Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis (original) (raw)
Abstract
Autophagy is a catabolic process involving self-digestion of cellular organelles during starvation as a means of cell survival; however, if it proceeds to completion, autophagy can lead to cell death. Autophagy is also a haploinsufficient tumor suppressor mechanism for mammary tumorigenesis, as the essential autophagy regulator beclin1 is monoallelically deleted in breast carcinomas. However, the mechanism by which autophagy suppresses breast cancer remains elusive. Here we show that allelic loss of beclin1 and defective autophagy sensitized mammary epithelial cells to metabolic stress and accelerated lumen formation in mammary acini. Autophagy defects also activated the DNA damage response in vitro and in mammary tumors in vivo, promoted gene amplification, and synergized with defective apoptosis to promote mammary tumorigenesis. Therefore, we propose that autophagy limits metabolic stress to protect the genome, and that defective autophagy increases DNA damage and genomic instability that ultimately facilitate breast cancer progression.
Keywords: Autophagy, Beclin1, apoptosis, breast cancer, DNA damage, genomic instability
Autophagy is an evolutionarily conserved catabolic process whereby cytoplasmic proteins and organelles are targeted to lysosomes for degradation (Klionsky and Emr 2000; Mizushima 2005; Yorimitsu and Klionsky 2005). During starvation autophagy is a temporary survival mechanism, as self-digestion provides an alternative energy source (Boya et al. 2005; Degenhardt et al. 2006). Upon growth factor deprivation, hematopoietic cells activate autophagy, which is essential for maintenance of ATP production and cellular survival (Lum et al. 2005). In normal mouse development, amino acid production by autophagic degradation of proteins allows maintenance of energy homeostasis and survival during neonatal starvation (Kuma et al. 2004). Chronically ischemic myocardium induces autophagy, which inhibits apoptosis and may function as a cardioprotective mechanism (Yan et al. 2005). Autophagy is not only involved in recycling of normal cellular constituents but is also essential for damaged protein and organelle removal, as defects in this process result in accumulation of ubiquitin-positive aggregates and deformed cellular structures that may promote cellular degeneration (Komatsu et al. 2005, 2006; Hara et al. 2006). Moreover, autophagy is involved in cellular development and differentiation (Levine and Klionsky 2004) and may have a protective role against aging (Melendez et al. 2003; Bergamini et al. 2004).
Autophagy is also a form of cell death when allowed to proceed to completion and when cells unable to undergo apoptosis are triggered to die. It is often unclear whether autophagy is directly involved in initiation and/or execution of cell death, or if it merely represents a failed or exhausted attempt to preserve cell viability. Recent studies indicate that autophagy may play an active role in programmed cell death, but the conditions under which autophagy promotes cell death versus cell survival remain to be resolved (Baehrecke 2005; Debnath et al. 2005).
Defective autophagy has been implicated in tumorigenesis, as the essential autophagy regulator beclin1 is monoallelically deleted in human breast, ovarian, and prostate cancers (Aita et al. 1999). Also, human breast carcinoma cell lines and tumor tissue show decreased Beclin1 levels (Liang et al. 1999). beclin1 is the mammalian ortholog of the yeast atg6/vps30 gene, which is required for autophagosome formation (Kametaka et al. 1998). Beclin1 expression in human MCF7 breast cancer cells suppresses tumorigenesis (Liang et al. 1999). _beclin1_−/− mice die early in embryogenesis, whereas aging beclin1+/− mice have an increased incidence of lymphoma and carcinomas of the lung and liver (Qu et al. 2003; Yue et al. 2003). In addition, mammary tissue from beclin1+/− mice shows hyperproliferative, preneoplastic changes (Qu et al. 2003). Tumors forming in beclin1+/− mice express wild-type beclin1 mRNA and protein, indicating that beclin1 is a haploinsufficient tumor suppressor (Yue et al. 2003). However, the mechanism by which Beclin1 and autophagy inhibit tumorigenesis is not known. Recent studies revealed that autophagy enables tumor cell survival in vitro and in vivo when apoptosis is inactivated (Degenhardt et al. 2006), as commonly occurs in human cancers. How inactivation of a survival pathway promotes tumorigenesis is hard to explain, and represents an area of great scientific interest with potentially significant clinical implications.
We investigated how beclin1 allelic loss promotes breast tumorigenesis and identified a mechanism that, for the first time, reconciles the prosurvival and tumor suppressor roles of autophagy. Defective autophagy renders mouse mammary epithelial cells (MMECs) susceptible to metabolic stress, and as such, prone to DNA damage and genomic instability via gene amplification, which is an important component of breast cancer pathophysiology (Burstein 2005; Chin et al. 2006). Understanding the role of autophagy in tumor progression may yield insight into how to best exploit this process for maximum therapeutic benefit and improved clinical outcome based on individual cancer genotype.
Results
A novel MMEC model for studying breast cancer-promoting functions
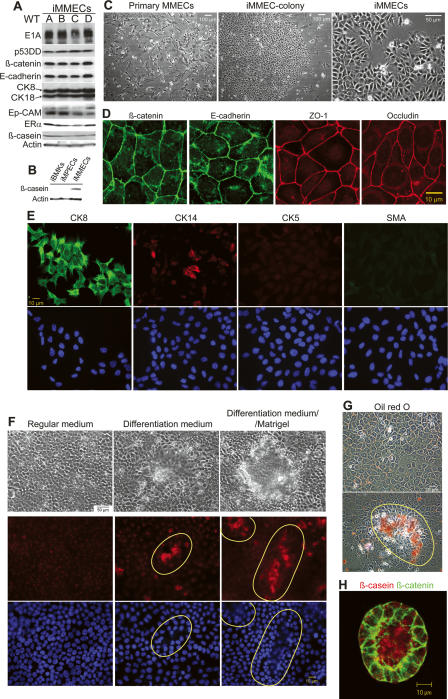
The role of beclin1 heterozygosity in mammary tumorigenesis was investigated using a novel mouse model for mammary epithelial tumor progression. Primary MMECs were isolated from young virgin female mice (Imagawa et al. 2000) and immortalized (iMMECs) by inactivation of the retinoblastoma and p53 pathways by stable expression of the adenovirus E1A protein and a dominant-negative p53 mutant (p53DD), as previously described for mouse kidney epithelial cells (Fig. 1A; Degenhardt et al. 2002a, b; Tan et al. 2005; Degenhardt and White 2006). In regular two-dimensional (2D) culture, iMMECs exhibited typical cuboidal epithelial morphology (Fig. 1C) and formed tight junctions (Fig. 1D). iMMECs expressed the epithelial cell markers β-catenin, E-cadherin, and epithelial cell adhesion molecule (Ep-CAM) (Fig. 1A) and the luminal epithelial cell markers cytokeratin (CK) 8 and 18 (Fig. 1A,E), but did not express several basal/myoepithelial cell markers, such as CK5, smooth muscle actin (SMA), vimentin, CD10, p63, and α6-integrin (Fig. 1E; some data not shown). Few iMMECs (<20%) expressed CK14 (Fig. 1E). ERα was expressed in iMMECs from virgin mice (Fig. 1A), and was highly up-regulated in iMMECs from pregnant mice (data not shown). iMMECs, but not similarly immortalized mouse kidney or prostate epithelial cells, expressed low levels of β-casein when cultured in regular growth medium (Fig. 1A,B). When exposed to lactogenic hormones, iMMECs grown on tissue culture plastic formed dome-like structures reminiscent of lactating mammary alveoli (Gordon et al. 2000) and exhibited β-casein up-regulation and secretion into the domes (Fig. 1F), which contained fat consistent with the presence of milk (Fig. 1G). In three-dimensional (3D) morphogenesis assays, iMMECs formed polarized acini that generated lumens via apoptosis (Fig. 2C), similarly to the immortalized, nontransformed human mammary epithelial cell line MCF-10A (Debnath et al. 2003), and secreted β-casein into the acinar lumen upon lactogenic stimulation (Fig. 1H). iMMECs were poorly tumorigenic, forming clonal adenocarcinomas with long latency (Fig. 2D,E).
Figure 1.
MMEC model. (A) Western blot analysis of E1A, p53DD, β-catenin, E-cadherin, CK8/18, Ep-CAM, ERα, β-casein, and actin in independently derived wild-type iMMEC cell lines (WTA-D) in regular growth medium. (B) Western blot analysis of β-casein and actin in iBMKs, immortalized mouse prostate epithelial cells (iMPECs), and iMMECs (WTA) in regular growth medium. (C) Phase contrast. (D) IF of WTA cells for tight junction proteins. (E) IF of WTA cells for luminal (CK8) and basal/myoepithelial (CK14, CK5, SMA) markers. (F) Lactogenic stimulation of WTA cells. (Top panel) Phase contrast. (Middle panel) β-casein IF counterstain. (Bottom panel) DAPI counterstain. (G) Red oil O staining. (H) Confocal cross-section through the center of WTA acinus grown for 12 d in regular 3D morphogenesis medium, stimulated for 2 d with lactogenic hormones, and immunostained with β-casein (red) and β-catenin (green). Domes formed upon lactogenic stimulation of WTA cells are encircled by a yellow line in F and G.
