Genome Dynamics in Aging Mice (original) (raw)
Abstract
Random spontaneous genome rearrangements are difficult to detect in vivo, especially in postmitotic tissues. Using a _lacZ_-plasmid reporter mouse model, we have previously presented evidence for the accumulation of large genome rearrangements in various tissues, including postmitotic tissues, during aging. These rearrangements, which were found to be organ-specific and to increase with age, have one breakpoint in the _lacZ_-reporter locus and the second elsewhere in the mouse genome. In this present work, we have used a mouse genome sequence database to physically characterize a total of 49 genome rearrangements in the brain, heart, and liver from young and old mice at two _lacZ_-plasmid reporter loci. Half of all breakpoints in the mouse genome occurred in chromosomes 3 and 4, each carrying a _lacZ_-reporter cluster, at distances varying from <100 kb to 66 Mb, indicating intrachromosomal deletions or inversions. The other half of the breakpoints in the mouse genome was found randomly on any of the other chromosomes, indicating translocations. Alternatively, part of the intra- and extrachromosomal events could involve transpositions. Regions of extended homology were not found at the breakpoints. These results lead us to postulate potential mechanisms for the origin of large genome rearrangements in mouse tissues and to predict their possible impact as a potential cause of aging.
Somatic mutations are thought to play a major role in cancer and possibly aging (DePinho 2000; Vijg 2000). To monitor tissue-specific patterns of somatic mutation accumulation during aging, a plasmid transgenic mouse model sensitive to a broad range of mutational events has been developed (Boerrigter et al. 1995). These mice harbor chromosomally integrated plasmids that can be efficiently recovered from genomic DNA and transferred into a suitable Escherichia coli host for mutant selection, quantitation, and characterization. Using this model, we have previously reported organ-specific differences in mutation accumulation with age (Dollé et al. 1997, 2000). A major advantage of this system is its sensitivity for genome rearrangements. Indeed, we have demonstrated that in addition to point mutations and small deletions, large rearrangements with one breakpoint in the _lacZ_-reporter gene and one in the mouse genome contribute significantly to the spontaneous mutant spectra (Dollé et al. 1997, 2000). Such large mutations are more likely to contribute to age-related cellular degeneration and death than randomly induced point mutations, for example, by haploidization of entire genomic regions or disruption of higher order genome organization (Vijg 2000; Vijg and Dollé 2002).
In view of the potentially high functional impact of genome rearrangements, characterization of such mutations could provide a better understanding of genome dynamics and its possible role in the aging process and in age-related diseases. Until recently, characterization of the rearrangements detected at the _lacZ_-reporter loci was limited to cumbersome genetic mapping, yielding little or no sequence information regarding the breakpoints in the mouse genome (Dollé et al. 2000). With the availability of a near complete mouse genome sequence database (Marshall 2001), physical locations and sequence information surrounding the breakpoints can now be readily obtained.
Here we present complete physical characterization of 49 spontaneous genome rearrangements recovered from somatic tissues of the mouse. Intrachromosomal mutations, up to 66 Mb in size, comprised 53% of all genome rearrangements analyzed. The remaining 47% may have resulted from translocations. No regions of extended homology were found at the breakpoints.
RESULTS
The _lacZ_-plasmid transgenic mice used in this study were homozygous for two integration sites, one on chromosome 3 and one on chromosome 4 (Dollé et al. 2000). The transgenes are integrated as arrays of multiple plasmids arranged in a head to tail fashion (Fig. 1). Previously, mutant frequencies and spectra at the _lacZ_-plasmid reporter loci were determined in the brain, liver, heart, and small intestine of mice of different ages (Dollé et al. 1997, 2000). Individual plasmids were obtained by digesting genomic DNA with Hin_dIII, cutting only once in the wild-type plasmid sequence (Fig. 1). Subsequently, the plasmids were purified using lacI-coated magnetic beads, circularized through self-ligation and electro-transferred into a Δ_lacZ, _galE_− E. coli host for mutant selection. The recovered mutant plasmids were characterized by restriction enzyme analysis on agarose gels. Mutant plasmids with similar restriction patterns as wild-type plasmids, such as point mutations and small deletions, were classified as no-change mutants, whereas plasmids with altered restriction patterns were termed size-change mutants. About 83% of the size-change mutants have been found to represent genome rearrangements with one breakpoint in the transgene and one in the mouse genome (Dollé et al. 2000). These genome rearrangements were recovered as truncated plasmids attached to a piece of mouse genomic sequence (Fig. 1).
