Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing (original) (raw)
Abstract
The spread of multidrug-resistant Staphylococcus aureus (MRSA) strains in the clinical environment has begun to pose serious limits to treatment options. Yet virtually nothing is known about how resistance traits are acquired in vivo. Here, we apply the power of whole-genome sequencing to identify steps in the evolution of multidrug resistance in isogenic S. aureus isolates recovered periodically from the bloodstream of a patient undergoing chemotherapy with vancomycin and other antibiotics. After extensive therapy, the bacterium developed resistance, and treatment failed. Sequencing the first vancomycin susceptible isolate and the last vancomycin nonsusceptible isolate identified genome wide only 35 point mutations in 31 loci. These mutations appeared in a sequential order in isolates that were recovered at intermittent times during chemotherapy in parallel with increasing levels of resistance. The vancomycin nonsusceptible isolates also showed a 100-fold decrease in susceptibility to daptomycin, although this antibiotic was not used in the therapy. One of the mutated loci associated with decreasing vancomycin susceptibility (the vraR operon) was found to also carry mutations in six additional vancomycin nonsusceptible S. aureus isolates belonging to different genetic backgrounds and recovered from different geographic sites. As costs drop, whole-genome sequencing will become a useful tool in elucidating complex pathways of in vivo evolution in bacterial pathogens.
S_taphylococcus aureus_ has remained one of the most frequent causes of a wide range of both hospital- and community-acquired infections, from superficial skin and other soft tissue infections to life threatening toxic shock, pneumonia, endocarditis, and septicemia. The spectacular adaptive capacity of this pathogen resulted in the emergence and worldwide spread of lineages that acquired resistance to the majority of available antimicrobial agents. The choice of therapy against such multidrug-resistant S. aureus (MRSA) strains has been narrowed to a few antibacterial agents, among them the glycopeptide antibiotic vancomycin, which has become the mainstay of therapy worldwide. MRSA strains with reduced susceptibility to vancomycin have been reported in clinical specimen since the late 1990s (1). In most of these so-called vancomycin intermediate-resistant S. aureus (VISA) isolates, decrease in drug susceptibility, as expressed by the increase in the minimal inhibitory concentration (MIC) of vancomycin, is sufficient to cause complications in therapy and treatment failure (2–7). VISA-type resistance has now been identified in each of the globally spread pandemic clones of MRSA (8).
The genetic basis of VISA-type resistance to vancomycin is unknown. Unlike the most recently described and currently still rare VRSA isolates which carry the Tn_1546_-linked resistance mechanism (9, 10), the VISA-type isolates do not seem to carry acquired genetic elements related to drug resistance: their reduced susceptibility to vancomycin appears to be based on a gradual adaptive process.
Examination of VISA-type isolates recovered from many parts of the world showed a number of different phenotypic alterations, including changes in cell morphology and changes in the composition, thickness, and/or turnover of cell walls (11, 12). Nevertheless, associating these altered properties with the mechanism of resistance has remained problematic because of the lack of availability of an isogenic vancomycin susceptible “parental” isolate that could be used as a valid comparison. For instance, comparing the sequences of the first clinical VISA isolate MU50 to the genetically related vancomycin susceptible strain N315 identified over 174 ORFs that carried nonsynonymous changes (13, 14). However, MRSA strain N315 was isolated 15 years earlier than strain Mu50 and from a different patient. Thus, it is not clear how many of the 174 mutations are related to the mechanism of drug resistance versus the different evolutionary history of the strains.
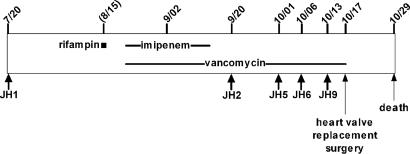
Recently we obtained a series of MRSA isolates from the blood stream of a patient with congenital heart disease who was treated extensively with vancomycin without success (15). Available clinical data suggests that the primary site of infection was endocarditis.‡‡ In addition to vancomycin, the patient also received a single dose of rifampin and a course of therapy with the β-lactam antibiotic imipenem. After ≈12 weeks of therapy and replacement of a heart valve, the patient died because of complications of the underlying disease.
The first isolate JH1 recovered before the beginning of chemotherapy was fully susceptible to vancomycin (MIC = 1 μg/ml). Vancomycin therapy was begun between the culture isolation of JH1 and JH2. The last isolate JH9 recovered at the end of chemotherapy showed decreased susceptibility to vancomycin (MIC = 8 μg/ml). Comparison of the series of JH isolates by several genetic typing techniques indicated that they were isogenic (15, 16). The JH lineage was also related, although more remotely, to the fully sequenced MRSA strains N315 and MU50 (17).
