | G.37 | C.39 | A.15 | ||||
| Hisp | 140 | C.25 | A.13 | C.28 | G.13 | T.23 | G.28 | C.26 | A.12 | ||||
| CEPH | 78 | C.27 | A.12 | C.36 | G.26 | T.26 | G.33 | C.33 | A.24 | ||||
| WRN SNPs | |||||||||||||
| Population | N | W1 | W5 | W12.1 | W12.2 | W15.3 | W18.1 | W18.2 | W20 | W22 | W23 | W26.1 | W26.2 |
| G/A | T/C | T/C | T/A | G/A | G/A | G/T | A/C | T/C | G/C | G/A | G/T | ||
| AfAm | 154 | G.02 | C.01 | C.17 | A.04 | A.12 | A.27 | G.34 | C.23 | T.44 | C.00 | A.00 | T.01 |
| Asian | 78 | G.12 | C.05 | C.13 | A.08 | A.24 | A.32 | T.45 | C.23 | T.30 | C.00 | A.00 | T.00 |
| Cauc | 158 | G.04 | C.06 | C.22 | A.20 | A.27 | A.30 | G.42 | C.25 | T.47 | C.03 | A.03 | T.01 |
| Hisp | 150 | G.15 | C.01 | C.15 | A.14 | A.29 | A.31 | G.50 | C.31 | T.43 | C.01 | A.01 | T.01 |
| CEPH | 78 | G.01 | C.04 | C.21 | A.21 | A.21 | A.31 | G.44 | C.37 | T.43 | C.01 | A.01 | T.01 |
| RECQL SNPs | |||||||||||||
| Population | N | R2 | R10 | R12.1 | R12.2 | R13.1 | R.13.2 | R15.1 | R15.2 | R19 | R25 | R27 | |
| A/C | T/C | G/C | G/T | G/A | G/A | T/C | G/A | G/A | T/C | T/C | |||
| AfAm | 156 | C.10 | C.38 | G.47 | G.47 | G.47 | A.19 | C.19 | G.49 | A.30 | C.47 | C.01 | |
| Asian | 74 | C.01 | C.30 | C.46 | T.46 | A.45 | G.45 | T.45 | A.45 | G.43 | C.05 | C.00 | |
| Cauc | 156 | C.07 | C.33 | C.47 | T.46 | A.46 | A.48 | A.48 | A.46 | C.48 | C.16 | C.04 | |
| Hisp | 152 | C.03 | C.34 | C.47 | T.46 | A.46 | A.45 | A.46 | A.42 | G.43 | C.17 | C.01 | |
| CEPH | 70 | C.06 | C.27 | C.40 | T.40 | A.39 | A.49 | A.47 | A.40 | G.50 | C.09 | C.04 |
Pairwise comparisons of allele frequencies between the different ethnic populations were carried out using the χ2 contingency test. The majority of alleles had similar frequencies in all groups. Our data, however, showed the African American sample group to be the only one to differ significantly (0.0001< P <0.001) in at least two SNPs per gene. This finding may be related to the greater allelic diversity among individuals of African origin (Clark et al. 1998; Nickerson et al. 1998; Cargill et al. 1999; Halushka et al. 1999).
Hardy-Weinberg Equilibrium and Nucleotide Diversity
Tests for deviation from Hardy-Weinberg equilibrium (HWE) were conducted for each locus-population combination of SNP genotypes of the three helicases. Within BLM, the observed frequencies in the African American sample group deviated from the expected values at one locus (locus 22, P ≅0.0006). Discrepancies were also observed at one locus of the WRN gene (W18.2) for the Caucasian population (P ≅0.0042). In both cases a deficit of heterozygotes was observed. Given that 2 of 118 tests showed deviation from HWE, these significant variations can be ascribed to chance alone (P_-values with the Bonferroni correction are 0.071 for B22 in African Americans and 0.496 for W18.2 in Caucasians). Also, no locus-specific trend of excess (or deficiency) of heterozygotes was found. Fisher's summed test statistic −2 Σ_i ln pi for the three helicases (summed over all locus-population tests) were equal to 66.79 (P > 0.25), 36.25 (P > 0.995), and 71.15 (P > 0.50) for BLM, RQL, and WRN, respectively (using the approximation of the distribution of the test statistic by the χ2 distribution with 2_k_ degrees of freedom, where k is the number of tests carried out).
Nucleotide diversity was measured by calculating π, the average heterozygosity per site in two sequences chosen from a randomly mating population (Nei 1987) in each ethnic group in each gene. Estimates of nucleotide diversity revealed no differences between populations at the levels of sequences spanned by the SNP sites. Mean values ranged from 0.0002 in BLM and WRN to 0.00033 in RECQL and were similar to those observed by Halushka et al. (1999) in a study of candidate genes for blood pressure homeostasis.
Haplotype Inference
EMHAPFRE (Excoffier and Slatkin 1995) was used to infer haplotypes and haplotype frequencies in the four ethnic groups (Caucasians, African American, Hispanics, and Asians) and in the members of the CEPH pedigrees. Taking into account inheritance patterns and assuming no recombination between generations, CEPH haplotypes were also determined manually and used as a control to test the accuracy of the EMHAPFRE algorithm. For the majority of the CEPH haplotypes, the two inference methods agreed. Small discrepancies were found among haplotypes of low frequencies. This is to be expected since EMHAPFRE is based on likelihood maximization and allows free recombination. Table 3 summarizes the results of the haplotype analysis in BLM, WRN, and RECQL.
Table 3.