Figure 1.
Recovery of integrated _LacZ_-plasmids. Each integration site comprises a concatamer of ∼10 head-to-tail organized plasmid copies (only three plasmid copies are depicted). Individual plasmid copies are released from mouse genomic DNA (red curving lines) by _Hin_dIII digestion (H indicates _Hin_dIII site). After purification, self-ligation, and transformation into Escherichia coli host cells, the plasmids are recovered in the form of ampicillin-resistant colonies. The dashed arrows represent the occurrence and detection of a hypothetical genome rearrangement with one breakpoint in a _lacZ_-reporter gene (lightning-sign) and one in the mouse genome 5′ (①) or 3′ (②) of the integration site or on another chromosome (③). When not deleted, only the upstream truncated plasmid sequence will result in a mutant colony after _Hin_dIII digestion, as the origin of replication (yellow ellipse) and the ampicillin-resistance gene (A) are needed for selective propagation of the transformed E. coli host. The orientation of the cloned 5′ sequences of the integration sites (Seq. Int.; orange curve), indicated by the arrow, corresponds with the direction of the blue arrowheads in Fig. 3 and is arbitrary within this scheme. The physical location and orientation of the cloned 5′ integration site sequence and the recovered mouse genomic sequence in the mutant plasmid allow for characterization of the recovered genome rearrangement. See text for more details.
As indicated in Table 1, the frequency of genome rearrangements was found to vary from organ to organ, being the lowest in the brain and the highest in heart. In addition, a significant increase in the frequency of genome rearrangements was found with age in the heart and liver but not in the brain or small intestine. Assuming that the _lacZ_-plasmid reporter locus is representative for the genome overall, we extrapolated the genome rearrangement frequencies from the 3-kb lacZ transgene to the entire 6 × 109-bp diploid genome. The outcome was subsequently divided by a factor of two, because only one of the two breakpoints needs to occur in a _lacZ_-reporter gene. The results indicate surprisingly high numbers of five to 37 genome rearrangements per cell, depending on the tissue of origin and donor age (Table 1), which prompted us to characterize a representative portion of these mutants in more detail.
Table 1.
Mutant Frequencies and Genome Rearrangements in Organs of Young and Old Mice
| Organ | Age (mo) | LacZ mutant frequency (×10−5)a | Genome rearrangements per cellb | Pc | |
|---|---|---|---|---|---|
| total | genome rearrangements | ||||
| Brain | 4 | 3.1 ± 0.3 | 0.5 ± 0.1 | 5 | 0.7333 |
| 32 | 5.0 ± 1.3 | 0.6 ± 0.4 | 6 | ||
| Liver | 4 | 3.1 ± 0.3 | 0.9 ± 0.2 | 9 | 0.0140 |
| 32 | 10.5 ± 2.4 | 2.7 ± 1.0 | 27 | ||
| Heart | 3 | 4.1 ± 1.4 | 1.9 ± 0.8 | 19 | 0.0262 |
| 32 | 10.1 ± 2.7 | 3.7 ± 2.0 | 37 | ||
| Small intestine | 3 | 6.1 ± 1.8 | 1.5 ± 0.5 | 15 | 0.9999 |
| 32 | 19.5 ± 1.0 | 2.4 ± 2.1 | 24 |
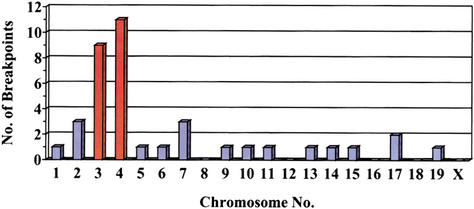
Mouse sequences present in 49 mutant _lacZ_-plasmids, recovered from the brain, liver, and heart of untreated mice of different ages, were sequenced. Subsequent homology searches with these sequence fragments against a mouse genome database led to the characterization of the breakpoints in the mouse genome (Table 2). For a total of 11 breakpoints, a physical location within the genome could not be established: Five mutant plasmids comprised L1-repeat sequences; five recovered mouse genomic fragments were homologous to sequence fragments in the database of unknown location; and one genomic mouse fragment was too short (11 bases) for unambiguous identification. Of the remaining 38 analyzed mutants, a total of 20 (53%) recovered mouse sequences were situated on the chromosomes containing the _lacZ_-plasmid integration sites of this reporter mouse strain: nine and 11 mouse sequences on chromosomes 3 and 4, respectively (Fig. 2). These mutant plasmids presumably represent intrachromosomal rearrangements. Mouse sequence fragments present in the remaining 18 (47%) mutant plasmids localized seemingly random to chromosomes other than 3 and 4 (Fig. 2) and were classified as translocations.
