Loss of Gcn5 Acetyltransferase Activity Leads to Neural Tube Closure Defects and Exencephaly in Mouse Embryos (original) (raw)
Abstract
Gcn5 was the first transcription-related histone acetyltransferase (HAT) to be identified. However, the functions of this enzyme in mammalian cells remain poorly defined. Deletion of Gcn5 in mice leads to early embryonic lethality with increased apoptosis in mesodermal lineages. Here we show that deletion of p53 allows _Gcn5_−/− embryos to survive longer, but _Gcn5_−/− _p53_−/− embryos still die in midgestation. Interestingly, embryos homozygous for point mutations in the Gcn5 catalytic domain survive significantly longer than _Gcn5_−/− or _Gcn5_−/− _p53_−/− mice. In contrast to _Gcn5_−/− embryos, _Gcn5_hat/hat embryos do not exhibit increased apoptosis but do exhibit severe cranial neural tube closure defects and exencephaly. Together, our results indicate that Gcn5 has important, HAT-independent functions in early development and that Gcn5 acetyltransferase activity is required for cranial neural tube closure in the mouse.
Histone acetyltransferases (HATs) serve as coactivators of transcription that are important for proper regulation of gene expression (32). Although much is known regarding the biochemical composition of HAT complexes, little is known regarding the functions of these enzymes in vivo. For example, few authentic gene targets have been identified for Gcn5, the prototypical transcription-related HAT (5), or for the highly related PCAF protein (41).
Gcn5 and PCAF are assembled into large, multisubunit complexes in Saccharomyces cerevisiae, flies, and mammalian cells (4, 13, 23, 29). The components of yeast Gcn5-containing complexes (SAGA, ADA, SALSA, and SLIK) have been characterized extensively (13, 14, 34). Each complex has unique components as well as some shared Spt, Ada, and TATA box-binding protein-associated factor (TAF) proteins. In Drosophila melanogaster, Gcn5 is part of at least two complexes that contain different Ada2 homologs (20). Mammalian Gcn5 is also part of multiple, related but distinct complexes, including STAGA and TFTC (4, 23, 29) that are remarkably similar to yeast SAGA in that they contain homologs of Ada2, Ada3, Spt3, Tra1 (PAF400), and several TAFs. STAGA and TFTC contain additional factors such as SAP130, an RNA splicing factor, and DDB1, which is involved in nucleotide excision repair, indicating that these complexes regulate processes other than transcription initiation (3, 7, 24). Indeed, TFTC exhibits increased binding to UV-damaged DNA in vitro, suggesting this complex may play a direct role in sensing or repair of DNA damage in vivo (3). Damage sensing might involve the p53 tumor suppressor, since both Gcn5 (our unpublished studies) and PCAF (21) acetylate p53 in vitro. Acetylation of p53 by these HATs and by p300/CBP is increased in vivo in response to DNA damage (1, 16). However, how p53 acetylation affects DNA repair or repair checkpoints is not yet clear.
Mammalian Gcn5 contains at least three important functional domains (22). An amino-terminal domain that is not present in the yeast enzyme mediates interactions with p300 and CBP (39). The conserved bromodomain binds to acetyl-lysine moieties and may help anchor the enzyme at chromatin targets (36). Critical conserved residues in the catalytic center of Gcn5 are required for acetyl coenzyme A binding and acetyltransferase activity (reviewed in reference 22). Mutation of a single glutamate (E173) in yeast Gcn5 that acts as a general base for catalysis reduces the specific activity of recombinant Gcn5 more than 300-fold (6, 35). Mutation of this glutamate or of other conserved residues in the catalytic center cripples the ability of yeast Gcn5 to function as a coactivator in vivo, even though the mutant Gcn5 proteins are still incorporated into multisubunit complexes (6, 35). Overexpression of these mutant forms of yeast Gcn5 are dominant negative relative to the wild-type protein, probably because SAGA subunits are sequestered into inactive HAT complexes (6).
Despite extensive biochemical and structural analyses of Gcn5 complexes (22, 32), the functions of this HAT in mammalian cells are not well defined. We and others have shown that deletion of Gcn5 in mice leads to embryonic death, indicating this HAT is essential for normal development (38, 40). Gcn5 null embryos complete gastrulation but do not form somites, a neural tube, or a notochord. The lack of somite formation stems from a paucity of paraxial mesoderm, which is specified normally in the Gcn5 null embryos but is subject to increased levels of apoptosis (38). In contrast, PCAF null mice are viable with no obvious abnormalities (38, 40). Embryos deficient for both Gcn5 and PCAF die much earlier than Gcn5 single mutants, however, indicating that PCAF does contribute to some early developmental processes (38).