EMHAPFRE Inferred Haplotypes for BLM, WRN, and RECQL
| BLM | WRN | RECQL | ||||
|---|---|---|---|---|---|---|
| H | N | H | N | H | N | |
| CEPH | 16(19) | 78(194) | 20(21) | 78(194) | 15(16) | 68(178) |
| Af-American | 22 | 146 | 23 | 154 | 21 | 156 |
| Asian | 16 | 78 | 14 | 78 | 10 | 74 |
| Caucasian | 25 | 152 | 29 | 158 | 20 | 156 |
| Hispanic | 22 | 144 | 21 | 150 | 27 | 152 |
| Combined | 50 | 598 | 56 | 618 | 47 | 616 |
| Shared Haplotypes | 5 | 4 | 6 |
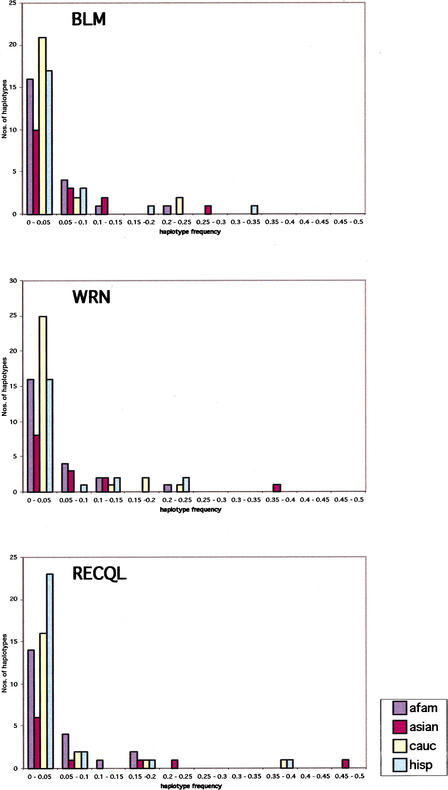
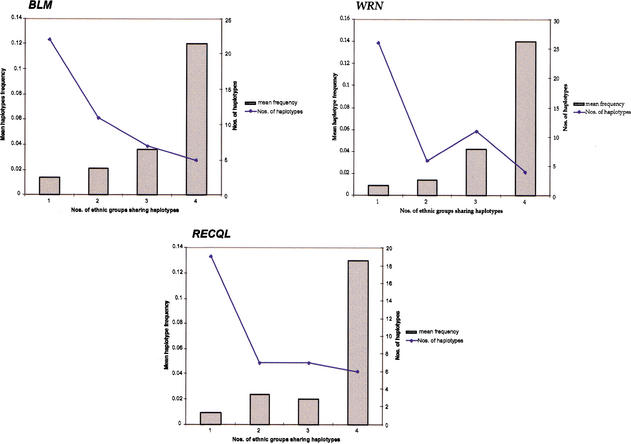
The expectation-maximization (EM) algorithm predicted 50, 56, and 47 different haplotypes in 598, 618, and 616 chromosomes in BLM, WRN, and RECQL, respectively. Figure 2 shows the haplotype distribution for each gene in the four different ethnic groups based on their frequencies. African American haplotypes are more evenly distributed, reflecting the diversity observed in that population. Approximately 10% of haplotypes for each gene were found shared among all four ethnic groups. The shared haplotypes were also those with the highest frequencies, ranging from 0.02 to 0.3, as estimated by EMHAPFRE when all samples were studied together. These shared haplotypes also account for the majority (54%–94%) of the Caucasian, Hispanic, and Asian chromosomes at the three loci. In African Americans, however, a smaller percentage of samples (36%–53%) harbored the same shared haplotypes. In Figure 3, mean haplotype frequencies and number of haplotypes are plotted based on whether these are shared by one, two, three, or all the ethnic groups studied here. A small number of haplotypes with very low frequencies (total frequency of 0.009 in BLM and 0.018 in WRN and RECQL) appear in the EMHAPFRE output when all samples are pooled together. These are not present in the individual ethnic lists of haplotypes and are due to the larger data set analyzed by the program. Their impact was not considered significant and thus these haplotypes were not included in the graphs. As can be seen, the number of haplotypes shared by all populations is inversely related to their mean frequency. In other words, the majority of haplotypes are not shared by all four ethnic groups but have low frequencies, ranging from 0.009 to 0.04.
Figure 2.
Haplotype distribution in the four ethnic groups for BLM, WRN, and RECQL. Haplotype numbers and their frequencies are plotted by ethnic background for the three genes. Frequencies are binned by .05 intervals. Afam, African American; asian, Asian; cauc, Caucasian; hisp, Hispanic.
Figure 3.
Ethnic sharing of haplotypes and their frequency for BLM, WRN, and RECQL. Haplotypes and their frequencies are plotted based on whether they are shared between one, two, three, or all four ethnic populations (_X_-axis). Bars indicate the mean frequencies of haplotypes shared; the line plots the number of haplotypes within each class.
Linkage Disequilibrium and Recombination
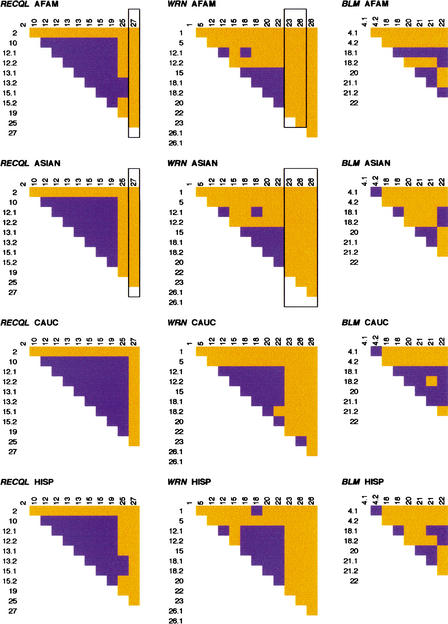
Linkage disequilibrium (LD) between polymorphic sites in each gene was assessed in each population separately. We used haplotypic data for each ethnic group as determined by EMHAPFRE to calculate the _D_-value (Lewontin and Kojima 1960) for each pairwise comparison in DnaSP. Figure 4 shows the results of pairwise tests, indicating the site pairs with a significant Fisher's exact test (P < 0.001). Site pairs with rare alleles failed to give a significant Fisher's exact test outcome, although disequilibrium may exist. Analyses of individual populations excluded disequilibria computation for site pairs that include sites monomorphic in individual populations but polymorphic in the total sample.
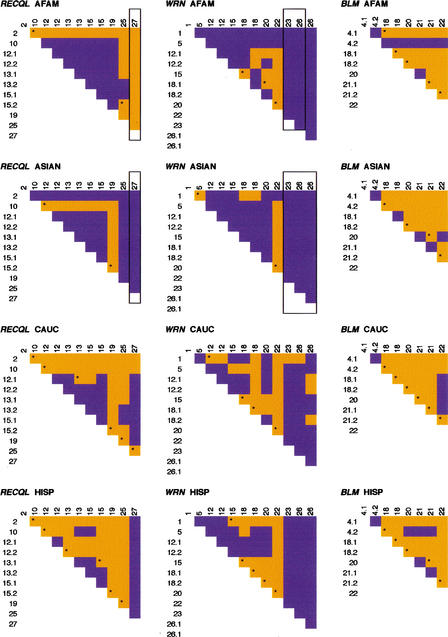
Figure 4.