Table 2.
Breakpoints in the pUR288 Transgene and the Mouse Genome of the Two Integration Sites and a Representative Selection of Genome Rearrangements
 |
|---|
Figure 2.
Chromosomal location of genomic mouse fragments recovered as parts of mutated plasmids from the brain, heart, and liver. The bars for chromosomes 3 and 4, containing the plasmid integration sites, are in orange.
None of the characterized rearrangements showed extended regions of homology between the breakpoints in the plasmid and the mouse genome, perhaps with the exception of liver mutant Li133 sharing a 50% homology with the first 38 bases directly 5′ of the breakpoints (Table 2). Occasionally, direct homology at the breakpoints was found for 1 to 6 bp (Table 2). Furthermore, we did not detect any obvious recurring sequence patterns between the breakpoints of different rearrangements.
For further characterization, we took into account that in this mouse model, genome rearrangements can only be detected if the upstream region of the plasmid, containing the ampicillin resistance gene (Ampr) and the origin of replication, remains intact (Fig. 1). The downstream region of a breakpoint in lacZ will not provide ampicillin resistance to the E. coli host cell during selection for transformed colonies (Fig. 1). The orientation of the integrated plasmids in combination with the location and orientation of the recovered mouse genomic sequence allowed us to deduce the simplest mutagenic event that could have resulted in the genome rearrangement detected.
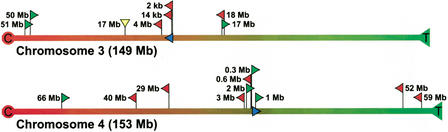
We first determined the physical location and orientation of the integrated _lacZ_-plasmid concatamers. Previously, the 5′ sequences of both integration sites (Fig. 1) had been cloned and localized to chromosomes 3 and 4 by both fluorescence in situ hybridization (FISH) and genetic mapping (Dollé et al. 2000). Part of these flanking sequences and the first 20 bases of the first plasmid copy of each integrated transgene cluster are given in Table 2. Homology searches with the 5′ flanking sequences of the integration sites against the mouse genomic database confirmed localization on chromosomes 3 and 4 (Table 2). The plasmid concatamers are positioned 59.5 and 87.2 Mb from the centromere on chromosome 3 and 4, respectively (Fig. 3). The orientation of the integrated plasmids could now be inferred from the orientation of the 5′ flanking sequence of the integration sites (Fig. 1). The blue arrowheads in Figure 3 indicate the order of Ampr, origin of replication, and lacZ: proximal (toward the centromere) on chromosome 3 and distal (toward the telomere) on chromosome 4.
Figure 3.
Schematic representation of physical locations and orientations of the integrated transgene clusters and genomic mouse fragments, recovered as parts of mutant plasmids, on chromosomes 3 and 4. The red (proximal) and green (distal) arrowheads indicate the direction of the recovered mouse fragments, such that the base and tip of the arrowhead represent the breakpoint and the _Hin_dIII site, respectively (yellow downward arrow indicates direction unknown). The blue arrow indicates the location and orientation of the integrated transgene cluster (see also Fig. 1). The rounded physical distances from the integration site to the localized breakpoints in the mouse genome are shown. C indicates centromere; T, telomere.
The physical locations and orientations of recovered mouse genomic sequences localized on chromosomes 3 and 4 relative to the integrated plasmids on the respective chromosome are also indicated in Figure 3. For both chromosomes, mouse sequences were recovered proximal and distal to the integration sites, with an average distance of ∼20 Mb and up to 66 Mb. The orientation of the recovered mouse sequences did not correlate with a proximal or distal location relative to the integrated plasmid cluster and its orientation on either chromosome (Fig. 3).