Our original Gcn5 null allele deleted all Gcn5 coding sequences, thereby completely abolishing Gcn5 functions (38). The early death of Gcn5 null embryos precluded determination of Gcn5 functions later in development. Here we report that deletion of the p53 tumor suppressor allows Gcn5 null embryos to survive longer. However, _Gcn5_−/− _p53_−/− double mutants still die by embryonic day 11.5 (E11.5) with abnormal morphology. Mouse embryos homozygous for point mutations in the predicted catalytic domain of the murine Gcn5 sequence develop much further than either _Gcn5_−/− or _Gcn5_−/− _p53_−/− null mice, surviving until E16.5. Cranial neural tube closure is defective in these Gcn5 HAT mutant mice, leading to exencephaly. Collectively, our data demonstrate that Gcn5 has important functions during development that are independent of its HAT activity, but Gcn5 HAT activity is critical for cranial neural tube closure.
MATERIALS AND METHODS
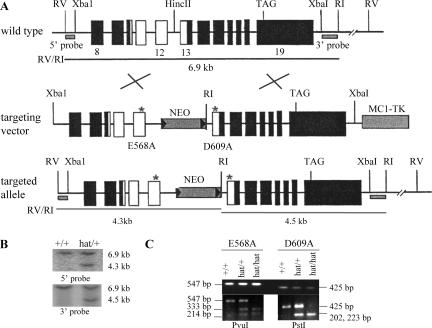
Creation of Gcn5 HAT mutation gene replacement vector.
Genomic sequences covering the entire Gcn5 coding region were isolated previously (39). The targeting vector was constructed by placing a 6.9-kb XbaI/EcoRV genomic fragment containing exons 8 to 19 of Gcn5 into Bluescript KS(+). The E568A mutation was introduced into exon 12, and D609A was introduced into exon 13 of Gcn5 by site-directed mutagenesis. Primer sequences used for mutagenesis will be provided upon request. A PGKneobpA neomycin resistance expression cassette was inserted into intron 12 accompanying an EcoRI site. A MC1-TK (thymidine kinase) cassette was added at the 3′ end for negative selection. For genotyping, genomic DNA was digested with EcoRI/EcoRV and probed with a 0.5-kb EcoRI/HindIII fragment (5′ probe) or a 0.6-kb XbaI/EcoRI fragment (3′ probe).
Creation of _Gcn5_hat/+ mice.
The Gcn5 targeting vector was linearized and electroporated into 129/SvEv-derived embryonic stem (ES) cells (AB1). Doubly resistant cells [resistant to G418 and 1-(2-deoxy-2-β-d-arabinofuranosyl)-_S_- iodouracid (FLAU)] were genotyped by Southern blot analyses using probes described above. Correctly targeted ES clones were microinjected into C57BL/6J blastocysts. Chimeras were mated with wild-type C57BL/6J mice, and heterozygotes were identified among agouti progeny by Southern blot analysis of tail genomic DNA using the probes described above. Heterozygous mice were intercrossed to generate mice homozygous for the Gcn5 point mutations.
Genotyping of embryos.
PCR-based genotyping was performed using primers specific for the Gcn5 wild-type allele, WL4 5′-TCACTATCTCGGATGGCTT-3′ and WL5Y 5′-ACAGCTACGGCACAACTC-3′ (800 bp), or the deletion allele, WL4 and WL6Y 5′-CTCTTCGCTATTACGCCAG-3′ (440 bp). Dimethyl sulfoxide (6%) was added to the PCR mixtures. PCR conditions were 1 cycle of 1 min at 94°C; 5 cycles of 30 s at 94°C, 40 s at 63°C, and 1 min at 68°C; 30 cycles of 30 s at 94°C, 40 s at 55°C, and 1 min at 72°C; and 1 cycle of 7 min at 72°C.
To identify embryos carrying the Gcn5 catalytic site mutations, PCR amplification of yolk sac DNA was performed with the following primers: for E568A mutation in exon 12, 5′-CTTTCAGGGTTCCGTTCCTG-3′ and 5′-TCTCACAGTAGACCAGGCTG-3′ (547 bp); for D609A mutation in exon 13, 5′-ACTCAAGTGCTGAGTCTGGC-3′, 5′-CCCCCAAAGGACCTTCCAAT-3′ (425 bp). PCR conditions were 1 cycle of 5 min at 95°C, 35 cycles of 1 min at 95°C, 1 min at 65°C, and 1 min at 72°C; and 1 cycle of 5 min at 72°C. The PCR products were then digested with PvuI to detect the E568A mutation (yielding 333-bp and 214-bp products) or PstI to detect the D609A mutation (yielding 223-bp and 202-bp products) as shown below (see Fig. 3C).