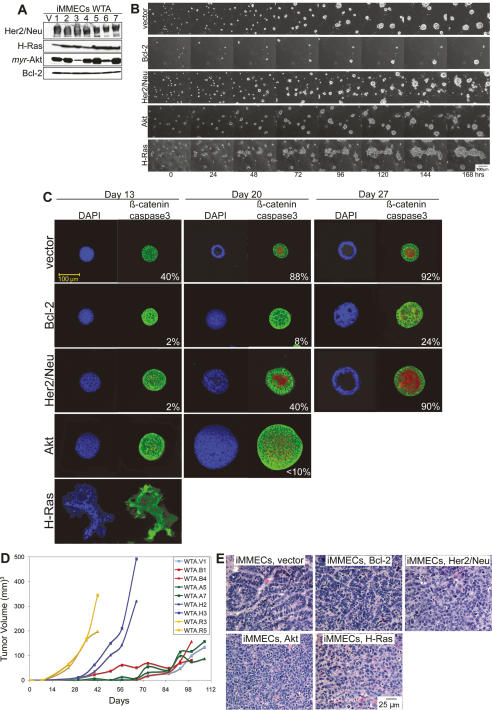
Figure 2.
MMEC model. (A) Western blots showing protein expression in iMMEC WTA-derived cell lines stably expressing vector control (V), wild-type HER2/neu (lanes 1–7), H-RasV12 (lanes 1–7), _myr_-Akt (lanes 1–7), and Bcl-2 (lanes 1–7). (B) Seven-day time course of 3D morphogenesis captured by time-lapse videos of mammary acini generated by WTA-derived cell lines expressing vector (WTA.V), Bcl-2 (WTA.B1), HER2/neu (WTA.H2), Akt (WTA.A5), and H-Ras (WTA.R3) cultured in Matrigel. (C) IF of mammary acini generated by the cell lines mentioned in B. Images are confocal cross-sections through the center of acini immunostained with β-catenin (green) and activated caspase-3 (red); nuclei were counterstained with DAPI (blue). Images are representative of the majority of acini for a specific genotype at a given time point, and the number at the bottom right corner of each frame is the percentage of acini with lumen formation. (D) Tumor growth kinetics of WTA-derived iMMEC cell lines. (E) H&E staining of tumors generated by the cell lines shown in B.
To study the effects of oncogene expression on 3D morphogenesis and tumorigenicity, iMMECs were engineered to stably express Bcl-2 or HER2/neu, or constitutively active Akt or Ras (Fig. 2A). Expression of different oncogenes affected 3D morphogenesis in an oncogene-dependent manner (Fig. 2B,C). Apoptosis blockade by Bcl-2 expression prevented lumen formation and increased acinar size, HER2/neu expression stimulated acinar growth without blocking apoptosis and delayed lumen formation, Akt activation stimulated proliferation and acinar growth while disrupting polarity without preventing apoptosis, and H-Ras activation stimulated growth and grossly disrupted acinar structure. H-Ras activation and HER2/neu expression rendered iMMECs highly tumorigenic, whereas Akt activation and apoptosis inhibition by Bcl-2 enhanced tumorigenesis to a lesser degree (Fig. 2D). Similarly to vector-expressing iMMECs, Bcl-2-expressing iMMECs formed adenocarcinomas, whereas Akt or H-Ras activation and HER2/neu expression resulted in less differentiated tumors (Fig. 2E), reminiscent of the tumors arising in bitransgenic mice expressing MMTV-neu and a dominant-negative p53 transgene, p53-172H, which are highly anaplastic (Li et al. 1997). Therefore, iMMECs represent an early stage in mouse mammary oncogenesis and provide a facile platform for studying mechanisms regulating breast cancer progression.
beclin1 heterozygosity impairs autophagy and increases susceptibility of mammary epithelial cells to metabolic stress in vitro
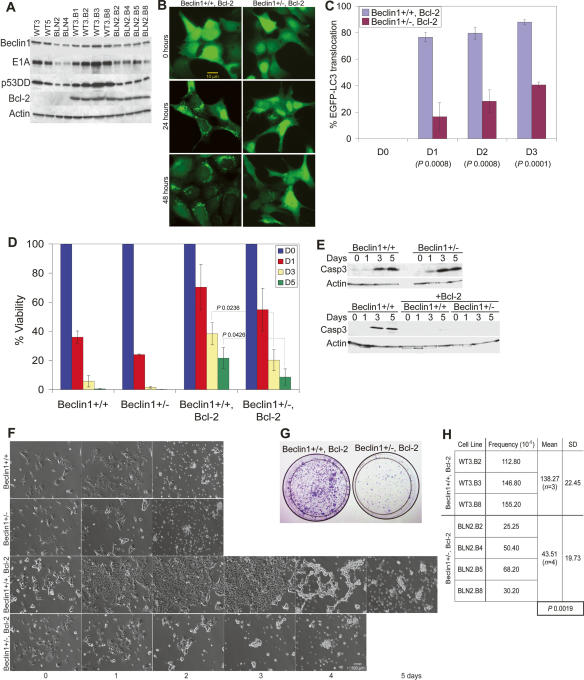
To determine the role of beclin1 heterozygosity in breast tumorigenesis, iMMECs were generated from beclin1+/+ and beclin1+/− mice (Fig. 3A). As previous work showed that apoptosis defects enable survival to metabolic stress through autophagy (Lum et al. 2005; Degenhardt et al. 2006), beclin1+/+ and beclin1+/− iMMECs were engineered to stably express Bcl-2 (Fig. 3A). The autophagy potential of apoptosis-competent and apoptosis-defective beclin1+/+ and beclin1+/− iMMECs under metabolic stress was investigated. In vitro ischemia (1% oxygen plus glucose deprivation) was previously shown to simulate metabolic stress in the tumor microenvironment (Nelson et al. 2004), which is where autophagy localizes in tumors in vivo (Degenhardt et al. 2006). To quantify autophagy induced by metabolic stress, cytosol-to-membrane translocation of the stably expressed autophagy marker EGFP-LC3 was monitored (Mizushima et al. 2004). Under normal conditions, beclin1+/+ and beclin1+/− iMMECs without and with Bcl-2 displayed diffuse EGFP-LC3 localization. Under metabolic stress, Bcl-2-expressing beclin1+/+ iMMECs showed robust autophagy activation, as EGFP-LC3 distribution became punctate, indicating LC3 lipidation and membrane translocation. These findings are similar to what was reported when apoptosis was disabled by bax and bak deficiency in immortalized baby mouse kidney (iBMK) cells (Degenhardt et al. 2006) or Bcl-2 expression in iBMK cells (Degenhardt et al. 2006; Mathew et al. 2007). Thus, autophagy is robustly induced by metabolic stress in apoptosis-deficient cells independently of the mechanism of apoptosis inactivation. Metabolically stressed beclin1+/− iMMECs with Bcl-2 exhibited attenuated and delayed autophagy induction (Fig. 3B,C). Membrane translocation of EGFP-LC3 under metabolic stress was not observed in apoptosis-competent beclin1+/+ and beclin1+/− iMMECs (data not shown), presumably due to apoptosis activation by metabolic stress (Fig. 3E). This result indicates that the role of autophagy may not be readily apparent when apoptosis is functional, as other studies have suggested (Lum et al. 2005; Degenhardt et al. 2006).
Figure 3.
beclin1 heterozygocity impairs autophagy and increases susceptibility of mammary epithelial cells to metabolic stress in vitro. (A) Western blot analysis of Beclin1, E1A, p53DD, Bcl-2, and actin in beclin1+/+ and beclin1+/− iMMEC cell lines. (B) Fluorescence of beclin1+/+ and beclin1+/− iMMECs with Bcl-2 (WT3.B3, BLN2.B4) and stably expressing EGFP-LC3 after 0, 24, and 48 h of metabolic stress. (C) Quantitation of autophagy (percentage EGFP-LC3 translocation) in WT3.B3 and BLN2.B4 stably expressing EGFP-LC3 following 0, 1, 2, and 3 d of metabolic stress. Three independent experiments were performed, and the mean values with standard deviation are presented. (D) Viability of beclin1+/+ and beclin1+/− iMMECs without and with Bcl-2 following 0, 1, 3, and 5 d of metabolic stress. Four independent cell lines per genotype were examined, and the mean values with standard deviation are presented. (E) Western blot analysis of activated caspase-3 in beclin1+/+ and beclin1+/− iMMECs without (WT3, BLN2) and with (WT3.B3, BLN2.B4) Bcl-2 in response to metabolic stress in vitro. (F) Identical frames from 2- to 5-d time-lapse videos of beclin1+/+ and beclin1+/− iMMEC cell lines without (WT3, BLN2) and with (WT3.B3, BLN2.B8) Bcl-2 under metabolic stress. (G,H) Clonogenic survival of beclin1+/+ and beclin1+/− iMMECs expressing Bcl-2 after exposure to metabolic stress for 12 d. (G) Representative plates (WT3.B3, BLN2.8). (H) Quantitation of clonogenic survival with standard deviation (SD). _P_-values were calculated by unpaired _t_-test and are provided for statistically significant results only.
To investigate the role of autophagy under metabolic stress, viability of beclin1+/+ and beclin1+/− iMMECs without and with Bcl-2 was assessed. Apoptosis-competent beclin1+/+ and beclin1+/− iMMECs were killed within 2–3 d (Fig. 2D,F), and cell death involved caspase-3 activation (Fig. 3E). Apoptosis-defective beclin1+/+ and beclin1+/− iMMECs survived longer, with beclin1+/+ cells being more resistant to cell death (Fig. 3D,F). Death of Bcl-2-expressing beclin1+/− iMMECs did not involve caspase-3 activation (Fig. 3E), suggesting that death was caspase independent. The increased susceptibility of Bcl-2-expressing beclin1+/− iMMECs to metabolic stress observed in short-term survival experiments was also apparent in clonogenic survival assays (Fig. 3G,H), indicating that beclin1 heterozygosity diminished the survival advantage provided by Bcl-2. The same phenotype was observed with Beclin1 down-regulation in Bax/Bak-deficient iBMK cells (Degenhardt et al. 2006) and Bcl-2 expression in beclin1+/− iBMK cells (Degenhardt et al. 2006; Mathew et al. 2007), and is thus independent of the means of apoptosis inactivation. Therefore, autophagy mitigates metabolic stress in mammary epithelial cells and is a survival mechanism upon nutrient and oxygen deprivation.