Linkage disequilibrium for each gene by ethnic group. Site pairs with significant Fisher's exact test (P <0.001) are shown in blue. Nonsignificant pairs are in orange. Loci are numbered in the 5′ to 3′; SNP order relative to the direction of transcription of each gene. “.1” and “.2” indicate SNPs found within the same PCR product. Framed loci were monomorphic and were not considered in the analysis. AFAM, African American; ASIAN, Asian; CAUC, Caucasian; HISP, Hispanic.
As expected, SNPs found within the same PCR product showed significant disequilibrium. For RECQL, all populations showed LD throughout the gene, with the exception of the outer-most markers. Similarly, in WRN all ethnic groups showed comparable patterns of disequilibrium in the middle section of the gene (between the fifth and eighth SNPs). In BLM, however, disequilibrium was not confined to a specific gene region. The significance plots of LD (Fig. 4) were different in each population and only the Hispanic and Asian sample groups shared a similar pattern. In fact, these plots nearly mimic the plot of the absolute value of the normalized LD, D', for all three helicases in the four samples studied. In particular, the regions of significant LD (dark areas of Fig. 4) coincide with the regions of D' = ±1. More quantitatively, the proportion of pairwise D' = ±1, is the lowest for BLM (17.9%–28.6%) and the highest for WRN (60.6%–73.3%) across the four samples.
Despite the similarities of regions of pairwise LD, LD values (magnitude, as well as sign) are not the same in all populations. One explanation is that different demographic histories of the populations may have erased the signature of recombinations at different sites that occurred prior to their separation. Inferences of recombination events were made based on the commonly used four-gamete test (Hudson and Kaplan 1985) and the DnaSP recombination module using EMHAPFRE-predicted haplotypes. Intragenic recombination was inferred in all three genes. Figure 5 is a plot showing the four-gamete test results of pairwise comparisons, indicating in orange the site pairs where all four possible gametic phases were present. The asterisk denotes intervals when historical recombination events are suggested to have taken place. As expected, their locations are highly correlated with the ones implied by the four-gamete test.
Figure 5.
Recombination for each gene by ethnic group. Orange indicates site pairs positive for the four-gamete test; blue indicates absence of one or more possible gametes. Presence of all four gametic phases suggests historical intragenic recombination. The minimum number of recombination events is indicated by asterisks. Loci are numbered in the 5′ to 3′ SNP order relative to the direction of transcription of each gene. “.1” and “.2” indicate SNPs found within the same PCR product. Framed loci were monomorphic and were not considered in the analysis. AFAM, African American; ASIAN, Asian; CAUC, Caucasian; HISP, Hispanic.
Within RECQL, recombination is practically absent in the middle region of the gene consistent with the increased linkage disequilibrium observed in the same area. In WRN, however, the region of high linkage disequilibrium coincides with that of recombination, suggesting that the recombination events are probably of a recent origin. In BLM, recombination seems to have occurred throughout the gene and population patterns are not significantly different. In contrast, in WRN and RECQL Caucasians and Hispanics exhibit more recombination events than African Americans. This is an interesting observation because the African haplotype pool is expected to be older and, therefore, African Americans should exhibit increased numbers of recombinants compared to the other ethnic populations (Kidd et al. 1998, 2000). Ascertainment bias could be one factor influencing this observation (Hispanics have 60%–70% of their genes of European origin; Cerda-Flores et al. 1992). However, because BLM does not exhibit this pattern, sampling bias of the SNPs cannot entirely explain this finding.
Primate Sequence Comparisons and Phylogenetic Tree Construction
To assess the age of the detected polymorphisms, we screened five primate samples (two chimpanzees, one bonobo, and two gorillas), both by ASO hybridization and by direct sequencing. In all cases, the primate sites were homozygous for only one of the human alleles (Table 4). The primate nucleotide coincided with the more common human allele in 24 of 31 sites tested. These findings suggest that the observed human variation occurred after the separation of the ancestors of modern humans and great apes and that for the majority of the polymorphisms studied, the common human allele is likely to represent the state of sequence in the last common ancestor. All primates shared one haplotype per gene for the human-specific SNPs. The primate haplotypes were also found in the human haplotype pool, albeit at low frequencies (Table 4).
Table 4.
Primate Haplotypes and Their Frequencies in Humans
| Primate haplotype | African American | Asian | Caucasian | Hispanic | All samples |
|---|---|---|---|---|---|
| _BLM_CGCCGCAG | 0.07 | 0.02 | NP | NP | 0.02 |
| _WRN_GTTTGGGATGGG | 0.016 | NP | NP | 0.021 | 0.001 |
| _RECQL_ATGGAGTGGTT | 0.006 | NP | NP | NP | 0.002 |
In the phylogenetic analysis of the haplotypes, we assumed the primate haplotype to be the ancestral one for each gene and used it as the outgroup (data not shown). The relatively large number of haplotypes for all three genes made it difficult to define haplo groups. RECQL haplotypes can be divided into two major clades, but the BLM and WRN trees are bushlike. Assessment of the haplotype clusters based on whether they were part of the African American haplotype pool did not result in any particular pattern (data not shown). It is noteworthy that the internal branch lengths in these trees are generally small. Hence, any single evolutionary factor (e.g., recent population expansion, early history of recombination, or natural selection) is not sufficient explanation because the topologies differ from gene to gene.
Pilot Association Studies
The ultimate goal of this line of experiments is to create resources for association and predisposition studies. To test the utility of the identified SNPs and predicted haplotypes, we carried out two pilot association studies. The _blm_Ash mutation (Ellis et al. 1995) is found in ∼97% of Bloom syndrome patients of Ashkenazi Jewish origin, patients of Spanish American ancestry (Ellis et al. 1998), and Jewish patients of Polish descent (Shahrabani-Gargir et al. 1998). The mutation has a carrier frequency of ∼1% among Ashkenazi Jews (Roa et al. 1999) and results in protein truncation due to 6-bp deletion/7-bp insertion in exon 10 (Ellis et al. 1995).