FIG. 3.
Creation of _Gcn5_hat/+ mice. (A) Strategy to introduce an allele of Gcn5 bearing two point mutations (E568A and D609A) in sequences encoding the catalytic center of the enzyme into the native Gcn5 chromosomal locus. The locations of the mutations (*) and EcoRI (RI) and EcoRV (RV) restriction sites used for genotyping by Southern blot analyses are shown together with the locations of 5′ and 3′ probes. Exons are depicted as boxes and introns as intervening lines. Note that only a portion (exons 8 to 19) of the Gcn5 gene is shown. The white boxes indicate the portion of the targeting vector that was subcloned for mutagenesis and then reinserted into the targeting vector. The positions of a PGKneobpA neomycin resistance expression cassette (NEO) and MC1-TK (thymidine kinase) cassette are also shown. (B) Southern blots to identify _Gcn5_hat/+ ES cells or mice. 5′ and 3′ probes (as indicated in panel A) were used to detect EcoRI/EcoRV digestion products corresponding to the wild-type (6.9-kb) or targeted (4.3- or 4.5-kb) Gcn5 alleles. (C) PCR genotyping to confirm the presence of the point mutations in _Gcn5_hat/hat embryos. PCR products flanking each mutation site were generated as described in Materials and Methods (top blot). The E568A mutation is linked to a PvuI restriction site, and the D609A mutation is linked to a PstI restriction site. Cleavage of the PCR products by these enzymes (bottom blots) confirms the presence of the mutations. The two PstI cleavage products are close in size (202 and 223 bp) and migrate as a single band.
Genotyping for the p53 allele was performed as described previously (27).
HAT assays.
Full-length murine Gcn5 cDNA sequences (wild type or with catalytic site mutations) were subcloned into pRSETB vector and expressed in Escherichia coli BL2l bacterial cells. His6-tagged proteins were induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). HAT assays were performed as described previously (26).
Staging of mouse embryos.
Mouse embryos were staged according to the time of maternal plugging and the observed number of somites in the embryos (28).
Sectioning of mouse embryos.
Mouse embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4°C, dehydrated in graded solutions of alcohol, cleared in xylene, and embedded in paraffin. Five- or 6-μm sections were generated from paraffin-embedded embryos using a Leica RM 2145 processor.
Section immunohistochemistry.
Section immunohistochemistry was performed on 5- or 6-μm sections of paraffin-embedded E8.5 embryos using the following antibodies: CM5 (NovoCastra NCL-p53-CM5p), cleaved caspase 3 (Asp175) (Cell Signaling Technology; 1:200 dilution), or phospho-histone H3 (Ser10) (Cell Signaling Technology; 1:50 dilution), according to the manufacturer's instructions as described in the mouse-on-mouse (M.O.M.) kit from Vector Labs. High-temperature antigen unmasking was performed for all slides. Briefly, slides were placed in coplin jars containing 0.01 M sodium citrate (pH 6.0). The citrate solution and slides were brought to a boil in a microwave. The coplin jar was then removed from the microwave and allowed to cool to room temperature before slides were removed for further processing. The 3,3′-diaminobenzidine (DAB) substrate kit (Vector Labs) was used for visualization of the peroxidase substrate.
TUNEL assays.
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed on 6-μm paraffin sections using the FragEL DNA fragmentation detection kit (Oncogene). The manufacturer's TdT-FragEL detailed protocol was followed, except that the volume of enzyme was halved and the termination of labeling reaction step was abridged.
Whole-mount in situ hybridizations.
Sense and antisense RNA probes were synthesized from linearized plasmids by in vitro transcription using digoxigenin-labeled dUTP (Epicenter Technologies). The cDNAs used to generate probes for Fgf8, Shh, En-1, and Twist were provided by R. Johnson (University of Texas M. D. Anderson Cancer Center [UTMDACC]) or R. Behringer (UTMDACC). Whole embryos were fixed overnight in phosphate-buffered saline (PBS) containing 4% PFA at 4°C and dehydrated in a graded methanol series prior to storage at −20°C. Whole-mount in situ hybridization was performed as described previously (38). Briefly, embryos were rehydrated in a graded methanol series and treated with 10 μg/ml proteinase K (QIAGEN). The embryos were refixed with 0.2% glutaraldehyde-4% PFA and washed in PBS containing 0.1% Tween 20 prior to hybridization overnight at 70°C. Embryos were washed three times in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% sodium dodecyl sulfate (pH 4.5) at 70°C, followed by three washes in 2× SSC at 65°C. Staining was visualized using a nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) (Roche) or BM purple (Roche) substrate.