The availability of these isogenic isolates offered a unique opportunity to identify steps in the in vivo evolution of drug resistance by sequencing the genomes of the initial (drug susceptible) and the terminal (drug resistant) isolates.
Results and Discussion
Thirty-Five Point Mutations Were Identified Between the First (JH1) and the Last (JH9) Blood Isolates from the Patient.
When the sequence data were analyzed, both JH1 and JH9 were found to contain one 2.9-Mb chromosome and a 30-kb plasmid, and the two isolates were predicted to differ by only 35 point mutations, 33 of which were on the chromosome, and two of which were on the plasmid. With the exception of two of the predicted mutations [see mutations numbered 34 and 35 in Table 1 and supporting information (SI) Appendix], all of the predicted mutations were confirmed by PCR sequencing. (In the two exceptional cases, the loci harboring the putative mutations could not be PCR sequenced because of technical reasons; see SI Appendix.) The list of 35 mutations is predicted to be nearly exhaustive, with <2 point mutations on the chromosome and zero mutations on the plasmid that are expected to have gone undetected (see SI Appendix).
Table 1.
Sequential appearance of 35 point mutations in the blood isolates
| Date of isolation, month/day/year | Isolate | MIC,* μg/ml | Numeric identifier of mutation† | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vancomycin | Rifampin | Oxacillin | Daptomycin | 1‡ | 2§ | 3–6¶ | 7‖ | 8** | 9†† | 10‡‡ | 11§§ | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19–33 | 34 and 35 | ||
| 7/20/2000 | JH1 | 1.0 | 0.012 | 0.75 | 0.01 | ● | ND | |||||||||||||||
| 9/20/2000 | JH2 | 4.0 | 16 | 25 | 0.05 | ○ | ● | ● | ● | ● | ND | |||||||||||
| 10/1/2000 | JH5 | 6.0 | 16 | 0.75 | 0.05 | ○ | ● | ● | ● | ● | ● | ND | ||||||||||
| 10/6/2000 | JH6 | 8.0 | 16 | 1.5 | 1.0 | ○ | ● | ● | ● | ● | ○ | ● | ● | ● | ● | ● | ● | ● | ● | ● | ND | |
| 10/13/2000 | JH9 | 8.0 | 16 | 0.75 | 1.0 | ○ | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ND |
These data confirm and refine the isogenic nature of the JH isolates. The 35 mutations identified in the terminal vancomycin nonsusceptible isolate were presumably generated by the selective pressures of antibiotic treatment with the three antibiotics in the complex in vivo environment of the invaded host.
Number of Loci Effected and Categories of Mutations.
The 35 point mutations that appeared in JH9 by the end of antibiotic treatment occurred in 31 distinct loci and included 25 substitutions, 1 insertion, and 9 deletions. The first 18 of these mutations are listed in Table 2 in the time order of their appearance. All 35 mutations are described in more detail in SI Appendix. Only 6 of the 35 mutations were synonymous substitutions.
Table 2.