Allelic loss of beclin1 accelerates death of central acinar cells in 3D morphogenesis assays and is associated with DNA damage response activation
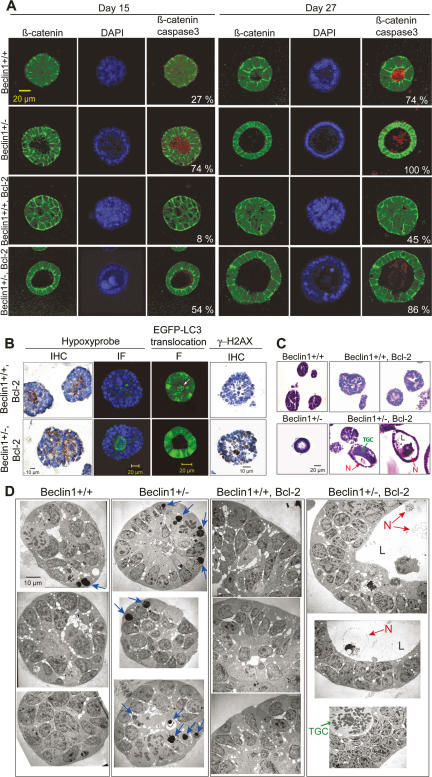
To assess the impact of beclin1 heterozygosity on ductal morphogenesis, 3D cultures of beclin1+/+ and beclin1+/− iMMECs without and with Bcl-2 were examined. Apoptosis-competent beclin1+/+ iMMECs formed polarized acini that generated lumens via apoptosis (Fig. 4A), similarly to other, independently derived wild-type iMMECs (Fig. 2C). In contrast, beclin1+/− iMMECs formed polarized acini with accelerated lumen formation (threefold) via apoptosis (Fig. 4A), indicating that autophagy plays a role in central acinar cell survival and may delay apoptosis. Bcl-2 expression substantially inhibited lumen formation in beclin1+/+ iMMECs (Fig. 4A), similarly to other, independently derived wild-type iMMECs expressing Bcl-2 (Fig. 2C). beclin1+/− iMMECs expressing Bcl-2 formed polarized acini with accelerated lumen formation (more than fivefold on day 15) compared with beclin1+/+ iMMECs expressing Bcl-2 (Fig. 4A). Therefore, defective autophagy abrogated survival conferred by Bcl-2 for central acinar cells, which were metabolically stressed, as indicated by hypoxyprobe staining (Fig. 4B). To localize autophagy, acini formed by EGFP-LC3-expressing beclin1+/+ and beclin1+/− iMMECs with Bcl-2 were examined. EGFP-LC3 membrane translocation was observed in centrally located beclin1+/+ acinar cells, where lumen formation occurred in beclin1+/− acini (Fig. 4B). Acini formed by beclin1+/+ and beclin1+/− iMMECs with Bcl-2 were also examined for evidence of DNA damage response activation, as a possible manifestation of metabolic stress. Acini from apoptosis-defective beclin1+/− iMMECs showed increased Ser139 phosphorylation of H2AX (γ-H2AX) (Fig. 4B). Taken together, these results indicated that autophagy was activated in the metabolically stressed, central acinar cells and promoted cell survival, presumably by mitigating metabolic stress and DNA damage.
Figure 4.
In 3D morphogenesis assays, allelic loss of beclin1 accelerates the death of central acinar cells, where metabolic stress and autophagy localize, and is associated with DNA damage response activation. (A) IF of mammary acini generated by beclin1+/+ and beclin1+/− iMMEC cell lines without (WT3, BLN2) and with Bcl-2 (WT3.B3, BLN2.B8) cultured in Matrigel. Acini were immunostained with β-catenin (green) and activated caspase-3 (red); nuclei were counterstained with DAPI (blue). Faint red color in lumens of acini formed by Bcl-2-expressing iMMECs represents background levels of activated caspase-3 staining. Images are representative of the majority of acini for a specific genotype at a given time point, and the number at the bottom right corner of each frame represents the percentage of acini with lumen formation. (B) Hypoxyprobe IHC and IF (green for hypoxyprobe, blue for nuclei) on acini formed by WT3.B3 and BLN2.B8 cells. Fluorescence (F) by confocal microscopy of acini formed by beclin1+/+ and beclin1+/− iMMECs with Bcl-2 (WT3.B3, BLN2.B8) and stably expressing EGFP-LC3. γ-H2AX IHC on acini formed by WT3.B3 and BLN2.B4 cells. (C,D) H&E (C) and electron microscopy (D) on acini formed by beclin1+/+ and beclin1+/− iMMECs without (WT3, BLN2) and with (WT3.B3, BLN2.B8) Bcl-2. (N) Necrotic cell; (TGC) aneuploid tumor giant cell; (L) lumen.
To further investigate lumen formation, acini formed by beclin1+/+ and beclin1+/− iMMECs without and with Bcl-2 were examined by histology and electron microscopy. Whereas Bcl-2 expression blocked lumen formation in beclin1+/+ acini, central acinar cells of Bcl-2-expressing beclin1+/− acini exhibited signs of necrotic cell death (Fig. 4C), suggesting that necrosis may be the default cell death mechanism upon concurrent apoptosis and autophagy inactivation, similarly to what was reported for kidney epithelial cells (Degenhardt et al. 2006). Electron microscopy showed increased numbers of engulfed apoptotic bodies in acini formed by beclin1+/− iMMECs, which may be due to increased apoptosis or defective corpse degradation. Necrosis was morphologically evident in the lumens of acini formed by Bcl-2-expressing beclin1+/− iMMECs (Fig. 4D), indicating that defective autophagy accelerated cell death by necrosis. Finally, beclin1+/+ iMMECs expressing Bcl-2 formed acini with centrally located, abnormally large cells (Fig. 4A), which resembled aneuploid tumor giant cells induced in immortal kidney epithelial cells by metabolic stress (Nelson et al. 2004). This phenotype was more pronounced in acini formed by Bcl-2-expressing beclin1+/− iMMECs, which had centrally located or lumen-associated abnormally large cells at increased frequency (Fig. 4C,D).
Allelic loss of beclin1 promotes mammary tumorigenesis and activation of a DNA damage response in tumors in vivo
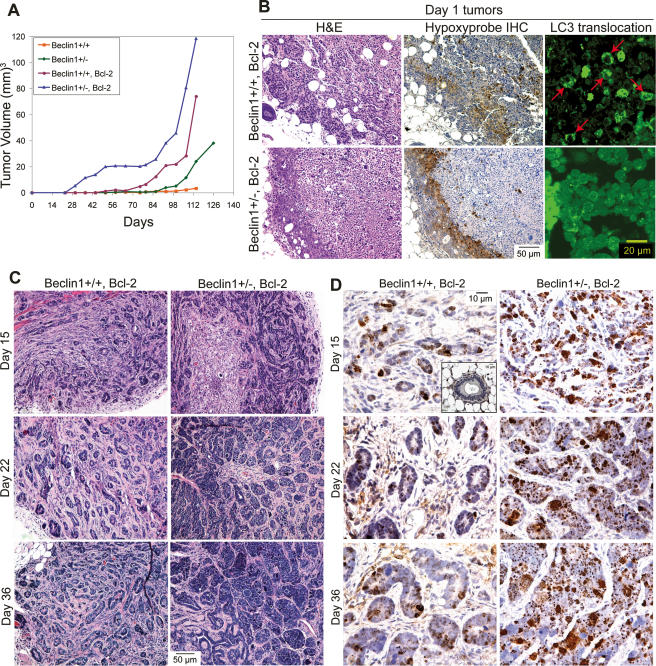
The tumorigenicity of beclin1+/+ and beclin1+/− iMMECs without and with Bcl-2 was determined by orthotopic injection in nude mice. beclin1+/− iMMECs were poorly tumorigenic, forming clonal adenocarcinomas marginally faster than beclin1+/+ iMMECs, whereas Bcl-2-expressing beclin1+/− iMMECs showed accelerated tumor kinetics compared with Bcl-2-expressing beclin1+/+ iMMECs and apoptosis-competent iMMECs (Fig. 5A).
Figure 5.
Allelic loss of beclin1 promotes mammary tumorigenesis and activation of the DNA damage response in tumors in vivo. (A) Tumor growth kinetics of beclin1+/+ and beclin1+/− iMMEC cell lines without and with Bcl-2. (B) Autophagy in tumors localizes to regions of metabolic stress. H&E (left column) and hypoxyprobe IHC (middle column) on day 1 tumors generated by beclin1+/+ and beclin1+/− iMMECs with Bcl-2 (WT3.B3, BLN2.B8). EGFP-LC3 membrane translocation was monitored with EGFP-LC3 fluorescence by confocal microscopy on day 1 tumors generated by beclin1+/+ and beclin1+/− iMMECs with Bcl-2 (WT3.B3, BLN2.B8) and stably expressing EGFP-LC3; the areas presented (right column) are within the hypoxyprobe-staining regions between the rim of healthy cells and the necrotic centers. (C,D) Tumors derived from beclin1+/+ and beclin1+/− iMMECs with Bcl-2 (WT3.B3, BLN2.B8) were excised at days 15, 22, and 36 post-orthotopic implantation, and were compared by H&E staining (C) and by γ-H2AX IHC (D). Insert in top left panel of D represents a normal mammary gland.