Preparation of mouse embryos for scanning electron microscopy.
Freshly dissected embryos were treated with a fixative containing 3% glutaraldehyde plus 2% PFA in 0.1 M cacodylate buffer, pH 7.3, for at least 1 h at ambient temperature. Embryos were then washed with 0.1 M cacodylate buffer (pH 7.3) three times for 5 min each time. The embryos were then fixed with 1% cacodylate-buffered osmium tetroxide for 1 h and washed with distilled water three times for 5 min each time. The embryos were sequentially treated with Millipore-filtered aqueous 1% thiocarbohydrazide for 10 min, aqueous 1% osmium tetroxide for 10 min, 1% aqueous tannic acid for 30 min, and 1% aqueous uranyl acetate overnight in the dark. The embryos were rinsed thoroughly with distilled water after every treatment to prevent any carryover of reagents. The samples were then dehydrated with a graded series of increasing concentrations of ethanol for 5 min each. The embryos were then transferred to graded series of increasing concentrations of hexamethyldisilazane (HMDS) for 5 min each and air dried overnight. Samples were mounted on double-stick carbon tabs (Ted Pella, Inc., Redding, CA), which had been previously mounted onto aluminum specimen mounts (Electron Microscopy Sciences, Ft. Washington, PA). The samples were then coated under vacuum using a Balzer MED 010 evaporator (Technotrade International, Manchester, NH) with platinum alloy for a thickness of 25 nm and then immediately flash carbon coated under vacuum. The samples were transferred to a desiccator until examination in a JSM-5900 scanning electron microscope (JEOL, USA, Inc., Peabody, MA) at an accelerating voltage of 5 kV.
Isolation of primary MEFs.
Brains, limbs, and internal organs were removed from freshly dissected E13.5 embryos, and the remaining tissues were transferred to a six-well plate, rinsed in 4 ml of PBS, and then transferred to new wells containing 1 ml of 0.25% (wt/vol) trypsin. Embryos were minced with a sterile razor blade and homogenized by repeated pipetting. After 10 min of incubation at 37°C, the cells were pipetted again to further separate clumps, and then 2 ml of mouse embryo fibroblast (MEF) medium (Dulbecco modified Eagle medium with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.1 mM β-mercaptoethanol) was immediately added to the well, and the cell suspension was divided among three 10-cm gelatinized culture plates. The medium was changed the next day, and the cells were grown to confluence at 37°C (passage 0) prior to splitting the cells 1:3 into fresh medium. After passage 6, cells were split 1:2. For the growth curve analyses shown below (see Fig. 7B), third passage MEFs were seeded at 1 × 105 cells per well of a six-well plate on day 0. The cells in duplicate plates were trypsinized, washed in PBS, and then stained in 0.4% trypan blue at the indicated time points. Trypan blue-negative cells were counted using a hemacytometer.
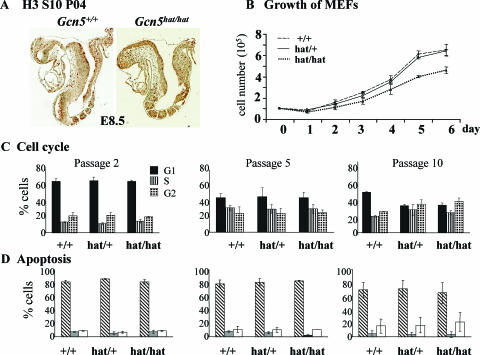
FIG. 7.