Descriptions of the point mutations in Table 1 found in JH1-JH6
| No.* | Isolate in which mutation appears first | Mutated locus† | Locus previously implicated in resistance to | Description of mutated locus | Mutation‡ |
|---|---|---|---|---|---|
| 1 | JH1 | SAP011 (blaR1) (on the plasmid) | β-lactams (21) | Involved in regulation of β-lactamase gene blaZ and broad spectrum β-lactam resistance gene mecA | Deletion in A× 8 frameshifted last 70% of gene. |
| 2 | JH2 | SA1702 | Vancomycin and β-lactams (23, 39, 46, 47) | In operon with gene vraR, which is possibly involved in the regulation of cell wall synthesis | Amino acid change H164R |
| 3 | JH2 | SA0500 (rpoB) | Rifampin (18) | Codes for β-subunit of RNA polymerase | Amino acid change D471Y |
| 4 | Amino acid change A473S | ||||
| 5 | Amino acid change A477S | ||||
| 6 | Amino acid change E478D | ||||
| 7 | JH2 | SA0501 (rpoC) | Daptomycin (38) | Codes for β'-subunit of RNA polymerase | Amino acid change E854K |
| 8 | JH2 | SA1129 | Contains match to RNA binding motif | Amino acid change D296Y | |
| 9 | JH5 | SA1249 | Putative gene possibly in an operon with the gene murG, which is involved in cell wall synthesis | Deletion in G×9 frameshifted last 80% of gene. | |
| 10 | JH6 | SA1843 (agrC) | Vancomycin (24, 25) | Part of agr locus involved in qorum sensing and regulation of the expression of virulence and cell surface proteins | Deletion in T×9 frameshifted last 70% of gene. |
| 11 | JH6 | SA0019 (yycH) | Daptomycin (38) | In a gene cluster with the gene yycF and possibly involved in the regulation of the autolysin gene lytM | Substitution introduced stop codon, truncating protein to 10% of its length. |
| 12 | JH6 | Between divergently transcribed genes SAS014 and SA0411 (ndhF) | NdhF is the F-subunit of NADH dehydrogenase. | Deletion in T×7 upstream of both SAS014 and SA0411 | |
| 13 | JH6 | SA0582 | Similar to Na+/H+ antiporter subunit MrpE in B. subtilis | Synonymous substitution | |
| 14 | JH6 | SA0980 (isdE) | Involved in passage of heme-iron to cytoplasm during pathogenesis | Amino acid change A84V | |
| 15 | JH6 | SA1659 (prsA) | Chaperone that assists post-translocational folding of proteins at the cytoplasmic/cell wall interface | Deletion in A×7 frameshifted last 15% of gene. | |
| 16 | JH6 | SA2094 | Similar to malic/Na+-lactate antiporter MleN in B. subtilis | Amino acid change A94T | |
| 17 | JH6 | Between divergently transcribed genes SA2125 and SA2126 | SA2125 matches protein family consisting of arginases, agmatinases, and formiminoglutamases. | Substitution upstream of both SA2125 and SA2126 | |
| 18 | JH6 | SA2320 (pfoR) | Contains a match to a domain of a sugar specific permease | Synonymous substitution |
Sequential Appearance of Antibiotic Resistance and Mutations in the JH Isolates During Antibiotic Treatment.
The time course of antibiotic treatment is shown in Fig. 1, and Table 1 summarizes the parallel changes observed in the antibiotic susceptibility profiles of the isolates. To explore parallels between the changes observed in antibiotic susceptibility and genetic changes, loci found mutated between JH1 and JH9 were PCR sequenced in the intermittent blood isolates JH2, JH5, and JH6. The same loci were also PCR sequenced in an additional fully vancomycin susceptible isolate JH15 that was recovered from the nares of a healthy contact of the patient (15).
Fig. 1.
Dates of antibiotic exposure and recoveries of MRSA isolates (month/day in year 2000).
The results summarized in Table 1 allowed us to make several conclusions.
- The first fully antibiotic susceptible blood isolate JH1 and the also fully susceptible contact isolate JH15 (data not shown) had identically low MICs to the antibiotics tested and carried only mutation 1.
- Once a mutation appeared in an early blood isolate, it was retained in all subsequent blood isolates (with the single exception of mutation 9). Thus, the genetic changes appeared in a sequential order in parallel with the increasing vancomycin MIC values.
- Following the brief rifampin therapy, the rifampin MIC of isolate JH1 (0.012 μg/ml) increased to 16 μg/ml in JH2 and remained at that level in all subsequent isolates.
- In parallel with the treatment with the beta-lactam antibiotic imipenem, the beta-lactam (oxacillin) MIC of strain JH1 (0.75 μg/ml) increased to 25 μg/ml in JH2 but then declined to between 0.75 and 1.5 μg/ml in all subsequent isolates.
- The vancomycin susceptibility of the isolates declined gradually in several discrete steps: the vancomycin MIC of strain JH1 (1 μg/ml) increased to 4 μg/ml in JH2, 6 μg/ml in JH5, and its maximum value of 8 μg/ml in JH6 and JH9.
- The MIC value for daptomycin also increased in the JH isolates, although this newly introduced antimicrobial agent had not been used in the therapy. The daptomycin MIC of JH1 (0.01 μg/ml) increased to 0.05 μg/ml in JH2 and JH5 and then underwent a further jump to 1.0 μg/ml in JH6 and JH9.
Appearance of Mutations Associated with Rifampin Resistance.