Since concurrent apoptosis and autophagy defects promoted tumorigenesis, tumors generated by Bcl-2-expressing beclin1+/+ and beclin1+/− iMMECs were investigated further. At day 1 post-implantation, tumors had a rim of healthy-appearing cells at the perimeter of large necrotic centers, presumably due to initial lack of blood supply (Fig. 5B). Autophagy, as measured by EGFP-LC3 translocation, was activated in tumors derived from Bcl-2-expressing beclin1+/+ iMMECs and localized at hypoxic regions adjacent to the necrotic centers (Fig. 5B), indicating that metabolic stress activated autophagy in vivo. Similarly to the results in vitro (Fig. 3B,C), beclin1+/− iMMECs showed reduced autophagy in tumors in vivo (Fig. 5B). To investigate how allelic loss of beclin1 affects tumor establishment and progression, tumors generated by apoptosis-defective beclin1+/+ and beclin1+/− iMMECs were excised at 15, 22, and 36 d post-implantation. Bcl-2-expressing beclin1+/− iMMECs generated tumors with altered glandular morphology and accelerated growth (Fig. 5C), confirming synergy between apoptosis and autophagy defects in mammary tumorigenesis.
Since beclin1 heterozygosity increased susceptibility to metabolic stress in vitro (Figs. 3D–H, 4A), tumors generated by beclin1+/+ and beclin1+/− iMMECs with Bcl-2 were examined for evidence of metabolic stress in vivo. Similarly to the in vitro data (Fig. 4B), beclin1 allelic loss resulted in activation of the DNA damage response in tumors in vivo, as monitored by increased γ-H2AX foci (Fig. 5D), suggesting that deficient autophagy may promote genome damage under metabolic stress.
Allelic loss of beclin1 promotes drug resistance mediated by gene amplification in vitro
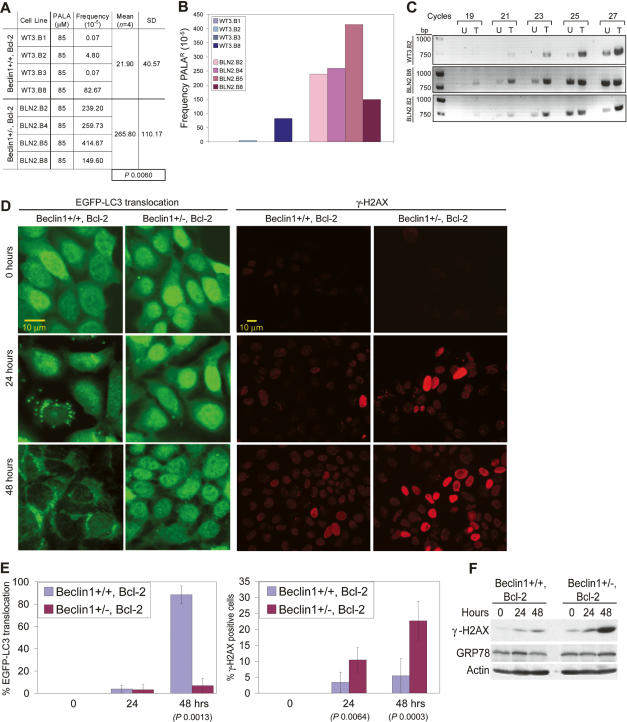
Since beclin1 heterozygosity rendered mammary epithelial cells prone to increased DNA damage (Figs. 4B, 5D), Bcl-2-expressing beclin1+/+ and beclin1+/− iMMECs were examined for evidence of drug resistance mediated by gene amplification, which requires DNA damage in the form of double-strand breaks. When subjected to _N_-(phosphonoacetyl)-L-aspartate (PALA), which inhibits the aspartate transcarbamylase activity of the CAD enzyme complex and selects for amplification of the CAD gene (Otto et al. 1989), beclin1+/− iMMECs gave rise to PALA-resistant (PALAR) colonies at much higher frequency compared with beclin1+/+ iMMECs (Fig. 6A,B). CAD gene amplification was verified by PCR of genomic DNA from several stable cell lines derived from independent PALAR colonies (Fig. 6C). Thus, autophagy defects are associated with genome damage and instability in the form of drug resistance linked to gene amplification.
Figure 6.
Allelic loss of beclin1 promotes drug resistance mediated by gene amplification in vitro. (A,B) Quantitation with standard deviation (SD) of PALA-resistance for beclin1+/+ and beclin1+/− iMMECs with Bcl-2 treated with PALA at 5× LD50. The _P_-value was calculated by unpaired _t_-test. (C) PCR of genomic DNA from several stable cell lines derived from independent PALAR colonies using primers for an 800-bp fragment of the CAD gene. (D) beclin1+/+ and beclin1+/− iMMECs with Bcl-2 (WT3.B3, BLN2.B4) and stably expressing EGFP-LC3 after 0, 24, and 48 h of PALA treatment at LD50. (Left panel) Fluorescence (F). (Right panel) γ-H2AX IF. (E) WT3.B3 and BLN2.B4 stably expressing EGFP-LC3 following 0, 24, and 48 h of PALA treatment at LD50. (Left panel) Quantitation of autophagy (percentage EGFP-LC3 translocation). (Right panel) Quantitation of γ-H2AX activation (percentage γ-H2AX-positive cells as number of cells with bright red nuclei/total number of cells). Three independent experiments were performed, and the mean values with standard deviation are presented. _P_-values were calculated by unpaired _t_-test. (F) Western blot analysis of γ-H2AX, GRP-78, and actin in WT3.B3 and BLN2.B4 stably expressing EGFP-LC3 following 0, 24, and 48 h of PALA treatment at LD50.
PALA inhibits de novo pyrimidine biosynthesis and results in depletion of intracellular deoxynucleotide triphosphate (dNTP) pools and inhibition of DNA synthesis (Moyer et al. 1982). DNA replication with limiting dNTP pools causes replication stress and DNA damage that can facilitate gene amplification. Autophagy-deficient cells may display increased replication stress resulting in promotion of CAD gene amplification. To examine if exposure of cells to PALA and the resultant pyrimidine starvation induce autophagy, beclin1+/+ and beclin1+/− iMMECs with Bcl-2 and stably expressing EGFP-LC3 were treated with PALA, and autophagy levels were quantified by EGFP-LC3 translocation. beclin1+/+ iMMECs with Bcl-2 showed robust autophagy induction by 48 h of PALA exposure, whereas beclin1+/− failed to do so (Fig. 6D,E). DNA damage accumulation (γ-H2AX foci) occurred in both beclin1+/+ and beclin1+/− iMMECs with Bcl-2 upon PALA treatment, but at significantly higher levels in beclin1+/− iMMECs (Fig. 6D–F), consistent with the increased DNA damage response in 3D morphogenesis (Fig. 4B) and in tumors in vivo (Fig. 5D). PALA exposure was specifically associated with DNA damage response activation and did not activate other forms of cellular stress, such as the unfolded protein response (GRP-78) (Fig. 6F). DNA damage upon treatment of MCF-7 cells with camptothecin was recently shown to induce autophagy and delay apoptotic cell death (Abedin et al. 2007), which raises the possibility that, in addition to pyrimidine starvation, DNA damage may also play a role in autophagy induction upon PALA exposure. In summary, PALA exposure triggers autophagy, which mitigates the metabolic stress imposed by depletion of dNTP pools and lessens replication stress. Defective autophagy creates sensitivity to pyrimidine starvation, which leads to increased replication stress and, in turn, to DNA damage and instability.
Discussion
Deficient autophagy and breast tumorigenesis
Several studies have indicated that defective autophagy in the form of beclin1 allelic loss may play a role in breast tumorigenesis. beclin1 maps to a region on chromosome 17q21 that is allelically deleted in 50% of breast cancers as detected by loss of heterozygosity (LOH) analysis (Futreal et al. 1992; Saito et al. 1993). Human breast carcinomas show decreased Beclin1 expression compared with normal adjacent tissue (Liang et al. 1999), and human breast cancer cell lines, although commonly polyploid for chromosome 17, exhibit deletions of one or more beclin1 alleles (Aita et al. 1999) and low protein levels (Liang et al. 1999). Beclin1 overexpression in MCF7 cells inhibits their tumorigenic potential (Liang et al. 1999), whereas mammary tissue from beclin1+/− mice shows hyperproliferative, preneoplastic changes (Qu et al. 2003). Similarly to what has been described for hematopoietic (Lum et al. 2005) and kidney epithelial cells (Degenhardt et al. 2006; Mathew et al. 2007), our results demonstrate that autophagy is a survival mechanism for mammary epithelial cells in response to metabolic stress, especially when apoptosis is disabled.
In 3D morphogenesis, autophagy localizes in the central acinar cells, which are under increased metabolic stress because of hypoxia (Fig. 4B), anoikis (Reginato et al. 2003; Collins et al. 2005; Carroll et al. 2006), and possibly diffusion-limited nutrient supply. beclin1+/− heterozygosity promotes apoptosis and abrogates the survival advantage conferred to central acinar cells by apoptosis defects, and is associated with increased DNA damage. The acceleration of lumen formation in mammary acini formed by beclin1+/− cells suggests that developmental regulation of autophagy may have a role in breast ductal morphogenesis.