Proliferation of _Gcn5_hat/hat cells. (A) Immunohistochemistry to detect phosphorylated H3 serine 10, which marks mitotic cells, to monitor cell proliferation in E8.5 Gcn5+/+ and _Gcn5_hat/hat embryos. No obvious differences in the numbers of mitotic cells were observed. (B) Equal numbers (1 × 105) of MEFs isolated from E13.5 Gcn5+/+, _Gcn5_hat/+, or _Gcn5_hat/hat embryos (as indicated) were plated after the third passage (day 0) and counted daily for 6 days (days 1 to 6) in duplicate to monitor the rate of cell division. Error bars indicate standard deviations from the means. (C) Flow cytometry profiles of propidium iodide-stained MEFs of the indicated genotypes at the indicated passage numbers. G1 cells, S-phase cells, and G2 cells are shown. (D) Flow cytometry profiles of cells stained with propidium iodide (PI) and annexin V antisera to detect live cells (annexin V and PI negative [hatched bars]) and cells in early (annexin V positive, PI negative [gray bars]) or late (annexin V and PI positive [white bars]) stages of apoptosis. The passage numbers are as indicated in panel C.
PI staining of MEFs.
MEFs (2 × 106 cells) were resuspended in 2 ml of PBS and vortexed gently prior to the addition of 5 ml of 95% ethanol. Cells were allowed to fix in the ethanol solution at room temperature for 30 min and then stored at 4°C. Just prior to flow cytometry, cells were collected by centrifugation at 3,000 × g for 5 min and stained with propidium iodide (PI) (50 μg/ml in PBS) (Sigma). RNase (100 μl of 1 mg/ml stock of DNase-Free RNase; Sigma) was then added, and cells were incubated for 30 min at 37°C. Flow cytometry was performed at the UTMDACC core facility.
Annexin V-FITC staining of MEFs.
MEFs were trypsinized, washed twice with cold PBS, and counted using a hemacytometer. Cells (1 × 106) were collected and then resuspended in binding buffer (annexin V-FITC apoptosis detection kit 1; BD Biosciences). Cells were vortexed gently, and 100 μl of the solution (1 × 105 cells) was transferred to a 5-ml tube, followed by addition of 5 μl of annexin V- fluorescein isothiocyanate (FITC) and 5 μl of PI. The cells were gently vortexed again and incubated for 15 min at room temperature in the dark. Binding buffer (400 μl) was added to each tube, and the stained cells were subjected to flow cytometry.
Preparation of MEF lysates and immunoblots.
Whole-cell lysates were prepared by lysing MEFs directly in Laemmli gel loading buffer with protease inhibitors, followed by sonication on ice three times for 3 seconds each time and centrifugation at 12,000 × g for 30 min at 4°C in a microcentrifuge to remove cellular debris. Nuclear extracts were prepared by lysing MEFs in buffer A (10 mM Tris, 1.5 mM MgCl2, 10 mM HCl. 0.5 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride). The lysate was held on ice for 10 min and homogenized 10 times. The nuclear pellet was collected by centrifugation as described above and then resuspended in buffer C (20 mM Tris, 0.42% NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride). After 30 min of rotation at 4°C, the extract was cleared by centrifugation as described above, and the supernatant (the nuclear extract) was removed and placed in a fresh tube. Proteins in whole-cell lysates or nuclear extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Histone H3 was detected using antisera specific for H3 acetylated at lysine 9 (K9) and lysine 18 (K18) (1:1,000 dilution [12]), acetylated at K14 (1:1,000; Upstate) or the C terminus of H3 (1:2,000 dilution; Abcam). Histone H4 was detected using an antisera specific for K12 acetylated isoforms (1:1,000; Upstate) or total H4 (1:2,000; Upstate). The anti-Gcn5 antiserum used in these experiments was from BioLegend (1:500 dilution), and the anti-β-actin antiserum was from Sigma (1:4,000 dilution).
RESULTS
Gcn5 mutants survive longer in the absence of p53.
Our previous studies revealed that _Gcn5_−/− mouse embryos are malformed by E8.5 and exhibit increased apoptosis relative to wild-type or heterozygous littermates as early as E7.5 (38). Other studies in our lab demonstrated that the increased apoptosis is p53 dependent, as it is not observed in _Gcn5_−/− _p53_−/− double mutants (P. Bu et al., unpublished data). However, deletion of p53 does not rescue the embryonic lethality caused by Gcn5 loss. No live _Gcn5_−/− _p53_−/− pups were observed among 121 12-day-old (P12) progeny obtained from intercrosses of Gcn5+/− _p53_−/− mice (Table 1). In contrast, both Gcn5+/+ _p53_−/− and Gcn5+/− _p53_−/− pups were observed at close to expected ratios (Table 1). Examination of embryos from Gcn5+/− _p53_−/− intercrosses revealed that the double null embryos were present at expected Mendelian ratios at E8.5 to E10.5 but were underrepresented by E12.5 (Table 1). Increased numbers of empty deciduas were observed from E12.5 onwards, consistent with the death of the _Gcn5_−/− _p53_−/− double mutants at these later time points.