We tested four DNA samples and five cell lines from the Bloom syndrome patient registry, available from the Coriell Cell Repository. Of these, six are of Ashkenazic ancestry, two are listed as Caucasians, and one is of Japanese origin. All samples were tested for the _blm_Ash mutation and the noncoding SNPs presented here by ASO analysis. As expected, all Ashkenazi Jewish samples were homozygous for the mutation, one Caucasian sample was heterozygous, and the remaining samples (one Caucasian and one Japanese) were negative. When we then studied haplotypes based on the BLM SNPs, we found the Ashkenazi Jews to be homozygous and share one haplotype (No. 25), which was also only present in a heterozygous state in the Caucasian sample heterozygous for the mutation. Based on the EMHAPFRE analysis, haplotype No. 25 is not seen in African Americans and its frequency ranges from 0.006 in Asians to 0.042 in Caucasians. When control samples of Caucasian origin carrying haplotype No. 25 in a heterozygous or homozygous state were tested for the _blm_Ash mutation, they were found to be negative, showing that this haplotype predated the _blm_Ash mutation. Our results are in agreement with previous haplotype studies (Ellis et al. 1998) that showed the presence of a predominant single haplotype (based on microsatellite markers surrounding BLM) considered to be the founder haplotype that _blm_Ash occurred in Ashkenazi Jews.
A second pilot association study utilized six previously described coding polymorphisms within the WRN gene (Meisslitzer et al. 1997; Ye et al. 1997; Vidal et al. 1998; Castro et al. 1999). Each of these was used separately in an attempt to establish association between any of the coding SNPs (cSNPs) and the WRN inferred haplotypes. A total of 136 individuals (40 African Americans, 40 Caucasians, 40 Hispanics, and 16 Asians) was assessed for noncoding SNP-based haplotypes and the presence or absence of coding variation. Allele frequencies were calculated for each cSNP in the four populations; the results are shown in Table 5.
Table 5.
Allele Frequencies of _SRN_-Coding SNPs
| cW1 | cW3 | cW4 | cW6 | cW7 | cW8 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | G | A | T | G | T | G | C | T | T | C | |
| AfAm (N = 80) | 0.54 | 0.46 | 0.97 | 0.03 | 0.51 | 0.49 | 0.64 | 0.36 | 0.85 | 0.15 | 0.86 | 0.14 |
| Asian (N = 32) | 0.8 | 0.2 | 1 | 0 | 0.54 | 0.46 | 0.5 | 0.5 | 0.82 | 0.18 | 0.87 | 0.13 |
| Caucasian (N = 80) | 0.72 | 0.28 | 0.91 | 0.09 | 0.47 | 0.53 | 0.47 | 0.53 | 0.77 | 0.23 | 0.69 | 0.31 |
| Hispanic (N = 80) | 0.75 | 0.25 | 0.91 | 0.09 | 0.54 | 0.46 | 0.54 | 0.46 | 0.72 | 0.28 | 0.82 | 0.18 |
Although allele frequencies differed among the four ethnic groups for some of the coding SNPs, no clear pattern could be observed between haplotype content and absence or presence of coding variation when all ethnic samples were pooled together. This is an apparent contradiction to our observation that the WRN haplotypes, in comparison to those in BLM gene, exhibit tighter LD (Fig. 4) and lesser recombination (Fig. 5). Recurrent mutations at the WRN cSNP sites may be one mechanism explaining the lack of association between cSNPs and noncoding SNPs at this gene. Two of the coding SNPs are in fact due to changes within CpG dinucleotides, supporting the possibility of recurrence.
Samples carrying coding SNPs cW4 or cW6 (c.2592T> G [Castro et al. 1999] and c.3453T> G [Meisslitzer et al. 1997], respectively) in a heterozygous or homozygous state for the low frequency allele (G) were studied based on their ethnic background. Hispanic individuals exhibited an increased frequency of haplotype No. 28 for both coding SNPs. This haplotype is shared among all ethnic groups showing the highest frequency in Hispanics (0.22 versus 0.16 in the pooled sample). A further increase of the frequency of haplotype No. 28 was noted among Hispanic individuals heterozygous and homozygous for the G allele at the cW4 (frequency 0.35) and cW6 (frequency 0.31) sites. These findings suggest an association between these polymorphisms and the chromosomal background of haplotype No. 28.
DISCUSSION
We chose SNPs to study variation because they are predominantly biallelic, abundant (1 in ∼1000 bp), and stable (Wang et al. 1998). We concentrated our efforts to detect variations in intronic/intragenic regions as these are expected to be more variable, while avoiding repeat sequences that would complicate analysis. Particular care was taken to disperse the PCR products/sequencing targets throughout the genomic regions surrounding each gene. A target of 8–12 SNPs per gene was set to identify adequate numbers of loci evenly distributed along the gene region and thus create informative haplotypes that would enable differentiation between chromosomes. Coding sequences were generally avoided to reduce the likelihood of bias toward functional variants.
Variation in BLM, WRN, and RECQL
From the observation that each of the 31 SNP sites examined showed no variation in five primate samples (two chimpanzees, two gorillas, and one bonobo), we postulate that the ages of the polymorphism in human are probably younger than the time of separation of human and ape lineage. Because the systematic search for SNPs in the noncoding regions of these helicases was made from sequence analysis of a small number (10) of Caucasian chromosomes, it is clear that we detected a small fraction of all polymorphic sites in the gene regions. Nevertheless, there are at least three lines of evidence suggesting that our observations are not severely affected by the ascertainment bias due to selection of sites.
One type of ascertainment bias might stem from the fact that the sequenced chromosomes belong to Caucasians. To determine if such bias is present in the data, we carried out computations of sequence diversities (Nei 1987) of the SNPs at the BLM, WRN, and RQL genes separately for each of the populations considered. In addition, we computed haplotype diversites at these genes in all populations. In this latter set of computations, each haplotype was treated as a different allele, identifiable by its SNP sequence. Detailed numerical result are depicted in Supplementary Table 3 available online at http://www.genome.org. The trends observed are as follows.