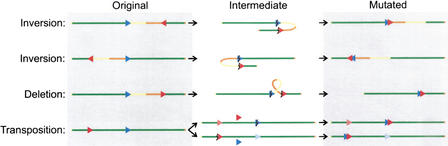
As mentioned above, by taking into account that for a genome rearrangement to be detected, the 5′ plasmid sequence of the breakpoint in lacZ must remain intact and end immediately in front of the recovered mouse sequence, the simplest intrachromosomal mutation that could have taken place was inferred (Fig. 4). Rearrangements with breakpoints in the mouse genome on either site of the integrated plasmid concatamer, but with reversely orientated sequences, could be inversions (Fig. 4). Rearrangements in the direction of the integrated plasmids, proximal for chromosome 3 and distal for chromosome 4 (Fig. 3), with similarly orientated breakpoints in the mouse genome, could be deletions (Fig. 4). Rearrangements in the reverse direction of the integrated plasmids, with reversely orientated mouse sequences, are more complicated and might be owing to transpositions (Fig. 4). According to these schemes, half of the intrachromosomal rearrangements would have been inversions, whereas deletions and transpositions each made up one fourth (Fig. 3). Alternatively, these rearrangements could be explained by translocations involving the transgene clusters integrated on either the homolog or the other chromosome.
Figure 4.
Schematic depiction of proposed mechanisms for observed intrachromosomal rearrangements. The blue and red arrows indicate the orientation of the integrated plasmid loci and the recovered mouse sequences, respectively, on the original non-rearranged chromosome (left column). All four combinations are given for an arbitrarily orientated chromosome (green line). The middle column shows how two breakpoints (lightning signs) could lead to the inversion or deletion of the encompassed chromosomal sequence (yellow-orange dual tone line) and result in a recoverable mutation in the right column. The last row indicates the two options for a transposition, in which either the transgene locus or the recovered mouse sequence is copied or excised (as indicated by the pink and light blue arrows) and integrates in the breakpoint at the other location.
The location of breakpoints on chromosomes other than 3 and 4 (Fig. 2, blue bars) were found throughout the chromosomes. There was no correlation between chromosomal location and orientation; that is, both distal and proximal orientated fragments were found on distal and proximal locations of the chromosomes (data not shown). Because of the contrasting orientation between integrated transgene clusters on chromosomes 3 and 4 (Fig. 3), and the inability to identify which transgene cluster was involved in a particular recovered translocation, we cannot infer the nature of the underlying mechanisms, such as chromosome arm exchanges or chromosome fusions.
DISCUSSION
The detection and characterization of somatic genome rearrangements occurring at random in vivo at low frequencies has been difficult, especially in postmitotic tissues. For example, previous attempts to demonstrate site-specific recombination in the mouse brain using _lacZ_-based reporter constructs yielded conflicting results and left the question open of whether such processes occur in postmitotic tissues (Matsuoka et al. 1991; Schatz and Chun 1992). In this present work, using a different type of transgenic reporter mouse in combination with the mouse genome sequence database, which has recently become available (Marshall 2001), we now provide sequence information on the breakpoints of rearrangements detected as part of mutant lacZ genes in a chromosomally integrated plasmid. The use of this system could help explain the mechanisms by which these events occur in different organs and tissues, why they are subject to organ-specificity, and why they increase with age in some but not in all organs. Surprisingly, the total number of rearrangements in the heart and liver at old age was found to be very high, that is, up to almost 40 events per cell in old heart (Table 1). Based on cytogenetic assays applied to metaphases in cultured cells, mostly lymphocytes, estimates of the frequency of chromosomal aberrations are generally 0.01 to 1 per cell, depending on cell type, age, and the laboratory where the analyses had been carried out. There are at least two explanations for this apparent discrepancy.