TABLE 1.
Progeny of _p53_−/− Gcn5 +/− intercrosses
| Time point and parameter | No. of progeny with the following genotype: | Total no. of progeny | ||
|---|---|---|---|---|
| Gcn5+/+_p53_−/− | Gcn5+/−_p5_−/− | _Gcn5_−/−_p53_−/− | ||
| P12 | ||||
| Observed no. | 44 | 77 | 0a | 121 |
| Expected no. | 31.5 | 63 | 31.5 | |
| E8.5 | ||||
| Observed no. | 25 | 44 | 16 | 85 |
| Expected no. | 21.25 | 42.5 | 21.25 | |
| E9.5 | ||||
| Observed no. | 4 | 3 | 5 | 12 |
| Expected no. | 3 | 6 | 3 | |
| E10.5 | ||||
| Observed no. | 4 | 9 | 4 | 17 |
| Expected no. | 4.25 | 8.5 | 4.25 | |
| E12.5b | ||||
| Observed no. | 8 | 16 | 0c | 24 |
| Expected no. | 6 | 12 | 6 | |
| E13.5b | ||||
| Observed no. | 4 | 14 | 0d | 18 |
| Expected no. | 4.5 | 9 | 4.5 |
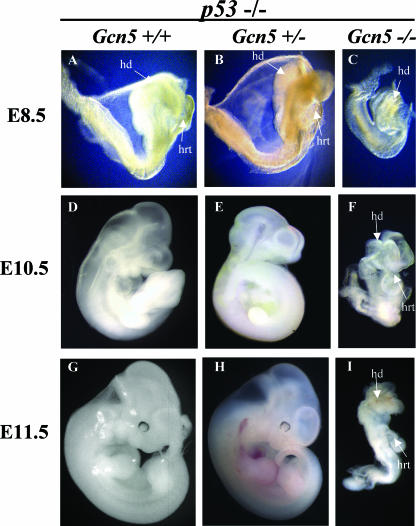
The _Gcn5_−/− _p53_−/− double mutant embryos were significantly smaller than their littermates at all times examined (Fig. 1). Unlike _Gcn5_−/− single mutants (38), _Gcn5_−/− _p53_−/− embryos had a distinct anterior-posterior axis, head folds, and somites (Fig. 1C, F, and I). However, the double mutants had fewer somites (1 to 5 somites) at E8.5 than their littermates did (7 to 13 somites). At later times, the double null embryos remained smaller than their littermates, were slow to turn, and appeared delayed in development (Fig. 1F and I). In contrast, Gcn5+/− _p53_−/− embryos were indistinguishable from their Gcn5+/+ _p53_−/− littermates (compare Fig. 1A, D, and G to Fig. 1B, E, and H).
FIG. 1.
Morphology of _Gcn5_−/− _p53_−/− embryos. Lateral views of E8.5 to E11.5 embryos indicate that Gcn5+/+ _p53_−/− and Gcn5+/− _p53_−/− embryos exhibit a wild-type phenotype, whereas _Gcn5_−/− _p53_−/− double null embryos show delayed development and exhibit abnormal morphologies at E10.5 and E11.5. hd, head; hrt, heart.
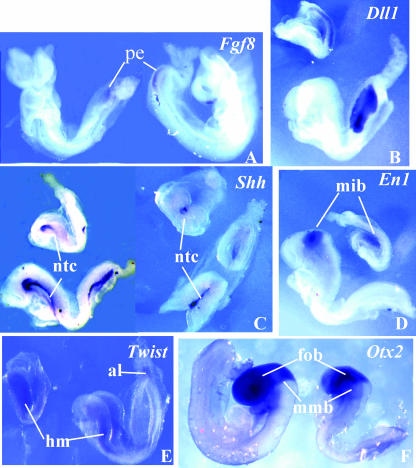
To further determine the extent of development achieved by the double mutants, we analyzed the expression of several marker genes at E8.5 to E9.0 (Fig. 2). The timing and extent of Fgf8 (fibroblast growth factor 8; Fig. 2A), Dll1 (delta1 like 1; Fig. 2B), and Tcf15 (paraxis; data not shown) expression in the E8.5 _Gcn5_−/− _p53_−/− embryos were unchanged relative to Gcn5 wild-type (Gcn5+/+ _p53_−/−) or heterozygous (Gcn5+/− _p53_−/−) littermates, consistent with the normal timing and spatial development of a primitive streak followed by formation of presomitic mesoderm and somites. Shh (sonic hedge hog) expression was observed along the anterior midline and floor plate of both Gcn5+/− _p53_−/− and _Gcn5_−/− _p53_−/− embryos (Fig. 2C), indicating normal initiation of notochord formation in the double mutants. Expression of _En_-1 (engrailed 1) and Twist also occurred with similar timing and placement in the double null embryos as in control littermates (Fig. 2D and E). Similarly, normal expression of Otx2 was observed from the anterior tip of the neural plate to the mesencephalon-metencephalon border of E9.0 _Gcn5_−/− _p53_−/− and Gcn5+/+ _p53_−/− or Gcn5+/− _p53_−/− embryos (Fig. 2F).