Sequence diversities (Nei 1987), in most cases, assume highest values for Caucasian or CEPH individuals, whereas African Americans, Asian Americans, and Hispanics are characterized by systematically lower values of this index. Although no formal test was carried out, the differences exceed the three-standard-deviations level. On the contrary, the haplotype diversities are systematically higher for African Americans and lower for the other subpopulations, again with the differences exceeding the fluctuation level.
Precise interpretation of these findings requires more study. However, it seems that although the effects of ascertainment bias are felt at the level of individual SNPs (Nei's sequence diversity is equal to the average of individual SNP diversities although these two indices have different variances; Nei 1987), they are not detectable at the level of haplotypes. This seems to constitute an argument for using haplotype-based statistics as being more resistant to ascertainment bias.
Another type of ascertainment bias might stem from the small number (10) of sequenced chromosomes, independently of the population from which they were taken. This type of bias was recently analyzed by us (A. Renwick et al., in prep.) and indicates that SNP data, collected with a screening protocol such as ours, results in a small (<10%) underestimation of nucleotide diversity.
Even though we did not detect the presence of more than two alleles in any of the SNP sites, we realize that individuals carrying a rare third or fourth allele would have been mistyped as homozygotes with our ASO-based genotyping assay. However, two features of our data rule out this as a major factor. First, the primate sequences revealed cross-species monomorphism for one of the human variants at each polymorphic site. Second, the statistical agreement of the observed and expected genotype frequencies (under HWE) also argues for the absence of a third or fourth allele. As mentioned before, the two departures from HWE expectation (B22 in African Americans and W18.2 in Caucasians,) can be ascribed to chance alone by Bonferroni adjustment of multiple testing, as well as Fisher's summed statistic of individual _P_-values. Further, there is no site-specific trend of heterozygote deficiencies, also suggestive of no allele drop-out effect of the ASO-based genotype assay.
Haplotype Prediction
The algorithm used (EMHAPFRE) for deconvoluting haplotypes from genotype data assumes that the genotype frequencies at every SNP site follow the expectations of Hardy-Weinberg equilibrium and that there is free recombination between sites. Although the first assumption appears approximately valid for our data, there is no direct validation for the second one. In fact, manual checking of the genotype data from the CEPH family samples showed a somewhat larger number of haplotypes (19 for BLM, 21 for WRN, and 16 for RECQL, instead of 16, 20, and 15, respectively, inferred by EMHAPFRE). The extra haplotypes detected through manual inference, however, are low frequency ones. We also verified the accuracy of haplotype inference by using a newly developed data-mining algorithm (N. Wang et al., in prep.) that depends on neither the assumption of HWE nor of free recombination. In general, common haplotypes inferred by the EM algorithm are also detected by the data-mining algorithm. The assumption of free recombination in EMHAPFRE results in a somewhat larger number of inferred haplotypes. The extra haplotypes are always rare. Hence, they contribute little to estimates of haplotype diversity and interpopulation variation. Despite these differences, the number of inferred haplotypes for each helicase is much larger than the number of polymorphic sites examined (50 haplotypes for the 8 BLM sites, 56 for the 12 WRN sites, and 47 for the 11 RECQL sites). This clearly indicates that recombination and/or recurrent mutation have been active in producing these haplotypes because unique mutations at these sites can only produce (s + 1) haplotypes with s polymorphic sites (Fu and Li 1993). From genotype data, we ruled out recurrent mutations producing more than two nucleotides as a major factor. However, the possibility of back mutation cannot be distinguished from recombination in the haplotype inference method. The number of segregating sites discovered is different among the helicases. Furthermore, the extent of haplotype variation, expressed in terms of number of haplotypes (Table 3), as well as haplotype frequencies (Fig. 2), differ among these loci.
Recombination Events and Linkage Disequilibrium
Despite the fact that we used a smaller number of segregating sites (8) for the BLM gene in comparison to the others, the resulting number of haplotypes for this helicase is nearly the same as for the others. Inference of recombination events (by the four-gamete test) and plotting significant LD values by pairwise location of sites (Figs. 4, 5) provide an explanation for this observation. In BLM, recombination appears to have occurred throughout the gene in each of the four ethnic groups. In RECQL, recombination events appear to occur predominantly at the 5′ and 3′ ends of the gene, leaving a middle section free of inferred recombination. The opposite (i.e., recombinations occurring in the center of the gene) is seen in WRN. This is consistent with the pattern of LD (Fig. 4) among the sites for these genes as well. For BLM, the tendency of more significant LD from the third SNP-extending 3′ is consistent with the physical location of the sites (Fig. 1; Table 1) because the second and third SNP are ∼100 kb apart.
The recombination patterns and linkage disequilibria observed in the African American sample are somewhat unexpected (Figs. 4, 5). For WRN and RECQL, the African American and the Asian samples exhibit similar patterns of recombination, which appear to be less when compared to the Caucasian and the Hispanic samples. Although not studied here, Africans are expected to have greater diversity reflecting their antiquity and, hence, recombinations should have been more prevalent in them. In all three genes, the Caucasian group shows the highest number of recombination events and haplotypes, with the exception of RECQL, in which the Hispanic sample has the maximum number of haplotypes. The African American and the Hispanic samples are populations of admixed origin, whereas the Caucasian sample is of primarily European ancestry. Therefore, these paradoxical observations cannot be readily explained by gene-specific recombination alone. Whether ascertainment bias (i.e., the SNP-screening protocol) or recurrent mutations are responsible should be studied in more detail.
Our observation that significant LD is found for all three genes in the four ethnic groups within 15 kb in BLM, ∼70 kb in WRN, and 100 kb in RECQL are relevant for the general utility of LD-based gene-mapping approaches. Recent studies suggest a range of 3 kb (Kruglyak 1999) to 500 kb (Thompson and Neel 1997) as target regions within which LD-based mapping approaches is feasible. Our data falls within this range, but the pattern of LD appears to be gene specific. Therefore, we argue that the genomic context should be considered as a factor in LD-based mapping strategies. We showed that the number, as well as the density of SNP markers that can produce similar haplotype diversity, can vary among loci. In a study similar to ours, Bonnen et al. (2000) showed that at the ATM gene region, a longer genomic region (142 kb) carries more restricted recombination and, consequently, tighter LD.