One possible explanation for the high number of genome rearrangements observed in this present study is that some or even most of the events scored by us as genome rearrangements are artifacts of the procedure applied to recover the mutant plasmids (Fig. 1). Although it is impossible to completely rule this out, we have addressed the possibility of artifacts extensively in a previous paper in which various control experiments had been performed on plasmids grown in E. coli, mixed with nontransgenic mouse genomic DNA, and mock-rescued into E. coli. Such experiments generally indicated significantly lower mutation frequencies in E. coli than in the mouse and no evidence for genome rearrangements as indicated by a mouse sequence at a lacZ breakpoint (Dollé et al. 1999b). Also, enhanced instability caused by the artificial nature of the _lacZ_-plasmid cluster in the mouse genome is unlikely to be responsible for the observed mutations. Indeed, neither the mutation frequencies nor their spectra are dramatically different from results reported with endogenous reporter genes such as HPRT, APRT, or HLA. Mutation frequencies at these loci were generally in the same range as our own values and also indicated a significant fraction of all mutations caused by genome rearrangements (Grist et al. 1992; Dempsey et al. 1993; Stambrook et al. 1996; Albertini 2001). In general, mutation frequencies at HPRT were among the lowest, possibly because of selection against HPRT mutant lymphocytes in vivo. Interestingly, although virtually all results obtained with HPRT and other endogenous reporters involved lymphocytes, in a study using human kidney cells, significantly higher mutation frequencies were found, that is, up to ∼4 × 10−4, than in lymphocytes (Martin et al. 1996; Colgin et al. 2002). This could reflect a significantly lower selection pressure operating on kidney cells than in lymphocytes. About 15% of these HPRT mutations were genome rearrangements such as deletions. Based on the 44-kb target size of HPRT, a similar extrapolation as performed for the _lacZ_-reporter gene resulted in up to four genome rearrangements per kidney cell, which might be an underestimate owing to the lethality of such events at this X-linked locus. Preliminary data on the same _lacZ_-reporter construct, but now integrated as a single copy transgene, in Drosophila show a similar or even higher frequency of genome rearrangements, also indicating that the concatamer of constructs in the current mouse model is not intrinsically less stable than a single copy transgene. Finally, the observed organ specificities and age-related increase make it highly unlikely that a significant fraction of the mutants scored in our system as genome rearrangements are artifacts.
A second possible explanation for the discrepancy between (1) genome rearrangements scored as lacZ mutants and (2) cytogenetic methods involves the nature of the different assays. Cytogenetic methods have thus far mainly been applied to cultured cells, such as lymphocytes. Although it has recently become possible to apply cytogenetic methods, such as FISH, on interphase cells (Slovak et al. 2001), no results are available for mouse organs and tissues to directly compare with our results. Indeed, the difficulty in analyzing genome rearrangements with cytogenetic methods or methods based on endogenous reporter genes was the reason to generate the lacZ plasmid model system in the first place (Gossen and Vijg 1993). Also the sensitivity of cytogenetic methods to detect rearrangements involving <2 Mb is low. Indeed, possibly the only types of events scored in our system that should be readily detectable by cytogenetic means are large deletions and translocations. However, because our scoring system is based on small mouse sequence fragments, we cannot distinguish deletions and translocations from transpositions. The latter would go undetected by cytogenetic methods. Although we cannot exclude that at least a part of the recovered mutants originated from transpositions, one would then also expect to find transposons to insert into the transgene cluster. Genomic insertion mutations have thus far not been detected in the _lacZ_-plasmid reporter (Martin et al. 2001) and appeared to be extremely rare in reporter gene mouse models in general (de Boer et al. 1997). Moreover, if most of the rearrangements that were scored in our system had resulted from transpositions, it is not clear why half of them occur at chromosomes 3 and 4, where our reporter gene cluster has been integrated (Fig 2). For these reasons, we believe that although part of our rearrangements might reflect transpositions, most of them are indeed large deletions, inversions, or translocations.
With respect to the mechanisms leading to the recovered genome rearrangements, it is probable that they resulted from double-strand breaks (DSBs) in the DNA. DSBs are inflicted by a variety of processes and agents from both endogenous and exogenous sources, such as background radiation, oxygen radicals, and DNA processing enzymes. It has been shown that two simultaneously inflicted DSBs can lead to a variety of recombination processes (Richardson and Jasin 2000). Two homology-dependent and one homology-independent DSB repair pathways have been described for mammalian cells: single-strand annealing (SSA), homologous recombination (HR), and nonhomologous end joining (NHEJ), respectively (for review, see Pastink et al. 2001). The absence of (extended) homology at the breakpoints (Table 2) of the genome rearrangements characterized in the present study implies that NHEJ could have been involved. Our observation that macrohomology was mainly limited to an occasional direct homology of one to six base pairs between breakpoints (Table 2) is in agreement with a recent study by Sandoval and Labhart (2002), which shows that macrohomology is not required for the end joining process.