The >1,000-fold increase in the rifampin MIC from 0.012 to 16 μg/ml between JH1 and JH2 is likely related to mutations 3–6 (Table 1), involving four amino acid changes (D471Y, A473S, A477S, and E478D) in the β-subunit RpoB of RNA polymerase (Table 2). The change D471Y has alone been shown to confer rifampin resistance, and all four changes occur in the region of amino acids 463–550 found to harbor the majority of mutations responsible for rifampin resistance in S. aureus (see ref. 18 for review). Most rifampin resistant S. aureus mutants are reported to have only one or two amino acid changes in RpoB. However, these changes frequently reduce fitness. It is possible that some of the four changes we report here in RpoB are compensatory mutations that helped to offset a loss in fitness. In prior work, rifampin resistant Escherichia coli mutants were passaged through hundreds of generations in vitro and were observed to evolve increased fitness by compensatory mutations in RpoB rather than by reversion to drug sensitivity (19).
Appearance of a Mutation Associated with β-Lactam Resistance.
The sharp rise in resistance to oxacillin (a β-lactam) between JH1 and JH2 is accompanied by the reversal of mutation 1. Found in both JH1 and JH15, mutation 1 involves a deletion of an adenine in a run of eight adenines that frameshifted 70% of blaR1 on the plasmid. The transmembrane sensor BlaR1 and its cognate cytosolic repressor BlaI are part of a signaling pathway that senses β-lactams and induces the expression of the β-lactamase gene blaZ and the broad spectrum β-lactam resistance gene mecA. It is known that β-lactams bind to and thereby activate BlaR1, which then in turn deactivates BlaI to alleviate the BlaI repression of blaZ and mecA (20). Therefore, the inactivation of BlaR1 would be expected to lead to the BlaI mediated constitutive repression of both blaZ and mecA. Hence, it is no surprise that a frameshift in blaR1 has been previously shown to abolish resistance to β-lactams (including to β-lactams, like imipenem and oxacillin, which are not susceptible to cleavage by β-lactamase) (21). The reversal of the frameshift in blaR1 may therefore be responsible for the increase in oxacillin resistance in JH2. Even though blaR1 remains in frame in JH2 onwards, the oxacillin resistance drops in subsequent isolates (beginning with JH5) as the vancomycin resistance increases. Such an inverse relationship between oxacillin and vancomycin resistance has been demonstrated in vitro (22). The mechanism of this effect is not known.
Stages in Vancomycin Resistance.
Between JH1 and JH2, the vancomycin MIC increases from 1.0 to 4.0 μg/ml. Along with this increase in resistance appears mutation 2, involving the amino acid change H164R in the protein SA1702. Although the gene SA1702 has an unknown function, it is in an operon with the gene vraR, which codes for a response regulator of a two-component system. Up-regulation of vraR has been shown to increase the vancomycin MIC by 4-fold in one S. aureus strain (23), and increased transcription of vraR in JH9 compared with JH1 was observed in a study with DNA microarrays (16). A further increase (from 4 to 6 μg/ml) in vancomycin MIC occurs between JH2 and JH5 and is accompanied by a single additional mutation in SA1249, a gene of unknown function.
Between JH5 and JH6.
The vancomycin MIC increases further from 6 to 8 μg/ml, associated with the appearance of mutations 10–18. Mutation 10 appears to frameshift 70% of agrC in the agr locus. Involved in quorum sensing, the agr locus regulates many virulence and cell surface genes. Loss of the locus has already been proposed to be associated with the VISA phenotype (24, 25). The increase in the vancomycin MIC between JH5 and JH6 is also linked to mutation 11, involving a substitution that introduces a premature stop codon in yycH, truncating the gene to 10% of its length. The gene yycH is in the yyc gene cluster with the gene yycF, which codes for the response regulator of a two-component system. In Bacillus subtilis, deletion of yycH results in the up-regulation of genes controlled by yycF (26). In S. aureus, the regulator YycF was shown to bind upstream of the gene lytM, coding for an autolysin (27). Hence, loss of yycH might be expected to lead to up-regulation of lytM, and indeed, transcriptome profiling has shown that the expression of lytM is 5-fold greater in JH9 compared with JH1 (16). Vancomycin blocks access of LytM to its cell-wall substrate (28), and the up-regulation of lytM may be a response to this inhibition. Although lytM is overexpressed in JH9, JH9 was shown to have decreased susceptibility to autolysis, which may be related to an abnormality and/or over-production of teichoic acids (11).
Reversions of Mutation 9 and the Potential Role of Homopolymeric Tracts in the Evolution of Resistance in the JH Isolates.