FIG. 2.
Normal expression of developmental markers in Gcn5 −/− p53 −/− embryos. Whole-mount in situ analyses of E8.5 to E9.0 embryos are shown for each of the indicated genes. The smaller embryo in each panel is _Gcn5_−/− _p53_−/−, and the larger embryo is a Gcn5+/+ _p53_−/− or Gcn5+/− _p53_−/− littermate. In each case, the timing and spatial distribution of the indicated marker gene were unchanged in the double null embryos. al, allantois; fob, forebrain; hm, head mesenchyme; mib, midbrain; mmb, mesencephalon-metencephalon border; ntc, notochord; pe, posterior end.
Altogether, these data indicate that elimination of p53 allows _Gcn5_−/− embryos to survive longer and to initiate somite and notochord formation. Our marker analyses indicate that anterior and midbrain development also initiates in the _Gcn5_−/− _p53_−/− embryos along the same time frame as in Gcn5+/+ _p53_−/− or Gcn5+/− _p53_−/− embryos. Gcn5, then, is not needed for these processes but is required for the normal patterning and survival of mouse embryos.
Gcn5 HAT mutations cause embryonic lethality.
To determine whether Gcn5 acetyltransferase activity is required for its functions during mouse development, we created mice bearing mutations in the Gcn5 catalytic center. We generated a gene replacement vector for Gcn5 in which the critical catalytic glutamate (E568; equivalent to E173 in yeast Gcn5) (22) as well as a conserved aspartate (D608) were converted to alanine (Fig. 3A). ES cell clones bearing the mutant allele integrated into the Gcn5 locus were identified by Southern blotting and PCR genotyping and were then used to generate mice heterozygous for the Gcn5 HAT mutation (_Gcn5_hat/+) (Fig. 3B and C).
_Gcn5_hat/+ mice appeared normal and were fertile. However, no live homozygous _Gcn5_hat/hat mice were identified among 237 progeny of _Gcn5_hat/+ intercrosses (Table 2), suggesting that loss of Gcn5 HAT activity causes embryonic lethality. Indeed, _Gcn5_hat/hat homozygotes were observed at expected frequencies at E8.5 to E11.5 but were found at reduced frequency at most later times of gestation (Table 2).
TABLE 2.
Progeny of _Gcn5_hat/+ intercrosses
| Time point and parameter | No. of progeny with the following genotype: | Total no. of progeny | ||
|---|---|---|---|---|
| Gcn5+/+ | _Gcn5_hat/+ | _Gcn5_hat/hat | ||
| P12 | ||||
| Observed no. | 88 | 149 | 0a | 237 |
| Expected no. | 59.25 | 118.5 | 59.25 | |
| E8.5b | ||||
| Observed no. | 41 | 77 | 44 (15)c | 162 |
| Expected no. | 40.5 | 81 | 40.5 | |
| E9.5 | ||||
| Observed no. | 7 | 24 | 15 | 46 |
| Expected no. | 11.5 | 23 | 11.5 | |
| E10.5b | ||||
| Observed no. | 4 | 6 | 5 | 15 |
| Expected no. | 3.75 | 7.5 | 3.75 | |
| E11.5b | ||||
| Observed no. | 7 | 10 | 5 | 22 |
| Expected no. | 5.5 | 11 | 5.5 | |
| E12.5b | ||||
| Observed no. | 15 | 32 | 7 | 54 |
| Expected no. | 13.5 | 27 | 13.5 | |
| E13.5b | ||||
| Observed no. | 11 | 26 | 5 | 42 |