SSA of direct repeat sequences around a breakpoint is thought to require a minimum of 30-bp homology (Sugawara et al. 2000). Clearly, such breakpoints have not been found in this study (Table 2). It is questionable whether the plasmid transgenic mouse model is suitable to detect SSA. Although regions of homology between lacZ and the mouse genome exist, the most obvious direct repeats around a breakpoint in lacZ are the surrounding plasmid copies of the integrated concatamer. Thus, SSA might resolve a breakpoint in one of the transgenes simply by deleting one or more plasmid copies, without creating a mutation in the remaining plasmid copies. Only when slippage or incorporation errors are made during the repair process might SSA be detected in the form of a point mutation, a small deletion, or insertion. From among the many lacZ point mutants that we characterized in the past (Dollé et al. 2002), evidence has been obtained that such events may indeed occur. The _lacZ_-plasmid mouse model carries multiple nonmutant single nucleotide polymorphisms (SNPs) among its wild-type integrated plasmid copies (Dollé et al. 1999b), which allow differentiation between plasmid copies. Although some of these SNPs are linked (i.e., are present within the same plasmid copy), others are not. Occasionally, point mutations flanked by SNPs that have not shown any linkage, or point mutations flanked by a single SNP that was linked to another, have been found (data not shown) and indicate that recombination between plasmid copies had occurred.
It is unlikely that the plasmid transgenic mouse model, certainly when homozygous for the transgenes, detects HR. Like SSA, only when errors occur during the repair event, might a mutated transgene result. In contrast, HR resulting in loss of heterozygosity has been detected in ∼48% of all spontaneous mutations at the Aprt locus in mouse lymphocytes (Liang et al. 2000). However, HR is a conservative pathway, essentially restoring the original organization of the chromosome. Hence, it seems that large structural changes in the genome mainly result from nonconservative pathways such as SSA and NHEJ.
An important question is whether the observed (up to threefold) induction in spontaneous genome rearrangements, found over the adult lifespan of the mouse (Table 1), could be a causative factor in the aging process (Warner and Johnson 1997). In this respect, it is important to realize that from an evolutionary perspective, maintenance of genome integrity after the period of first reproduction offers no selective advantage (Kirkwood and Austad 2000). It is therefore likely that already at reproductive maturity, the maximum number of somatic mutations, still compatible with optimal fitness, has been reached. Even slight increases above this threshold are likely to adversely impact on the structure and function of an organism. Although the accumulation of point mutations is unlikely to have adverse effects, even a limited number of genome rearrangements, such as the ones described in this paper, could severely affect patterns of gene regulation, namely, through gene dosage or position effects (Vijg 2000; Vijg and Dollé 2002).
METHODS
Plasmid Recovery
Aging cohorts of male C57Bl/6 pUR288-lacZ mice of founder-line 60 were maintained in the animal facilities of the Beth Israel Deaconess Medical Center as described previously (Dollé et al. 1997). The animals were killed by decapitation following asphyxiation by CO2. Organs and tissues were removed, rinsed in PBS, placed in 1.5-mL microcentrifuge tubes, and frozen on dry ice. Any macroscopic lesions observed during tissue collection were excised and stored separately. The tissues were maintained at −80°C until used. DNA was extracted by routine phenol/chloroform extractions.
Complete protocols for plasmid rescue and mutant frequency determinations with this model are given elsewhere (Dollé et al. 1996). Briefly, between 10 and 20 μg genomic DNA was digested with Hind_III for 1 h in the presence of magnetic beads (Dynal) precoated with lacI-lacZ fusion protein. The beads were washed 3 times to remove the unbound mouse genomic DNA. Plasmids were subsequently eluted from the beads by Isopropylthio-β-D-glastocide (IPTG). After circularization of the plasmids with T4 DNA ligase, they were ethanol-precipitated and used to electro-transform E. coli C (Δ_lacZ, galE−) cells. One thousandth of the transformed cells was plated on a titer plate (with X-gal); the remainder, on a selective plate (with p-gal). The plates were incubated for 15 h at 37°C. Mutant frequencies were determined as the number of colonies on the selective plates versus the number of colonies on the titer plate (times the dilution factor of 1000).