There are two possible explanations why mutation 9 (involving a frameshift in SA1249) reverts between the blood isolates JH5, JH6, and JH9 (Tables 1 and 2). It is possible that the population of S. aureus in the blood of the patient was heterogeneous, with some individuals carrying the wild-type allele of SA1249 and other individuals carrying the mutant allele. Alternatively, mutation 9 may have appeared and fixed in all of the bacteria, then disappeared entirely, and finally reappeared and fixed again. Both scenarios are plausible, because mutation 9 is a deletion of a cystine in a string of nine cystines. Because of slippage of the DNA polymerase, a homopolymeric tract can rapidly increase or decrease in length (29). In our list of 33 confirmed point mutations, changes in runs of identical nucleotides are statistically overrepresented (P = 10−7) (SI Appendix). Our 33 confirmed point mutations include 1-bp expansions or contractions of eight distinct homopolymeric segments, all initially ≥6 bp (mutations 1, 9, 10, 12, 15, 21, 22, and 33). Because rapid changes in phenotype have been associated with variable length simple repeats (e.g., phase variation in Neisseria meningitidis) (30), it is tempting to speculate that runs of identical nucleotides in S. aureus can promote quick switching between resistant phenotypes. In particular, the VISA phenotype has been noted to be unstable (31).
Sequential Appearance of Mutations.
The data summarized in Table 1 strongly suggest that we are observing genetic alterations in a single S. aureus lineage under the selective pressure of the antimicrobial agents used for the therapy. The selection for these mutations and the emergence of genetically altered bacterial populations most likely occurred at the primary site of infection: in the endocarditic vegetation of the infected heart valve, which is known to be capable of carrying large (109–1011 bacteria per g of tissue) and physiologically heterogeneous populations of bacteria (32), which then “seed” the blood stream followed by rapid clearance (33).
We also had access to a single additional isolate (JH14), which was recovered from the heart-valve after replacement surgery performed on 17 November 2000 (Fig. 1). The heart valve isolate JH14 had exactly the same antibiotic resistance and mutational profiles as JH6: resistance to rifampin, decreased susceptibility to daptomycin (MIC = 1 μg/ml) and to vancomycin (MIC = 8 μg/ml), and the same set of confirmed mutations (2–8 and 10–18) identified in isolate JH6 and also carried in JH9. These observations suggest that the “extra” 16 confirmed mutations (9 and 19–33) identified only in JH9 reflect some unknown selective pressure operative in the complex and heterogeneous environment of the endocarditic lesion but are not essential for the decreased susceptibility to vancomycin. Isolate JH9 may have originated from an area of the endocarditic vegetation where growth and replication of bacteria can be slow (32), leading to the appearance of traits, such as antibiotic tolerance and/or persistence, which are often associated with slow growth. Decreased growth rate and slow loss of viability during antibiotic treatment has been documented in JH9 (11).
Decreased Susceptibility to Daptomycin in the JH Isolates.
We were surprised to find that the MIC value for daptomycin also increased in the JH isolates, although this antibiotic was not used in the therapy.
The antibiotic daptomycin was introduced in 2003 as a therapeutic agent against MRSA infections, including strains with decreased susceptibility to vancomycin (34). The current clinical breakpoint for resistance to daptomycin is defined as an MIC ≥ 2 μg/ml (7). Although the effectiveness of daptomycin against a number of clinical VISA strains was demonstrated (35), more recent studies found a strong positive correlation between reduced susceptibility to vancomycin and daptomycin among VISA isolates (36, 37). In one of these studies (36), 70 independent clinical and laboratory VISA isolates with vancomycin MICs between 4–16 μg/ml were tested, and >80% were observed to have daptomycin MICs ≥ 2 μg/ml.
The genetic mechanisms underlying decreased susceptibility to both vancomycin and daptomycin are unknown. It was therefore of interest that the late JH isolates harbor mutation 7 in rpoC coding for the β′-subunit of RNA polymerase and mutation 11 in the yyc gene cluster (Table 1). These loci were two of only several loci found genome-wide to be mutated in laboratory generated S. aureus mutants with reduced susceptibility to daptomycin (38). In our study, the daptomycin MIC of JHI (0.01 μg/ml) increased to 0.05 μg/ml in JH2-JH5 followed by a sharp increase to 1 μg/ml in JH6 onwards. These increases in resistance coincided with the mutations in rpoC and the yyc gene cluster.
A Limited Number of Mutations Can Cause Extensive Changes in the Gene Expression Profile.