Autophagy also localizes to regions of metabolic stress in mammary tumors in vivo. beclin1 heterozygosity greatly compromises the autophagy potential of mammary cancer cells and is associated with accumulation of γ-H2AX foci in tumors. Thus, autophagy may be critical for preservation of cellular fitness in the tumor microenvironment by temporarily sustaining cellular function under metabolic stress, which is a common occurrence in solid tumors (Folkman 2003). By promoting breast cancer cell survival, autophagy limits genome damage and instability and ultimately inhibits mammary tumorigenesis. This finding is consistent with the role of DNA repair proteins in limiting cancer progression (D’Andrea and Grompe 2003; Risinger and Groden 2004) and reconciles the intuitively contradictory roles of autophagy as both a survival and a tumor suppressor mechanism.
Deficient autophagy, DNA damage, and genomic instability
Recent studies showed that autophagy defects in normal cells result in accumulation of ubiquitinated and likely damaged proteins that promote cellular degeneration (Komatsu et al. 2005, 2006; Hara et al. 2006). Our results indicate that cancer cells with autophagy defects accumulate γ-H2AX foci, and thus DNA damage, in 3D morphogenesis and upon PALA treatment in vitro and in tumors in vivo. Impaired survival and activation of the DNA damage response are also observed in _atg5_−/− iBMK cells upon exposure to metabolic stress (Mathew et al. 2007), indicating that these activities result from defective autophagy rather than low Beclin1 levels.
Defective autophagy compromises the ability of cells to adapt to metabolic stress, which may lead to insufficient ATP generation (Jin and White 2007) and accumulation of damaged mitochondria with excessive reactive oxygen species (ROS) (Jin 2006). Limiting ATP levels and/or direct DNA insult by ROS may cause replication stress and DNA damage response activation, which normally arrests cell cycle progression or triggers apoptosis, and can therefore act as a barrier to early carcinogenesis (Bartkova et al. 2005; Gorgoulis et al. 2005). In our model system, inactivation of p53, a critical cell cycle checkpoint regulator, is inherent to the immortalization process, and therefore, autophagy-deficient cells expressing Bcl-2 lack the capacity to undergo p53-mediated checkpoint activation and cell cycle arrest or apoptosis in response to DNA damage. Instead, concurrent inactivation of autophagy and apoptosis leads to accumulation of DNA damage and double-strand breaks under metabolic stress, enabling gene amplification and, thus, creating a permissive environment for genomic instability and cancer progression.
Deficient autophagy, gene amplification, and breast cancer
As a means of measuring genomic instability in autophagy-deficient mammary epithelial cells, we used selection for PALA resistance, which involves CAD gene amplification. PALA exposure induced autophagy in wild-type cells and was associated with increased DNA damage and drug resistance involving CAD amplification in autophagy-deficient cells. Similar results were obtained with beclin1+/− kidney epithelial cells (Mathew et al. 2007) and _atg5_−/− MEFs (S. Jin, pers. comm.), indicating that defective autophagy can promote genomic instability in the form of gene amplification in response to metabolic and replication stress independent of cell type or means of autophagy inactivation.
Gene amplification is common in solid tumors (Albertson et al. 2003; Gebhart 2005; Albertson 2006) and is likely initiated by DNA double-strand breaks in cells lacking robust checkpoints (Chernova et al. 1998; Paulson et al. 1998). Breast cancers frequently show genome copy number aberrations, including high-level amplifications that correlate with worse prognosis (Kallioniemi et al. 1994; Al-Kuraya et al. 2004; Naylor et al. 2005; Chin et al. 2006). HER2/neu amplification, which is associated with aggressive breast cancer, poor clinical outcome (Slamon et al. 1987; Burstein 2005; Chin et al. 2006), and chemotherapy resistance (Modi et al. 2005; Shin et al. 2006), has been successfully exploited therapeutically (Baselga et al. 2006) and is the paradigm for the identification of other therapeutic targets within the 66 genes associated with poor outcome that were recently reported as amplified in breast carcinomas (Chin et al. 2006). Whether autophagy defects segregate with specific gene amplifications in solid tumors, and particularly in breast cancer, remains to be investigated.
Therapeutic implications
Autophagy-deficient mammary cancer cells are susceptible to DNA damage in response to metabolic and replication stress, and this finding may have therapeutic implications for the rational design of breast cancer treatments. Whereas promoting autophagy in breast cancer may be an effective chemopreventive strategy, reasonable predictions are that breast cancer cells with autophagy defects may be particularly sensitive to agents exacerbating metabolic stress, like anti-angiogenic drugs, and to DNA damaging agents, particularly drugs that exert their anti-tumor effects during DNA replication. Treatment with these drugs may enhance killing of autophagy-deficient cancer cells that have intact p53 and apoptosis, and cannot therefore evade senescence or cell death upon massive DNA damage. Combinatorial treatment may prove more efficacious than single agent therapy, especially if the two classes of anti-cancer drugs have a synthetic lethal interaction in autophagy-deficient cells (Kaelin 2005). On the contrary, treatment of cancer cells defective in both autophagy and apoptosis with metabolic or replication stress-inducing agents may prove detrimental, as infliction of further DNA damage may promote genomic instability with resultant enhanced tumorigenic potential and development of drug resistance. Thus, the same mechanism driving cancer progression in an autophagy-deficient tumor may actually represent the tumor’s “Achilles heel” worth exploring therapeutically.
Materials and methods
MMEC isolation and immortalization
The fourth inguinal mammary glands were excised from 6- to 8-wk-old virgin wild-type and littermate beclin1+/+ and beclin1+/− C57Bl/6 female mice. Mammary tissue was mechanically minced and digested with 0.1% collagenase A (Roche Molecular Biochemicals) (Imagawa et al. 2000). Primary MMECs were washed and transfected with cytomegalovirus (CMV)-driven adenovirus type 5 E1A and dominant-negative mouse p53 (C-terminal fragment, termed p53DD) plasmids by electroporation, as previously described for BMK epithelial cells (Degenhardt et al. 2002b; Degenhardt and White 2006). Cells were plated in plates precoated with fetuin (Rijnkels and Rosen 2001) and were allowed to settle for 2 d in 2× hormone plating medium (F12, 10 μg/mL insulin, 2 μg/mL hydrocortisone, 10 ng/mL EGF) with 10% fetal bovine serum (FBS) before switching to 1× hormone medium with 5% FBS (to minimize fibroblast growth). Colonies of doubly transfected, iMMECs formed by 6 wk. Independent clones were isolated and expanded to stable cell lines in regular growth medium (F12, 10% FBS, 5 μg/mL insulin, 1 μg/mL hydrocortisone, 5 ng/mL EGF).
Lactogenic stimulation
Cells were grown on plastic culture dishes until confluent and were subsequently induced with differentiation medium (F12, 5 μg/mL insulin, 1 μg/mL hydrocortisone, 3 μg/mL prolactin) ± 2% Matrigel (Streuli et al. 1995) for 6 d, with medium changes every 2 d.
Generation of stable cell lines
iMMECs stably expressing human Bcl-2, H-RasV12, _myr_-Akt, wild-type human HER2/neu, or the vector control were derived by electroporation with pcDNA3.1-hBcl-2 (Nelson et al. 2004), pcDNA3.H-RasV12 (Lin et al. 1995), pcDNA3.Myr-Akt (Plas et al. 2001), pcDNA3.wild-type-HER2/neu, or pcDNA3.1 vector (Invitrogen), followed by selection with geneticin. beclin1+/+ and beclin1+/− iMMECs stably expressing EGFP-LC3 were derived by electroporation with pcDNA3.EGFP-LC3 (Mizushima et al. 2004), followed by selection with geneticin, whereas Bcl-2-expressing beclin1+/+ and beclin1+/− iMMECs stably expressing EGFP-LC3 were derived by electroporation with pcDNA3.EGFP-LC3 and pcDNA3.1zeo (Invitrogen), followed by double selection with geneticin and zeocin. Geneticin and zeocin were used at 300 and 100 μg/mL, respectively.
Western blotting, immunofluorescence (IF), and immunohistochemistry (IHC)
Antibodies against the following antigens were used: Beclin1, Bcl-2, ERα, β-casein, integrin α6, CD10, vimentin, Ep-CAM (Santa Cruz Biotechnology); p53, E1A, Ras, actin (Oncogene); activated caspase-3, HER2/neu, Akt (Cell Signaling); β-catenin, ZO-1, occludin (Zymed); E-cadherin (RDI); CK8, CK5, CK14 (Covance); SMA (Sigma); p63 (Lab Vision); Ser139-phosphorylated H2AX (γ-H2AX) (Upstate Biotechnology); GRP-78 (Stressgen); hypoxyprobe (Chemicon). Western blotting, IF, and IHC were performed as previously described (Cuconati et al. 2003; Nelson et al. 2004).