Pilot Association Studies
Our pilot association studies support the utility of SNP-based haplotypes in identifying mutations of disease relevance. In the first pilot study, we identified a haplotype (No. 25) in the BLM gene, which appears to be the ancestral haplotype on which the _blm_Ash mutation occurred in the Ashkenazi Jewish population. Ellis et al. (1998) using microsatellite data obtained similar association results. However, the stability of SNP sites offers opportunities to estimate the age of such disease mutations more precisely.
The importance of ethnically matched controls in disease-gene association studies is revealed in the data of our second pilot study. In particular, the increased frequency of WRN haplotype No. 28 (0.35 compared to 0.22) seen in the Hispanic individuals who are either homozygous or heterozygous for one or both of the two cSNP mutations (c.2592T > G and c.3453T > G) was no longer found in the pooled sample of the four ethnic groups.
Primate Variation in the Respective Loci
Studying the sequence variation in five primate samples at these gene regions served two purposes. First, we observed no variation at any of the SNP sites for all three helicases in the primates. Further, for all three genes, the primate haplotypes were indeed observed in the human populations, albeit in low frequencies (0.02, 0.001, and 0.002 for BLM, WRN, and RECQL, respectively). These helped us infer the ancestral haplotypes in human, allowing rooting of the haplotype phylogeny in our tree reconstruction effort. Second, based on the approximate time of divergence of chimpanzees and human (5 million years, or ∼200,000 generations) and the average frequency of SNP occurrence per base-pair distance (1 in 1 kb), we can estimate the average rate of nucleotide substitution in these three genes. The result (5 × 10−9 per base pair per generation) coincides with the generally accepted value (Nei 1987).
In conclusion, our data show that even with nearly equal nucleotide diversity in different gene regions, haplotype diversity based on several SNP sites may have a very different pattern across genes. Differences of demographic history of populations, superimposed on different patterns of recombination among genes, can result in different haplotype distributions across populations. Therefore, careful ethnic matching and/or family-based sampling designs such as the TDT (Spielman et al. 1993) should be a critical aspect of SNP-based disease-gene association studies.
METHODS
DNA Samples
Lymphoblastoid cell line samples from five unrelated Caucasians were used in screening the genomic regions of BLM, WRN, and RECQL for single nucleotide polymorphisms. ASO analysis was carried out in ten three-generation CEPH families (parents, four children, and paternal and maternal grandparents) and samples from four ethnic groups: Caucasians, African Americans, Hispanics, and Asians. The ethnic samples were a kind gift from Drs. Michael Weil and Michael Story (MD Anderson, Houston, TX). Random, ethnically self-described blood donors in Houston were asked to contribute under informed consent. Blood samples were anonymized and DNAs were coded with ethnic identifiers. A minimum of 70 individuals was screened for each gene, with the exception of the Asian group, which had only 39 samples available. DNA samples and cell lines used in the association studies for the _blm_Ash mutation were obtained from Coriell Cell Repositories (GM00811, GM01492, GM03510, GM02085, GM02520, NA04408, NA09960, NA03403, and NA05289). Of these, six are from individuals of Ashkenazi Jewish origin, homozygous for the mutation. Primate genotypes were ascertained by screening two chimpanzee (Pan troglodytes, PTR), one bonobo (Pan paniscus, PPA), and two gorilla (Gorilla gorilla, GGO) cell lines for the human SNPs. Primate cell lines were kindly provided by Dr. E. Nickerson, Cold Spring Harbor Laboratories.
Analysis of Genomic Sequences
BLM and WRN genomic sequences were obtained from GenBank (accession nos. AC002312 for BLM and AF181896 and AF181897 for WRN). Complete genomic sequence for the RECQL locus was developed in the course of this study from a bacterial artificial chromosome clone from a human library (RPCI11–501E26) and deposited into GenBank under accession no. AC006559.
In all three genes, the genomic sequence was masked for known repetitive elements with RepeatMasker (RepeatMasker available at http://ftp.genome.washington.edu/RM/RepeatMasker.html). The remaining unmasked sequence was used with the MacVector software package to design PCR primers that would amplify single-copy regions of the genes. Exons were avoided where possible.
SNP Identification and Detection
PCR products from five unrelated Caucasians (10 chromosomes) were sequenced using 33P terminators (ThermoSequenase Radiolabeled Terminator Cycle Sequencing kit, USB Pharmaceuticals). Reaction products were visualized by autoradiography of dried polyacrylamide gels. Polymorphic PCR products were combined into multiplex PCR groups for the screening of the ethnic samples. The PCR conditions and the primer sequence for the multiplex groups for the three genes are listed in Supplementary Table 1 available online at http://www.genome.org. Only the conditions for the SNPs used in the haplotype analysis (see Results) are given.
PCR reactions were carried out in a GeneAmp PCR System 9700 Thermocycler (Perkin Elmer) for 35 cycles at 50 μL final volume with 1× reaction buffer (10× PCR Buffer, Perkin Elmer), 250 μM each dNTP (Amersham), 0.5U Taq (Perkin Elmer), and 1 μM each primer, except where otherwise stated. In some reactions 4%–8% DMSO was added to facilitate the annealing of the primer to the template. Six known coding polymorphisms within the WRN gene (Meisslitzer et al. 1997; Ye et al. 1997; Vidal et al. 1998; Castro et al. 1999) were used in an association pilot study. PCR products spanning the polymorphisms were pooled together into two multiplex groups (Supplementary Table 1 available online at http://www.genome.org.) and amplified in 40 Caucasians, 40 African Americans, 40 Hispanics, and 16 Asians. Numbering of coding SNPs reflects their order from 5′ to 3′ of the gene. Prior to sequencing, all PCR products were purified with the PCR product presequencing kit (Amersham). Unwanted remnants of PCR reactions were enzymatically removed using equal volumes of Shrimp alkaline phosphatase (200U) and Exonuclease I (1000U) and incubating samples at 37°C for 15 min and then at 80°C for an additional 15 min.