Although only 35 point mutations in 31 loci are observed between JH1 and JH9, prior transcriptome profiling work found 224 genes that were differentially expressed by 2-fold or more between the isolates (16). We assessed the overlap between this list of 224 genes and three other lists of transcriptional changes described in the literature: the changes in the expression of 48 genes controlled by vraR (39); 244 genes controlled by positive regulators of the agr locus (40–42); and 32 genes controlled by yycF (27). Each of the loci vraR, agr, and yycF codes for a transcriptional regulator that appears to be involved with decreased susceptibility to vancomycin. In each case, the overlap was statistically significant (P = 10−11, 10−2, and 10−2, respectively) (see SI Appendix). Thus, many of the mRNA expression changes between JH1 and JH9 are likely due to the mutations 2, 10, and 11 (Table 1). Additionally, mutations 3–6 (Table 1) may effect the expression of many genes, because nonsynonymous substitutions in rpoB conferring rifampin resistance in B. subtilis were shown to have pleitropic effects (43).
The Relationship of the JH Isolates to Previously Sequenced S. aureus.
In prior work (15), multilocus sequence typing (MLST) (44) showed that the JH isolates (each with an MLST 1-4-1-4-12-1-28) were closely related to two sequenced clinical S. aureus isolates: the MRSA strain N315 and the rifampin resistant VISA MU50 (both with a MLST 1-4-1-4-12-1-10). Similar to JH1 and JH9, N315 and MU50 were each found when sequenced to contain one 2.8- to 2.9-Mb chromosome and a 20- to 30-kb plasmid (17). In this work, multialignments of orthologous JH1, N315, and MU50 chromosomal sequence were constructed, and a randomly chosen subset of 50 cases were examined where the N315 and MU50 sequences differed from each other by an isolated nucleotide difference. Among these cases, JH1 was found to agree with N315 three-fourths of the time, indicating that JH1 is more closely related to N315 than MU50. Despite the similar MLSTs of JH1 and N315, a full genome-wide comparison indicated that the two isolates differ on the chromosome by >500 mutations ranging in size from 1 to 50,000 bp (see SI Appendix). Almost all of the >500 reported mutations are expected to be real, with at most only a very small fraction representing sequencing errors (see SI Appendix). As for the JH1 plasmid, it was found to be a chimera of the N315 and MU50 plasmids (SI Appendix). The sheer number of mutations between JH1 and N315 again highlights the need for the careful selection of isolates for an intergenomic comparison to be informative.
Common Mutations in Geographically Diverse VISA Isolates.
It was of interest to check whether any of the loci mutated in JH9 were also mutated in the fully sequenced rifampin resistant VISA strain Mu50. MU50 was found to harbor mutations in the genes vraS in the vraR operon, agrA in the agr locus, and rpoB. In MU50, the mutation in rpoB (a single amino acid change H481Y in RpoB) was shown to confer rifampin resistance (18). It has already been shown that many geographically diverse VISA isolates carry loss of function mutations in the agr locus (24, 25).
The Status of the vraR Operon in Geographically Diverse VISA Isolates.
Because the vraR operon was mutated in both JH9 and MU50, we proceeded to PCR sequence the vraR operon in other VISA isolates. In total, we knew or determined the sequence of the vraR operon in six geographically diverse clinical VISA isolates (vancomycin MICs = 4–8 μg/ml) from different genetic backgrounds: JH9 isolated in Maryland,‡‡ MU50 from Japan (17), PC3 from New York (3), VNJ from New Jersey (45), VMI from Michigan (45), and HSMB1 from Portugal (H.d.L., unpublished data). We also determined the sequence of the vraR operon in a laboratory mutant VM3 (vancomycin MIC = 6 μg/ml) (22). In all seven VISA isolates, a mutation was found in the vraR operon, and this was shown to be highly statistically significant [P < 0.001 (possibly much less)] (see SI Appendix).
In Table 1, mutation 2 in SA1702 in the vraR operon is one of only eight genetic changes that occur between JH1 and JH2, during which time the vancomycin MIC value increases from 1 to 4 μg/ml. Of the other mutations, mutation 1 in blaR1 and mutations 3–6 in rpoB are likely to be related to β-lactam and rifampin resistance respectively. Thus, the preponderance of evidence strongly suggests that genetic alterations in the vraR operon, a key regulatory system involved with monitoring cell-wall biosynthesis in S. aureus (39, 46, 47), plays an important role in the VISA type vancomycin resistance.
From Genetic Change to Altered Phenotype.