3D morphogenesis
3D culture of iMMECs on a reconstituted basement membrane was performed according to the protocol previously described for the immortalized, nontransformed human mammary epithelial cell line MCF-10A (Debnath et al. 2003). For lactogenic stimulation, mammary acini were grown for 12 d, and then the medium was changed to F12, 5 μg/mL insulin, 1 μg/mL hydrocortisone, and 3 μg/mL prolactin with 2% Matrigel for 2 d. Mammary acini were fixed and processed for IF as previously described (Debnath et al. 2003). Acini were incubated with primary antibodies for 90 min at 37°C, washed, and then incubated with fluorescein- or rhodamine-coupled secondary antibodies for 40 min at room temperature. Finally, acini were stained with DAPI (4′,6′-diamidino-2-phenylindole; Sigma), washed, and mounted with Prolong anti-fade (Molecular Probes). Confocal laser scanning microscopy was done with a Zeiss LSM510-META confocal microscope system at the W.M. Keck Center for Collaborative Neuroscience, Rutgers University. The percentage of acini with lumen formation was the mean of two independent experiments (for each experiment, 100 acini were scored for each cell line at each time point). For histology, mammary acini were grown for 12 d, fixed in 10% formalin, scraped from the glass slide with a razor blade, pelleted, embedded in paraffin, and processed for H&E staining. For EM, acini were grown for 12 d and processed as previously described (Mills et al. 2004). Electron microscopy was performed with a JEOL 1200EX electron microscope.
Detection and quantitation of autophagy
beclin1+/+ and beclin1+/− iMMECs expressing Bcl-2 (WT3.B3 and BLN2.B4) were stably transfected with the EGFP-LC3 expression vector (Mizushima et al. 2004) and several independent clones were isolated. Two clones (one per genotype) were subjected to metabolic stress (i.e., ischemia defined as 1% oxygen in the absence of glucose) for 0, 1, 2, and 3 d, as previously described (Nelson et al. 2004), and autophagy was quantified by EGFP-LC3 membrane translocation by fluorescence microscopy, as previously described (Degenhardt et al. 2006). Three-hundred cells were evaluated for EGFP-LC3 translocation for each clone at each time point. Three independent experiments were performed, and the mean percentage EGFP-LC3 translocation (n = 3) with standard deviation for each genotype at each time point is presented. For localization of autophagy in mammary acini, EGFP-LC3 fluorescence was monitored by confocal microscopy on acini formed by beclin1+/+ and beclin1+/− iMMECs with Bcl-2 and stably expressing EGFP-LC3 (Mizushima et al. 2004). For localization of autophagy in vivo, tumors formed by beclin1+/+ and beclin1+/− iMMECs with Bcl-2 (WT3.B3, BLN2.B8) and stably expressing EGFP-LC3 were excised at day 1 post-implantation and snap-frozen. Cryosections were mounted and EGFP-LC3 membrane translocation was monitored with EGFP-LC3 fluorescence by confocal microscopy.
Clonogenic survival assays
beclin1+/+ and beclin1+/− iMMECs expressing Bcl-2 were plated at a density of 2 × 106 cells per 10 cm dish, allowed to settle for 24 h, and then subjected to metabolic stress for 12 d. Colony formation was assessed by restoration of normal growth conditions, followed by 2% Giemsa staining. Survival frequency was calculated as the number of colonies/2 × 106 cells plated, since the plating efficiency was the same for all cell lines.
Detection of metabolic stress in mammary acini and tumors
Detection of areas of increased metabolic stress in mammary acini was performed by hypoxyprobe IHC and IF. Mammary acini formed by WT3.B3 and BLN2.B8 cells and grown in Matrigel for 12 d were incubated with 200 μM hypoxyprobe (pimonidazole) in tissue culture medium for 2 h at 37°C. For IHC, acini were fixed in 10% formalin, paraffin-embedded, and processed as previously described (Nelson et al. 2004). For IF, acini were fixed and processed as described above. Detection of areas of increased metabolic stress in tumors was performed by hypoxyprobe IHC. Mice were injected with hypoxyprobe at a dose of 60 μg/kg body weight 30 min prior to sacrifice. Tumors were excised, paraffin-embedded, and processed as previously described (Nelson et al. 2004).
Tumorigenicity assays
Cells were harvested by trypsinization, washed, and resuspended in PBS (107 cells/mL). Orthotopic mammary gland implantation of iMMECs was performed using Institutional Animal Care and Use Committee-approved protocol. Five-week-old to 8-wk-old NCR nude female mice were anesthetized with ketamine and xylazine. A small incision was made to reveal the right third mammary gland, and 106 cells were injected into the mammary fat pad. The incision was closed with surgical clips that were removed 2 d later. Tumor outgrowth was monitored by weekly measurements of tumor length (L) and width (W). Tumor volume was calculated as π_LW_2/6. For the tumorigenicity assays described in Figure 2D, five mice per cell line and two independently derived cell lines per genotype (one for vector control) were used. For the tumorigenicity assays described in Figure 5A, four mice per cell line, three cell lines per genotype for beclin1+/+ and beclin1+/− iMMECs, and four cell lines per genotype for beclin1+/+ and beclin1+/− iMMECs with Bcl-2 were used.
PALA selection
PALA was obtained from the Drug Synthesis and Chemistry Branch (Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute). For each cell line, the PALA LD50 was determined. There was minimal variation between the LD50 values for the four beclin1+/+ and the four beclin1+/− iMMEC cell lines with Bcl-2 that were examined, and thus LD50 of 17 μM PALA was used for all cell lines. The plating efficiency was the same for all cell lines. Selections of 5 × 105 cells per 10-cm plate were done in triplicate in the presence of 10% (v/v) dialyzed FBS and PALA at 5× LD50. Drug-resistant colonies, detectable after 2 wk, were fixed with 3:1 methanol:acetic acid, stained with 2% Giemsa, and counted. The frequency of PALA resistance was calculated as the number of resistant colonies/5 × 105 cells plated.
PCR for CAD gene amplification
An 800-base-pair (bp) fragment from the CAD gene was amplified by PCR from genomic DNA obtained from PALAR colonies using the following set of primers: 4-fwd (5′-GGAGCGGA GACTCCGACG-3′) and 4-rev (5′-CTAATGAACAGGAAGAT CCGGTATC-3′).
Time-lapse microscopy
Time-lapse microscopy was performed as previously described (Degenhardt et al. 2006). In summary, cells were plated in T25 tissue culture flasks equipped with tubing to allow cell culture in an ischemic gas environment, and placed in a time-lapse environmental chamber. Phase-contrast images (100×) at 10 different fields were obtained at 10-min intervals for up to 5 d.
Acknowledgments
We thank Dr. Kurt Degenhardt for expert advice during the early stages of the MMEC model development, T. Sharkey for assistance with preparation of the manuscript, the members of the Tissue Analytical Services at The Cancer Institute of New Jersey for assistance with histology, Dr. Noriko Kane-Goldsmith for assistance with confocal microscopy, R. Patel for assistance with electron microscopy, Drs. Nathaniel Heintz and Zhenyu Yue for providing the beclin1+/− mice, Dr. Noboru Mizushima for providing the EGFP-LC3 plasmid, Dr. Michael Reiss for technical assistance with the orthotopic mammary fat pad injections, and Dr. Edmund Lattime for providing the HER2/neu plasmid. V.K.-W. is a Stacy Goldstein Medical Oncology Fellow at UMDNJ-Robert Wood Johnson Medical School and The Cancer Institute of New Jersey, and was supported by a training grant from the National Institute of Health (T32 CA99946). This work was also supported by grants from the National Institutes of Health and the Howard Hughes Medical Institute (to E.W.), and the New Jersey Commission on Cancer Research (to S.P. and O.K.).