ASO Hybridization
The protocol that was followed for the ASO design, dot blot of PCR samples, and ASO hybridization has been previously described in DeMarchi and colleagues (1994). Multiplexed PCR products were spotted onto GeneScreen Plus nylon membranes (New Life Science). All ASO probes were 19mers with the variable site usually at the tenth position. As suggested by DeMarchi et al. (1994), to avoid false-positives, the DNA strand from which the ASO was chosen was the one that avoided G:T and G:A mismatches between mutant oligonucleotide and wild-type DNA template. SNPs that did not result in robust PCR products or hybridization signals were not included in the ASO analysis. A total of 16, 24, and 22 ASOs (two oligonucleotides per SNP) were designed and used in the analysis for BLM, WRN, and RECQL, respectively (Supplementary Table 2 available online at http://www.genome.org.).
Forty picomoles of oligonucleotide were 5′-end labeled with γ-32P ATP and purified through G25 Sephadex Quick Spin columns (Boehringer) each time. Labeling reactions were at 100 μL final volume. PCR products were spotted onto membranes which were prehybridized in 5 mL hybridization buffer (DeMarchi et al. 1994) at 65°C for at least 30 min. Hybridization was carried out in the same buffer after adding 25 μg/mL sheared salmon sperm DNA, 80 pmoles the alternative nonradioactive oligo, and one-fifth labeling reaction. Hybridization bags containing the membranes were submerged in a water bath for 30 min at 65°C. The temperature was then adjusted to 34°C and the bags were left to cool overnight with shaking. Membranes were then washed collectively at room temperature in 5× SSC for 5 min twice. Background was removed with an additional wash in 2× SSC for 30 min at a temperature that was empirically determined for each probe. Membranes were exposed to X-omat AR film (Kodak) overnight.
Screening of the samples for the coding SNPs was carried out by ASO hybridization as for the noncoding SNPs. ASO sequences and wash conditions for each gene are shown in Supplementary Table 2 available online at http://www.genome.org. Amplification of the region spanning the _blm_Ash mutation and screening of patients by ASOs were carried out as described by Roa et al (1999).
Haplotype Inference
Haplotypes were ascertained using EMHAPFRE, a computer-assisted statistical analysis based on maximum likelihood. EMHAPFRE infers haplotypes and haplotype frequencies under the assumption of Hardy-Weinberg equilibrium and implementing an expectation-maximization (EM) algorithm. For the CEPH pedigrees, haplotypes were also assigned by inspection as follows. Once a complete homozygote was identified within a family, a haplotype was unambiguously assigned. If an individual had a single heterozygous site, then two haplotypes were unambiguously identified. The remaining unresolved sequences were determined based on the known haplotypes and the inheritance pattern under the assumption of zero recombination events between generations. This method is easily applicable to related samples and the haplotypes of the 10 families studied could be unambiguously assigned. The four ethnic groups (Caucasians, African American, Hispanics, and Asians), however, comprised unrelated individuals and therefore the deduced haplotypes were the result of the EMHAPFRE-based analysis.
Assessing Nucleotide Diversity, Linkage Disequilibrium, and Recombination
DnaSP version 3.14 software package (Rozas and Rozas 1995, 1999) was used to estimate nucleotide variation and test for linkage disequilibrium (LD) and recombination between polymorphic sites in each gene locus. The measure of sequence variability that was calculated in this study is π (Nei 1987), the average number of differences per site between two sequences. The degree of LD is estimated by computing D (Lewontin and Kojima 1960) and D' (Lewontin 1964), and Fisher's exact test was used to determine significance of associations between polymorphic sites. Positions for recombination events were determined using the recombination module of DnaSP. This module calculates the minimum number of recombination events based on the four-gamete test, as described by Hudson and Kaplan (1985).
Acknowledgments
We are indebted to Drs. Michael Weil and Michael Story for providing the ethnic samples and Dr. Elizabeth Nickerson for providing the primate cell lines. We also thank Dr. Ning Wang for providing the Data Mining software package and Dr. Nessan A. Bermingham for constructive comments throughout the preparation of this paper. This work was supported by a grant from the National Cancer Institute of the NIH (CA75432).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nelson@bcm.tmc.edu; FAX (713) 798-5386.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.176702.
REFERENCES
- Adhoc Committee on Mutation Nomenclature. Update on human gene mutations. Hum Mutat. 1996;8:197–202. doi: 10.1002/humu.1380080302. [DOI] [PubMed] [Google Scholar]
- Bonnen PE, Story MD, Ashorn CL, Buchholz TA, Weil MM, Nelson DL. Haplotypes at ATM identify coding-sequence variation and indicate a region of extensive linkage disequilibrium. Am J Hum Genet. 2000;67:1437–1451. doi: 10.1086/316908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Castro E, Ogburn CE, Hunt KE, Tilvis R, Louhija J, Penttinen R, Erkkola R, Panduro A, Riestra R, Piussan C, et al. Polymorphisms at the Werner locus: I. Newly identified polymorphisms, ethnic variability of 1367Cys/Arg, and its stability in a population of Finnish centenarians. Am J Med Genet. 1999;82:399–403. [PubMed] [Google Scholar]
- Cerda-Flores RM, Kshatriya GK, Berion TK, Hewett-Emmett D, Hanis CL, Chakraborty R. Gene diversity and estimation of genetic admixture among Mexican-Americans of Starr County, Texas. Ann Hum Biol. 1992;19:347–360. doi: 10.1080/03014469200002222. [DOI] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengard J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, et al. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet. 1998;63:595–612. doi: 10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarchi JM, Richards CS, Fenwick RG, Pace R, Beaudet AL. A robotics-assisted procedure for large scale cystic fibrosis mutation analysis. Hum Mutat. 1994;4:281–290. doi: 10.1002/humu.1380040409. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Ciocci S, Proytcheva M, Lennon D, Groden J, German J. The Ashkenazic Jewish Bloom syndrome mutation blmAsh is present in non-Jewish Americans of Spanish ancestry. Am J Hum Genet. 1998;63:1685–1693. doi: 10.1086/302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Foucault F, Vaury C, Barakat A, Thibout D, Planchon P, Jaulin C, Praz F, Amor-Gueret M. Characterization of a new BLM mutation associated with a topoisomerase II α defect in a patient with Bloom's syndrome. Hum Mol Genet. 1997;6:1427–1434. doi: 10.1093/hmg/6.9.1427. [DOI] [PubMed] [Google Scholar]