Our discovery of an ordered series of mutations leading to a level of VISA type vancomycin resistance that can compromise therapy provides a short list of mutations that should enable us to design experiments to reconstruct the vancomycin resistant phenotype in susceptible strains of S. aureus. Polygenic traits are very hard to trace by association studies, and even expression array data may be hard to interpret when regulators with pleiotropic functions are involved. Our short list of mutant loci can easily be screened in other VISA strains to determine their prevalence.
With many new inexpensive differential sequencing technologies (48) on the horizon, subsequent clinical studies could collect more samples to temporally resolve the appearance of single mutations and the population structure of the bacteria as has been done for HIV (49). The spread and emergence of virulent strains among patients could be followed.
Methods
For full details, see SI Appendix.
For JH1 (JH9), the whole-genome shotgun sequencing was carried out to a mean coverage of 8.5× (9.5×), with 98.5% (97.7%) of the chromosome having a coverage ≥1×. The JH1 and JH9 assembled sequences and reads were multialigned, and a Bayesian probabilistic model was used to identify mutations in the multialignment. In 94% of the genome, the read coverage and quality was sufficient to call mutations between JH1 and JH9 with an expected error rate (both false positive and negative) of zero. The number of mutations estimated to have gone undetected in the remaining 6% of the genome is <2 point mutations. PCR sequencing was done to confirm the model's predictions and to rule out marginal cases.
Supplementary Material
Supporting Appendix
Acknowledgments
We thank Seth Darst, Lu Bai, and Alex Morozov for valuable discussions on the genes rpoB and rpoC and Neal Steigbigel for discussion on medical/microbiological aspects of the paper. This work was supported by United States Public Health Service Grant RO1 AI37275NIH (to A.T.) and National Science Foundation Grant DMR 129848 (to E.D.S.).
Abbreviations
MIC
minimal inhibitory concentration
MRSA
multidrug-resistant S. aureus
VISA
vancomycin intermediate-resistant S. aureus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
‡‡
Flayhart, D., Hanlon, A., Wakefield, T., Ross, T., Borio, L., Dick, J. (2001) in Abstracts of the 101st General Meeting of the American Society of Microbiology, May 20–24, 2001, Orlando, FL, Abstr. A-39.
References
- 1.Appelbaum PC. Clin Microbiol Infect. 2006;12(Suppl 1):16–23. doi: 10.1111/j.1469-0691.2006.01344.x. [DOI] [PubMed] [Google Scholar]
- 2.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 3.Sieradzki K, Roberts RB, Haber SW, Tomasz A. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 4.Charles PG, Ward PB, Johnson PD, Howden BP, Grayson ML. Clin Infect Dis. 2004;38:448–451. doi: 10.1086/381093. [DOI] [PubMed] [Google Scholar]
- 5.Howden BP, Ward PB, Charles PG, Korman TM, Fuller A, du Cros P, Grabsch EA, Roberts SA, Robson J, Read K, et al. Clin Infect Dis. 2004;38:521–528. doi: 10.1086/381202. [DOI] [PubMed] [Google Scholar]
- 6.Howden BP. Intern Med J. 2005;35(Suppl 2):S136–S140. doi: 10.1111/j.1444-0903.2005.00986.x. [DOI] [PubMed] [Google Scholar]
- 7.Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 16th informational supplement M100–S16. Wayne, PA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- 8.Howe RA, Monk A, Wootton M, Walsh TR, Enright MC. Emerg Infect Dis. 2004;10:855–857. doi: 10.3201/eid1005.030556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 10.Severin A, Tabei K, Tenover F, Chung M, Clarke N, Tomasz A. J Biol Chem. 2004;279:3398–3407. doi: 10.1074/jbc.M309593200. [DOI] [PubMed] [Google Scholar]
- 11.Sieradzki K, Tomasz A. J Bacteriol. 2003;185:7103–7110. doi: 10.1128/JB.185.24.7103-7110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeltz RF, Wilkinson BJ. Curr Drug Targets Infect Disord. 2004;4:273–294. doi: 10.2174/1568005043340470. [DOI] [PubMed] [Google Scholar]
- 13.Avison MB, Bennett PM, Howe RA, Walsh TR. J Antimicrob Chemother. 2002;49:255–260. doi: 10.1093/jac/49.2.255. [DOI] [PubMed] [Google Scholar]
- 14.Ohta T, Hirakawa H, Morikawa K, Maruyama A, Inose Y, Yamashita A, Oshima K, Kuroda M, Hattori M, Hiramatsu K, et al. DNA Res. 2004;11:51–56. doi: 10.1093/dnares/11.1.51. [DOI] [PubMed] [Google Scholar]