Mutant Classification
Mutant colonies were taken from the selective plates and grown overnight in 3 mL Luria-Bertani medium. Then, 1 μL was directly plated on X-gal–supplemented LB-agar medium to screen for galactose insensitive host cells (Dollé et al. 1999a). The remainder of the cell culture was used for plasmid mini preparation (Wizard 9600, Promega). The purified plasmids were digested with _Pst_I and _Ava_I and size-separated on 1% agarose gels. Mutant plasmids with restriction patters resembling and deviating from the wild-type restriction pattern were classified as “no-change” and “size-change” mutants, respectively. Size-change mutants with a fragment of the mouse genome as part of their mutated lacZ gene were called genome rearrangements, as identified by sequencing and/or a polymerase chain reaction screening method as described before (Dollé et al. 2000). To exclude possible cloning artifacts as a result of the accidental inclusion of mouse sequence fragments during plasmid recovery, size-change mutants were routinely digested with _Hin_dIII; only mutant plasmids with a unique _Hin_dIII site were considered in vivo derived genome rearrangements (Dollé et al. 1999b).
Sequencing
Sequence reactions were performed with the CEQ dye terminator cycle sequencing kit (Beckman), according to the manufacturer's standard protocol, and were analyzed with a CEQ 2000 DNA analysis system (Beckman). The primers used were the same as described earlier (Dollé et al. 1999a).
Physical Characterization of Genome Rearrangements
Physical locations, directionality, and missing breakpoint sequence information of the recovered mouse sequences within these mutant plasmids were obtained through blastn searches against the entire mouse genome in the Celera Discovery System and Celera Genomics' associated databases (www.celera.com). The cloned 5′ sequences of the integration sites (Dollé et al. 2000) were used as physical locations for the breakpoints in the lacZ transgene to calculate sizes of intrachromosomal rearrangements.
WEB SITE REFERENCE
www.celera.com; Celera Discovery System and Celera Genomics' associated databases.
Acknowledgments
This work was supported by grant 1PO1AG17242.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dolle@uthscsa.edu; FAX (210) 562-5028.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.125502.
REFERENCES
- Albertini RJ. HPRT mutations in humans: Biomarkers for mechanistic studies. Mutat Res. 2001;489:1–16. doi: 10.1016/s1383-5742(01)00064-3. [DOI] [PubMed] [Google Scholar]
- Boerrigter ME, Dollé ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377:657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- Colgin LM, Hackmann AF, Emond MJ, Monnat RJ., Jr The unexpected landscape of in vivo somatic mutation in a human epithelial cell lineage. Proc Natl Acad Sci. 2002;99:1437–1442. doi: 10.1073/pnas.032655699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer JG, Suri A, Walsh D, Glickman BW. The recovery of a B2 insertion in the lacI gene of a rat cell line containing a λ/LIZ shuttle vector. Mutat Res. 1997;375:79–82. doi: 10.1016/s0027-5107(96)00255-2. [DOI] [PubMed] [Google Scholar]
- Dempsey JL, Pfeiffer M, Morley AA. Effect of dietary restriction on in vivo somatic mutation in mice. Mutat Res. 1993;291:141–145. doi: 10.1016/0165-1161(93)90153-q. [DOI] [PubMed] [Google Scholar]
- DePinho RA. The age of cancer. Nature. 2000;408:248–254. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- Dollé ME, Martus HJ, Gossen JA, Boerrigter ME, Vijg J. Evaluation of a plasmid-based transgenic mouse model for detecting in vivo mutations. Mutagenesis. 1996;11:111–118. doi: 10.1093/mutage/11.1.111. [DOI] [PubMed] [Google Scholar]