- Fu Y, Li WH. New statistical tests of neutrality for DNA samples from population. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom syndrome: A mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- Goddard KA, Hopkins PJ, Hall JM, Witte JS. Linkage disequilibrium and allele-frequency distributions for 114 single-nucleotide polymorphisms in five populations. Am J Hum Genet. 2000;66:216–234. doi: 10.1086/302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Tanimoto K, Horiuchi Y, Sasazuki T. Family analysis of Werner's syndrome: A survey of 42 Japanese families with a review of the literature. Clin Genet. 1981;19:8–15. doi: 10.1111/j.1399-0004.1981.tb00660.x. [DOI] [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Helland A, Langerod A, Johnsen H, Olsen AO, Skovlund E, Borresen-Dale AL. p53 polymorphism and risk of cervical cancer. Nature. 1998;396:530–531. doi: 10.1038/25034. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson AM, Magnusson PK, Ylitalo N, Quarforth-Tubbin P, Ponten J, Adami HO, Gyllensten UB. p53 polymorphism and risk of cervical cancer. Nature. 1998;396:531. doi: 10.1038/25037. [DOI] [PubMed] [Google Scholar]
- Karow JK, Chakraverty RK, Hickson ID. The Bloom's syndrome gene product is a 3′–5′ DNA helicase. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Morar B, Castiglione CM, Zhao H, Pakstis AJ, Speed WC, Bonne-Tamir B, Lu RB, Goldman D, Lee C, et al. A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet. 1998;103:211–227. doi: 10.1007/s004390050809. [DOI] [PubMed] [Google Scholar]
- Kidd JR, Pakstis AJ, Zhao H, Lu RB, Okonofua FE, Odunsi A, Grigorenko E, Tamir BB, Friedlaender J, Schulz LO, et al. Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet. 2000;66:1882–1899. doi: 10.1086/302952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. Hereditary cancer: Two hits revisited. J Cancer Res Clin Oncol. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat Genet. 1997;17:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- ————— Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Kwok PY, Deng Q, Zakeri H, Taylor SL, Nickerson DA. Increasing the information content of STS-based genome maps: Identifying polymorphisms in mapped STSs. Genomics. 1996;31:123–126. doi: 10.1006/geno.1996.0019. [DOI] [PubMed] [Google Scholar]
- Lai E, Riley J, Purvis I, Roses A. A 4-Mb high-density single nucleotide polymorphism–based map around human APOE. Genomics. 1998;54:31–38. doi: 10.1006/geno.1998.5581. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Kojima K. The evolutionary dynamics of complex polymorphisms. Evolution. 1960;14:450–472. [Google Scholar]
- Li A, Huang Y, Swift M. Neutral sequence variants and haplotypes at the 150 kb ataxia-telangiectasia locus. Am J Med Genet. 1999;86:140–144. [PubMed] [Google Scholar]
- Long AD, Langley CH. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 1999;9:720–731. [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Gilbert JR, Lai EH, Riley J, Rogala AR, Slotterbeck BD, Sipe CA, Grubber JM, Warren LL, Conneally PM, et al. Analysis of association at single nucleotide polymorphisms in the APOE region. Genomics. 2000;63:7–12. doi: 10.1006/geno.1999.6057. [DOI] [PubMed] [Google Scholar]
- Meisslitzer C, Ruppitsch W, Weirich-Schwaiger H, Weirich HG, Jabkowsky J, Klein G, Schweiger M, Hirsch-Kauffmann M. Werner syndrome: Characterization of mutations in the WRN gene in an affected family. Eur J Hum Genet. 1997;5:364–370. [PubMed] [Google Scholar]
- Moser MJ, Oshima J, Monnat RJ., Jr WRN mutations in Werner syndrome. Hum Mutat. 1999;13:271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nickerson DA, Taylor SL, Weiss KM, Clark AG, Hutchinson RG, Stengard J, Salomaa V, Vartiainen E, Boerwinkle E, Sing CF. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat Genet. 1998;19:233–240. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- Ober C, Leavitt SA, Tsalenko A, Howard TD, Hoki DM, Daniel R, Newman DL, Wu X, Parry R, Lester LA, et al. Variation in the interleukin 4-receptor α gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet . 2000;66:517–526. doi: 10.1086/302781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Roa BB, Savino CV, Richards CS. Ashkenazi Jewish population frequency of the Bloom syndrome gene 2281 delta 6ins7 mutation. Genet Test. 1999;3:219–221. doi: 10.1089/gte.1999.3.219. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP, DNA sequence polymorphism: An interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci. 1995;11:621–625. doi: 10.1093/bioinformatics/11.6.621. [DOI] [PubMed] [Google Scholar]
- ————— DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Shahrabani-Gargir L, Shomrat R, Yaron Y, Orr-Urtreger A, Groden J, Legum C. High frequency of a common Bloom syndrome Ashkenazi mutation among Jews of Polish origin. Genet Test. 1998;2:293–296. doi: 10.1089/gte.1998.2.293. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- Thompson EA, Neel JV. Allelic disequilibrium and allele frequency distribution as a function of social and demographic history. Am J Hum Genet. 1997;60:197–204. [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal V, Bay JO, Champomier F, Grancho M, Beauville L, Glowaczower C, Lemery D, Ferrara M, Bignon YJ. The 1396del A mutation and a missense mutation or a rare polymorphism of the WRN gene detected in a French Werner family with a severe phenotype and a case of an unusual vulvar cancer. Hum Mutat. 1998;11:413–414. doi: 10.1002/(SICI)1098-1004(1998)11:5<413::AID-HUMU16>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Wagner TM, Hirtenlehner K, Shen P, Moeslinger R, Muhr D, Fleischmann E, Concin H, Doeller W, Haid A, Lang AH, et al. Global sequence diversity of BRCA2: Analysis of 71 breast cancer families and 95 control individuals of worldwide populations. Hum Mol Genet. 1999;8:413–423. doi: 10.1093/hmg/8.3.413. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Ye L, Miki T, Nakura J, Oshima J, Kamino K, Rakugi H, Ikegami H, Higaki J, Edland SD, Martin GM, et al. Association of a polymorphic variant of the Werner helicase gene with myocardial infarction in a Japanese population. Am J Med Genet. 1997;68:494–498. doi: 10.1002/(sici)1096-8628(19970211)68:4<494::aid-ajmg30>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]