- 15.Sieradzki K, Leski T, Dick J, Borio L, Tomasz A. J Clin Microbiol. 2003;41:1687–1693. doi: 10.1128/JCM.41.4.1687-1693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAleese F, Wu SW, Sieradzki K, Dunman P, Murphy E, Projan S, Tomasz A. J Bacteriol. 2006;188:1120–1133. doi: 10.1128/JB.188.3.1120-1133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 18.O'Neill AJ, Huovinen T, Fishwick CW, Chopra I. Antimicrob Agents Chemother. 2006;50:298–309. doi: 10.1128/AAC.50.1.298-309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds MG. Genetics. 2000;156:1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 21.Hackbarth CJ, Miick C, Chambers HF. Antimicrob Agents Chemother. 1994;38:2568–2571. doi: 10.1128/aac.38.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieradzki K, Tomasz A. J Bacteriol. 1999;181:7566–7570. doi: 10.1128/jb.181.24.7566-7570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda M, Kuwahara-Arai K, Hiramatsu K. Biochem Biophys Res Commun. 2000;269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 24.Sakoulas G, Eliopoulos GM, Moellering RC, Jr, Wennersten C, Venkataraman L, Novick RP, Gold HS. Antimicrob Agents Chemother. 2002;46:1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakoulas G, Eliopoulos GM, Fowler VG, Jr, Moellering RC, Jr, Novick RP, Lucindo N, Yeaman MR, Bayer AS. Antimicrob Agents Chemother. 2005;49:2687–2692. doi: 10.1128/AAC.49.7.2687-2692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szurmant H, Nelson K, Kim EJ, Perego M, Hoch JA. J Bacteriol. 2005;187:5419–5426. doi: 10.1128/JB.187.15.5419-5426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubrac S, Msadek T. J Bacteriol. 2004;186:1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sieradzki K, Tomasz A. Antimicrob Agents Chemother. 2006;50:527–533. doi: 10.1128/AAC.50.2.527-533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaaper RM, Dunn RL. Proc Natl Acad Sci USA. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moxon ER, Rainey PB, Nowak MA, Lenski RE. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 31.Boyle-Vavra S, Berke SK, Lee JC, Daum RS. Antimicrob Agents Chemother. 2000;44:272–277. doi: 10.1128/aac.44.2.272-277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durack DT, Beeson PB. Br J Exp Pathol. 1972;53:50–53. [PMC free article] [PubMed] [Google Scholar]
- 33.Beeson PB, Brannon ES, Warren JV. J Exp Med. 1945;81:9–23. doi: 10.1084/jem.81.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alder JD. Drugs Today (Barc) 2005;41:81–90. doi: 10.1358/dot.2005.41.2.882660. [DOI] [PubMed] [Google Scholar]
- 35.Petersen PJ, Bradford PA, Weiss WJ, Murphy TM, Sum PE, Projan SJ. Antimicrob Agents Chemother. 2002;46:2595–2601. doi: 10.1128/AAC.46.8.2595-2601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel JB, Jevitt LA, Hageman J, McDonald LC, Tenover FC. Clin Infect Dis. 2006;42:1652–1653. doi: 10.1086/504084. [DOI] [PubMed] [Google Scholar]
- 37.Cui L, Tominaga E, Neoh HM, Hiramatsu K. Antimicrob Agents Chemother. 2006;50:1079–1082. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman L, Alder JD, Silverman JA. Antimicrob Agents Chemother. 2006;50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. Mol Microbiol. 2003;49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 40.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korem M, Gov Y, Kiran MD, Balaban N. Infect Immun. 2005;73:6220–6228. doi: 10.1128/IAI.73.10.6220-6228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, Ji Y. J Bacteriol. 2005;187:5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maughan H, Galeano B, Nicholson WL. J Bacteriol. 2004;186:2481–2486. doi: 10.1128/JB.186.8.2481-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Centers for Disease Control Prevention. Morbid Mortal Wkly Rep. 1997;46:813–815. [Google Scholar]
- 46.Yin S, Daum RS, Boyle-Vavra S. Antimicrob Agents Chemother. 2006;50:336–343. doi: 10.1128/AAC.50.1.336-343.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardete S, Wu SW, Gill S, Tomasz A. Antimicrob Agents Chemother. 2006;50:3424–3434. doi: 10.1128/AAC.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Service RF. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 49.Rambaut A, Posada D, Crandall KA, Holmes EC. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Appendix