Footnotes
References
- Abedin M.J., Wang D., McDonnell M.A., Lehmann U., Kelekar A., Wang D., McDonnell M.A., Lehmann U., Kelekar A., McDonnell M.A., Lehmann U., Kelekar A., Lehmann U., Kelekar A., Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- Aita V.M., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Kalachikov S., Gilliam T.C., Levine B., Gilliam T.C., Levine B., Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Albertson D.G. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Collins C., McCormick F., Gray J.W., Collins C., McCormick F., Gray J.W., McCormick F., Gray J.W., Gray J.W. Chromosome aberrations in solid tumors. Nat. Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Al-Kuraya K., Schraml P., Torhorst J., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Schraml P., Torhorst J., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Torhorst J., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Spichtin H., Maurer R., Mirlacher M., Kochli O., Maurer R., Mirlacher M., Kochli O., Mirlacher M., Kochli O., Kochli O., et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534–8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- Baehrecke E.H. Autophagy: Dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Guldberg P., Sehested M., Nesland J.M., Lukas C., Sehested M., Nesland J.M., Lukas C., Nesland J.M., Lukas C., Lukas C., et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Baselga J., Perez E.A., Pienkowski T., Bell R., Perez E.A., Pienkowski T., Bell R., Pienkowski T., Bell R., Bell R. Adjuvant trastuzumab: A milestone in the treatment of HER-2-positive early breast cancer. Oncologist. 2006;11 (Suppl. 1):4–12. doi: 10.1634/theoncologist.11-90001-4. [DOI] [PubMed] [Google Scholar]
- Bergamini E., Cavallini G., Donati A., Gori Z., Cavallini G., Donati A., Gori Z., Donati A., Gori Z., Gori Z. The role of macroautophagy in the ageing process, anti-ageing intervention and age-associated diseases. Int. J. Biochem. Cell Biol. 2004;36:2392–2404. doi: 10.1016/j.biocel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Boya P., Gonzalez-Polo R.A., Casares N., Perfettini J.L., Dessen P., Larochette N., Metivier D., Meley D., Souquere S., Yoshimori T., Gonzalez-Polo R.A., Casares N., Perfettini J.L., Dessen P., Larochette N., Metivier D., Meley D., Souquere S., Yoshimori T., Casares N., Perfettini J.L., Dessen P., Larochette N., Metivier D., Meley D., Souquere S., Yoshimori T., Perfettini J.L., Dessen P., Larochette N., Metivier D., Meley D., Souquere S., Yoshimori T., Dessen P., Larochette N., Metivier D., Meley D., Souquere S., Yoshimori T., Larochette N., Metivier D., Meley D., Souquere S., Yoshimori T., Metivier D., Meley D., Souquere S., Yoshimori T., Meley D., Souquere S., Yoshimori T., Souquere S., Yoshimori T., Yoshimori T., et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein H.J. The distinctive nature of HER2-positive breast cancers. N. Engl. J. Med. 2005;353:1652–1654. doi: 10.1056/NEJMp058197. [DOI] [PubMed] [Google Scholar]
- Carroll D.K., Carroll J.S., Leong C.O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W., Carroll J.S., Leong C.O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W., Leong C.O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W., Brown M., Mills A.A., Brugge J.S., Ellisen L.W., Mills A.A., Brugge J.S., Ellisen L.W., Brugge J.S., Ellisen L.W., Ellisen L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- Chernova O.B., Chernov M.V., Ishizaka Y., Agarwal M.L., Stark G.R., Chernov M.V., Ishizaka Y., Agarwal M.L., Stark G.R., Ishizaka Y., Agarwal M.L., Stark G.R., Agarwal M.L., Stark G.R., Stark G.R. MYC abrogates p53-mediated cell cycle arrest in N-(phosphonacetyl)-L-aspartate-treated cells, permitting CAD gene amplification. Mol. Cell. Biol. 1998;18:536–545. doi: 10.1128/mcb.18.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K., DeVries S., Fridlyand J., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., DeVries S., Fridlyand J., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., Fridlyand J., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., Lapuk A., Neve R.M., Qian Z., Ryder T., Neve R.M., Qian Z., Ryder T., Qian Z., Ryder T., Ryder T., et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Collins N.L., Reginato M.J., Paulus J.K., Sgroi D.C., Labaer J., Brugge J.S., Reginato M.J., Paulus J.K., Sgroi D.C., Labaer J., Brugge J.S., Paulus J.K., Sgroi D.C., Labaer J., Brugge J.S., Sgroi D.C., Labaer J., Brugge J.S., Labaer J., Brugge J.S., Brugge J.S. G1/S cell cycle arrest provides anoikis resistance through Erk-mediated Bim suppression. Mol. Cell. Biol. 2005;25:5282–5291. doi: 10.1128/MCB.25.12.5282-5291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuconati A., Mukherjee C., Perez D., White E., Mukherjee C., Perez D., White E., Perez D., White E., White E. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes & Dev. 2003;17:2922–2932. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea A.D., Grompe M., Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., Brugge J.S., Muthuswamy S.K., Brugge J.S., Brugge J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Debnath J., Baehrecke E.H., Kroemer G., Baehrecke E.H., Kroemer G., Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., White E., White E. A mouse model system to genetically dissect the molecular mechanisms regulating tumorigenesis. Clin. Cancer Res. 2006;12:5298–5304. doi: 10.1158/1078-0432.CCR-06-0439. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., Chen G., Lindsten T., White E., Chen G., Lindsten T., White E., Lindsten T., White E., White E. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cell. 2002a;2:193–203. doi: 10.1016/s1535-6108(02)00126-5. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., Sundararajan R., Lindsten T., Thompson C., White E., Sundararajan R., Lindsten T., Thompson C., White E., Lindsten T., Thompson C., White E., Thompson C., White E., White E. Bax and Bak independently promote cytochrome c release from mitochondria. J. Biol. Chem. 2002b;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Mukherjee C., Shi Y., Gelinas C., Fan Y., Shi Y., Gelinas C., Fan Y., Gelinas C., Fan Y., Fan Y., et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J. Angiogenesis and apoptosis. Semin. Cancer Biol. 2003;13:159–167. doi: 10.1016/s1044-579x(02)00133-5. [DOI] [PubMed] [Google Scholar]
- Futreal P.A., Soderkvist P., Marks J.R., Iglehart J.D., Cochran C., Barrett J.C., Wiseman R.W., Soderkvist P., Marks J.R., Iglehart J.D., Cochran C., Barrett J.C., Wiseman R.W., Marks J.R., Iglehart J.D., Cochran C., Barrett J.C., Wiseman R.W., Iglehart J.D., Cochran C., Barrett J.C., Wiseman R.W., Cochran C., Barrett J.C., Wiseman R.W., Barrett J.C., Wiseman R.W., Wiseman R.W. Detection of frequent allelic loss on proximal chromosome 17q in sporadic breast carcinoma using microsatellite length polymorphisms. Cancer Res. 1992;52:2624–2627. [PubMed] [Google Scholar]
- Gebhart E. Double minutes, cytogenetic equivalents of gene amplification, in human neoplasia—A review. Clin. Transl. Oncol. 2005;7:477–485. doi: 10.1007/BF02717000. [DOI] [PubMed] [Google Scholar]
- Gordon K.E., Binas B., Chapman R.S., Kurian K.M., Clarkson R.W., Clark A.J., Lane E.B., Watson C.J., Binas B., Chapman R.S., Kurian K.M., Clarkson R.W., Clark A.J., Lane E.B., Watson C.J., Chapman R.S., Kurian K.M., Clarkson R.W., Clark A.J., Lane E.B., Watson C.J., Kurian K.M., Clarkson R.W., Clark A.J., Lane E.B., Watson C.J., Clarkson R.W., Clark A.J., Lane E.B., Watson C.J., Clark A.J., Lane E.B., Watson C.J., Lane E.B., Watson C.J., Watson C.J. A novel cell culture model for studying differentiation and apoptosis in the mouse mammary gland. Breast Cancer Res. 2000;2:222–235. doi: 10.1186/bcr57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B., Liloglou T., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B., Ditullio R.A., Kastrinakis N.G., Levy B., Kastrinakis N.G., Levy B., Levy B., et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Yokoyama M., Mishima K., Saito I., Okano H., Mishima K., Saito I., Okano H., Saito I., Okano H., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Imagawa W., Yang J., Guzman R.C., Nandi S., Yang J., Guzman R.C., Nandi S., Guzman R.C., Nandi S., Nandi S. Collagen gel method for the primary culture of mouse mammary epithelium. In: Ip M.M., Asch B.B., Asch B.B., editors. Methods in mammary gland biology and breast cancer research. Kluwer Academic/Plenum Publishers; New York: 2000. pp. 111–123. [Google Scholar]
- Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2:80–84. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- Jin S., White E., White E. Role of autophagy in cancer: Management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Pinkel D., Gray J.W., Waldman F.M., Gray J.W., Waldman F.M., Waldman F.M. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc. Natl. Acad. Sci. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y., Okano T., Ohsumi M., Ohsumi Y., Ohsumi M., Ohsumi Y., Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Emr S.D., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Mizushima N., Ohsumi Y., Uchiyama Y., Ohsumi Y., Uchiyama Y., Uchiyama Y., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Ueno T., Koike M., Uchiyama Y., Kominami E., Koike M., Uchiyama Y., Kominami E., Uchiyama Y., Kominami E., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Ohsumi Y., Tokuhisa T., Mizushima N., Tokuhisa T., Mizushima N., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J., Klionsky D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Li B., Rosen J.M., McMenamin-Balano J., Muller W.J., Perkins A.S., Rosen J.M., McMenamin-Balano J., Muller W.J., Perkins A.S., McMenamin-Balano J., Muller W.J., Perkins A.S., Muller W.J., Perkins A.S., Perkins A.S. neu/ERBB2 cooperates with p53-172H during mammary tumorigenesis in transgenic mice. Mol. Cell. Biol. 1997;17:3155–3163. doi: 10.1128/mcb.17.6.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B., Brown K., Kempkes B., Hibshoosh H., Levine B., Kempkes B., Hibshoosh H., Levine B., Hibshoosh H., Levine B., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lin H.J., Eviner V., Prendergast G.C., White E., Eviner V., Prendergast G.C., White E., Prendergast G.C., White E., White E. Activated H-ras rescues E1A-induced apoptosis and cooperates with E1A to overcome p53-dependent growth arrest. Mol. Cell. Biol. 1995;15:4536–4544. doi: 10.1128/mcb.15.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum J.J., Bauer D.E., Kong M., Harris M.H., Li C., Lindsten T., Thompson C.B., Bauer D.E., Kong M., Harris M.H., Li C., Lindsten T., Thompson C.B., Kong M., Harris M.H., Li C., Lindsten T., Thompson C.B., Harris M.H., Li C., Lindsten T., Thompson C.B., Li C., Lindsten T., Thompson C.B., Lindsten T., Thompson C.B., Thompson C.B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Mathew R., Kongara S., Beaudoin B., Karp C.M., Bray K., Degenhardt K., Chen G., Jin S., White E., Kongara S., Beaudoin B., Karp C.M., Bray K., Degenhardt K., Chen G., Jin S., White E., Beaudoin B., Karp C.M., Bray K., Degenhardt K., Chen G., Jin S., White E., Karp C.M., Bray K., Degenhardt K., Chen G., Jin S., White E., Bray K., Degenhardt K., Chen G., Jin S., White E., Degenhardt K., Chen G., Jin S., White E., Chen G., Jin S., White E., Jin S., White E., White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes & Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Eskelinen E.L., Hall D.H., Levine B., Hall D.H., Levine B., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mills K.R., Reginato M., Debnath J., Queenan B., Brugge J.S., Reginato M., Debnath J., Queenan B., Brugge J.S., Debnath J., Queenan B., Brugge J.S., Queenan B., Brugge J.S., Brugge J.S. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc. Natl. Acad. Sci. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005;12 (Suppl. 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y., Matsui M., Yoshimori T., Ohsumi Y., Yoshimori T., Ohsumi Y., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S., DiGiovanna M.P., Lu Z., Moskowitz C., Panageas K.S., Van Poznak C., Hudis C.A., Norton L., Tan L., Stern D.F., DiGiovanna M.P., Lu Z., Moskowitz C., Panageas K.S., Van Poznak C., Hudis C.A., Norton L., Tan L., Stern D.F., Lu Z., Moskowitz C., Panageas K.S., Van Poznak C., Hudis C.A., Norton L., Tan L., Stern D.F., Moskowitz C., Panageas K.S., Van Poznak C., Hudis C.A., Norton L., Tan L., Stern D.F., Panageas K.S., Van Poznak C., Hudis C.A., Norton L., Tan L., Stern D.F., Van Poznak C., Hudis C.A., Norton L., Tan L., Stern D.F., Hudis C.A., Norton L., Tan L., Stern D.F., Norton L., Tan L., Stern D.F., Tan L., Stern D.F., Stern D.F., et al. Phosphorylated/activated HER2 as a marker of clinical resistance to single agent taxane chemotherapy for metastatic breast cancer. Cancer Invest. 2005;23:483–487. doi: 10.1080/07357900500201301. [DOI] [PubMed] [Google Scholar]
- Moyer J.D., Smith P.A., Levy E.J., Handschumacher R.E., Smith P.A., Levy E.J., Handschumacher R.E., Levy E.J., Handschumacher R.E., Handschumacher R.E. Kinetics of N-(phosphonacetyl)-L-aspartate and pyrazofurin depletion of pyrimidine ribonucleotide and deoxyribonucleotide pools and their relationship to nucleic acid synthesis in intact and permeabilized cells. Cancer Res. 1982;42:4525–4531. [PubMed] [Google Scholar]
- Naylor T.L., Greshock J., Wang Y., Colligon T., Yu Q.C., Clemmer V., Zaks T.Z., Weber B.L., Greshock J., Wang Y., Colligon T., Yu Q.C., Clemmer V., Zaks T.Z., Weber B.L., Wang Y., Colligon T., Yu Q.C., Clemmer V., Zaks T.Z., Weber B.L., Colligon T., Yu Q.C., Clemmer V., Zaks T.Z., Weber B.L., Yu Q.C., Clemmer V., Zaks T.Z., Weber B.L., Clemmer V., Zaks T.Z., Weber B.L., Zaks T.Z., Weber B.L., Weber B.L. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–R1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.A., Tan T.T., Rabson A.B., Anderson D., Degenhardt K., White E., Tan T.T., Rabson A.B., Anderson D., Degenhardt K., White E., Rabson A.B., Anderson D., Degenhardt K., White E., Anderson D., Degenhardt K., White E., Degenhardt K., White E., White E. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes & Dev. 2004;18:2095–2107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E., McCord S., Tlsty T.D., McCord S., Tlsty T.D., Tlsty T.D. Increased incidence of CAD gene amplification in tumorigenic rat lines as an indicator of genomic instability of neoplastic cells. J. Biol. Chem. 1989;264:3390–3396. [PubMed] [Google Scholar]
- Paulson T.G., Almasan A., Brody L.L., Wahl G.M., Almasan A., Brody L.L., Wahl G.M., Brody L.L., Wahl G.M., Wahl G.M. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol. Cell. Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas D.R., Talapatra S., Edinger A.L., Rathmell J.C., Thompson C.B., Talapatra S., Edinger A.L., Rathmell J.C., Thompson C.B., Edinger A.L., Rathmell J.C., Thompson C.B., Rathmell J.C., Thompson C.B., Thompson C.B. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J. Biol. Chem. 2001;276:12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Eskelinen E.L., Mizushima N., Ohsumi Y., Mizushima N., Ohsumi Y., Ohsumi Y., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato M.J., Mills K.R., Paulus J.K., Lynch D.K., Sgroi D.C., Debnath J., Muthuswamy S.K., Brugge J.S., Mills K.R., Paulus J.K., Lynch D.K., Sgroi D.C., Debnath J., Muthuswamy S.K., Brugge J.S., Paulus J.K., Lynch D.K., Sgroi D.C., Debnath J., Muthuswamy S.K., Brugge J.S., Lynch D.K., Sgroi D.C., Debnath J., Muthuswamy S.K., Brugge J.S., Sgroi D.C., Debnath J., Muthuswamy S.K., Brugge J.S., Debnath J., Muthuswamy S.K., Brugge J.S., Muthuswamy S.K., Brugge J.S., Brugge J.S. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat. Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- Rijnkels M., Rosen J.M., Rosen J.M. Adenovirus–Cre-mediated recombination in mammary epithelial early progenitor cells. J. Cell Sci. 2001;114:3147–3153. doi: 10.1242/jcs.114.17.3147. [DOI] [PubMed] [Google Scholar]
- Risinger M.A., Groden J., Groden J. Crosslinks and crosstalk: Human cancer syndromes and DNA repair defects. Cancer Cell. 2004;6:539–545. doi: 10.1016/j.ccr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Saito H., Inazawa J., Saito S., Kasumi F., Koi S., Sagae S., Kudo R., Saito J., Noda K., Nakamura Y., Inazawa J., Saito S., Kasumi F., Koi S., Sagae S., Kudo R., Saito J., Noda K., Nakamura Y., Saito S., Kasumi F., Koi S., Sagae S., Kudo R., Saito J., Noda K., Nakamura Y., Kasumi F., Koi S., Sagae S., Kudo R., Saito J., Noda K., Nakamura Y., Koi S., Sagae S., Kudo R., Saito J., Noda K., Nakamura Y., Sagae S., Kudo R., Saito J., Noda K., Nakamura Y., Kudo R., Saito J., Noda K., Nakamura Y., Saito J., Noda K., Nakamura Y., Noda K., Nakamura Y., Nakamura Y. Detailed deletion mapping of chromosome 17q in ovarian and breast cancers: 2-cM region on 17q21.3 often and commonly deleted in tumors. Cancer Res. 1993;53:3382–3385. [PubMed] [Google Scholar]
- Shin I., Miller T., Arteaga C.L., Miller T., Arteaga C.L., Arteaga C.L. ErbB receptor signaling and therapeutic resistance to aromatase inhibitors. Clin. Cancer Res. 2006;12:1008s–1012s. doi: 10.1158/1078-0432.CCR-05-2352. [DOI] [PubMed] [Google Scholar]
- Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L., Wong S.G., Levin W.J., Ullrich A., McGuire W.L., Levin W.J., Ullrich A., McGuire W.L., Ullrich A., McGuire W.L., McGuire W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Streuli C.H., Schmidhauser C., Bailey N., Yurchenco P., Skubitz A.P., Roskelley C., Bissell M.J., Schmidhauser C., Bailey N., Yurchenco P., Skubitz A.P., Roskelley C., Bissell M.J., Bailey N., Yurchenco P., Skubitz A.P., Roskelley C., Bissell M.J., Yurchenco P., Skubitz A.P., Roskelley C., Bissell M.J., Skubitz A.P., Roskelley C., Bissell M.J., Roskelley C., Bissell M.J., Bissell M.J. Laminin mediates tissue-specific gene expression in mammary epithelia. J. Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.T., Degenhardt K., Nelson D.A., Beaudoin B., Nieves-Neira W., Bouillet P., Villunger A., Adams J.M., White E., Degenhardt K., Nelson D.A., Beaudoin B., Nieves-Neira W., Bouillet P., Villunger A., Adams J.M., White E., Nelson D.A., Beaudoin B., Nieves-Neira W., Bouillet P., Villunger A., Adams J.M., White E., Beaudoin B., Nieves-Neira W., Bouillet P., Villunger A., Adams J.M., White E., Nieves-Neira W., Bouillet P., Villunger A., Adams J.M., White E., Bouillet P., Villunger A., Adams J.M., White E., Villunger A., Adams J.M., White E., Adams J.M., White E., White E. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Yan L., Vatner D.E., Kim S.J., Ge H., Masurekar M., Massover W.H., Yang G., Matsui Y., Sadoshima J., Vatner S.F., Vatner D.E., Kim S.J., Ge H., Masurekar M., Massover W.H., Yang G., Matsui Y., Sadoshima J., Vatner S.F., Kim S.J., Ge H., Masurekar M., Massover W.H., Yang G., Matsui Y., Sadoshima J., Vatner S.F., Ge H., Masurekar M., Massover W.H., Yang G., Matsui Y., Sadoshima J., Vatner S.F., Masurekar M., Massover W.H., Yang G., Matsui Y., Sadoshima J., Vatner S.F., Massover W.H., Yang G., Matsui Y., Sadoshima J., Vatner S.F., Yang G., Matsui Y., Sadoshima J., Vatner S.F., Matsui Y., Sadoshima J., Vatner S.F., Sadoshima J., Vatner S.F., Vatner S.F. Autophagy in chronically ischemic myocardium. Proc. Natl. Acad. Sci. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D.J., Klionsky D.J. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005;12 (Suppl. 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N., Jin S., Yang C., Levine A.J., Heintz N., Yang C., Levine A.J., Heintz N., Levine A.J., Heintz N., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]