- Dollé ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- Dollé ME, Martus HJ, Novak M, van Orsouw NJ, Vijg J. Characterization of color mutants in lacZ plasmid–based transgenic mice, as detected by positive selection. Mutagenesis. 1999a;14:287–293. doi: 10.1093/mutage/14.3.287. [DOI] [PubMed] [Google Scholar]
- Dollé ME, Snyder WK, van Orsouw NJ, Vijg J. Background mutations and polymorphisms in lacZ-plasmid transgenic mice. Environ Mol Mutagen. 1999b;34:112–120. [PubMed] [Google Scholar]
- Dollé ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc Natl Acad Sci. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé ME, Snyder WK, Dunson DB, Vijg J. Mutational fingerprints of aging. Nucleic Acids Res. 2002;30:545–549. doi: 10.1093/nar/30.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen J, Vijg J. Transgenic mice as model systems for studying gene mutations in vivo. Trends Genet. 1993;9:27–31. doi: 10.1016/0168-9525(93)90069-T. [DOI] [PubMed] [Google Scholar]
- Grist SA, McCarron M, Kutlaca A, Turner DR, Morley AA. In vivo human somatic mutation: Frequency and spectrum with age. Mutat Res. 1992;266:189–196. doi: 10.1016/0027-5107(92)90186-6. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- Liang L, Deng L, Shao C, Stambrook PJ, Tischfield JA. In vivo loss of heterozygosity in T-cells of B6C3F1 Aprt+/− mice. Environ Mol Mutagen. 2000;35:150–157. [PubMed] [Google Scholar]
- Marshall E. Genome sequencing: Celera assembles mouse genome; public labs plan new strategy. Science. 2001;292:822. doi: 10.1126/science.292.5518.822. [DOI] [PubMed] [Google Scholar]
- Martin GM, Ogburn CE, Colgin LM, Gown AM, Edland SD, Monnat RJ., Jr Somatic mutations are frequent and increase with age in human kidney epithelial cells. Hum Mol Genet. 1996;5:215–221. doi: 10.1093/hmg/5.2.215. [DOI] [PubMed] [Google Scholar]
- Martin SL, Hopkins CL, Naumer A, Dollé ME, Vijg J. Mutation frequency and type during ageing in mouse seminiferous tubules. Mech Ageing Dev. 2001;122:1321–1331. doi: 10.1016/s0047-6374(01)00267-6. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Nagawa F, Okazaki K, Kingsbury L, Yoshida K, Muller U, Larue DT, Winer JA, Sakano H. Detection of somatic DNA recombination in the transgenic mouse brain. Science. 1991;254:81–86. doi: 10.1126/science.1925563. [DOI] [PubMed] [Google Scholar]
- Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat Res. 2001;480/481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- Sandoval A, Labhart P. Joining of DNA ends bearing non-matching 3′-overhangs. DNA Repair. 2002;1:397–410. doi: 10.1016/s1568-7864(02)00013-7. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Chun JJ. V(D)J recombination and the transgenic brain blues. New Biol. 1992;4:188–196. [PubMed] [Google Scholar]
- Slovak ML, Tcheurekdjian L, Zhang FF, Murata-Collins JL. Simultaneous detection of multiple genetic aberrations in single cells by spectral fluorescence in situ hybridization. Cancer Res. 2001;61:831–836. [PubMed] [Google Scholar]
- Stambrook PJ, Shao C, Stockelman M, Boivin G, Engle SJ, Tischfield JA. APRT: A versatile in vivo resident reporter of local mutation and loss of heterozygosity. Environ Mol Mutagen. 1996;28:471–482. doi: 10.1002/(SICI)1098-2280(1996)28:4<471::AID-EM25>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J. Somatic mutations and aging: A re-evaluation. Mutat Res. 2000;447:117–135. doi: 10.1016/s0027-5107(99)00202-x. [DOI] [PubMed] [Google Scholar]
- Vijg J, Dollé ME. Large genome rearrangements as a primary cause of aging. Mech Ageing Dev. 2002;123:907–915. doi: 10.1016/s0047-6374(02)00028-3. [DOI] [PubMed] [Google Scholar]
- Warner HR, Johnson TE. Parsing age, mutations and time. Nat Genet. 1997;17:368–370. doi: 10.1038/ng1297-368. [DOI] [PubMed] [Google Scholar]