| Expected no. | 10.5 | 21 | 10.5 | |
| E14.5b | ||||
| Observed no. | 6 | 17 | 4 | 27 |
| Expected no. | 6.75 | 13.5 | 6.75 | |
| E16.5b | ||||
| Observed no. | 6 | 8 | 2 | 16 |
| Expected no. | 4 | 8 | 4 | |
| E18.5b | 2 | 5 | 0 | 7 |
| Observed no. | ||||
| Expected no. |
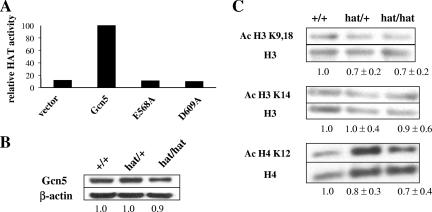
The negative effects of the E568A and D608A mutations on Gcn5 catalytic activity were confirmed in vitro by HAT assays using mutant and wild-type recombinant Gcn5 with isolated histones as the substrate (Fig. 4A). Expression of the mutant Gcn5 protein in primary fibroblasts isolated from embryos (MEFs) was not significantly affected by the HAT mutations (Fig. 4B), so the lethality of the _Gcn5_hat/hat is not likely due to alterations in Gcn5 expression levels. Global levels of H3 acetylation at K9/K18 were slightly reduced in both _Gcn5_hat/+ and _Gcn5_hat/hat MEFs, but no reduction was seen in the levels of H3 K14 acetylation (Fig. 4C). Levels of H4 K12 acetylation were also slightly reduced, consistent with reports that Gcn5 complexes in Drosophila acetylate this site (9). The overall persistence of H3 and H4 acetylation in the _Gcn5_hat/hat MEFs likely reflects the activities of several other HATs in these cells.
FIG. 4.
Acetylation of histones H3 and H4 in MEFs. (A) HAT assays to measure acetyltransferase activity of equal amounts of wild-type or mutant versions of Gcn5 expressed in bacteria. The activity of wild-type Gcn5 was set at 100%. (B) Immunoblots to compare expression of wild-type and mutant Gcn5 proteins in MEFs isolated from E13.5 embryos. Gcn5 levels were normalized to levels of β-actin, and Gcn5 expression in wild-type cells was set at 1.0. (C) Representative immunoblots comparing levels of H3 and H4 acetylation at the indicated sites in wild-type, _Gcn5_hat/+, and _Gcn5_hat/hat MEFs are shown. Numbers underneath the blots represent averages of signals from multiple blots. Acetylated H3 (Ac H3) or H4 levels were normalized to total H3 or H4, and levels in wild-type cells were set at 1.0.
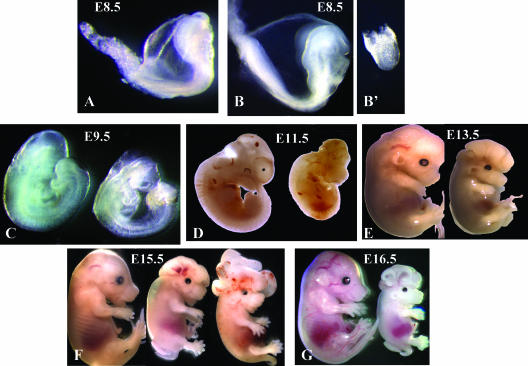
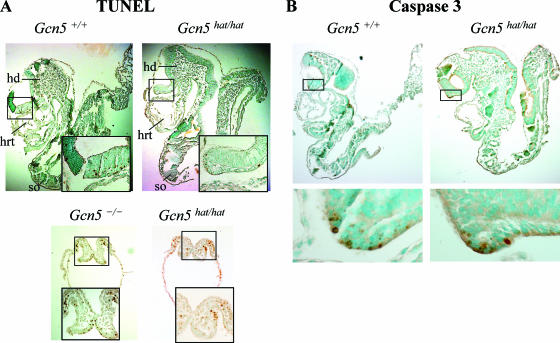
At E8.5, the _Gcn5_hat/hat embryos exhibited two distinct morphologies (Fig. 5B and B'; Fig. 6A). Some of these embryos (15 of 44 [34%]) were quite small and did not develop somites (Table 2 and Fig. 5B'), a phenotype very similar to that of embryos homozygous for a complete deletion of Gcn5 (38). Other _Gcn5_hat/hat embryos (29 of 44 [66%]) were larger (Fig. 5B) and were more similar to their wild-type littermates (Fig. 5A), developing somites and head folds, structures not observed in the Gcn5 deletion mutants. PCR genotyping and restriction digestions confirmed that embryos with both phenotypes contained both the E568A and D609A mutations (data not shown). The molecular basis for the differences in phenotype among the _Gcn5_hat/hat embryos at E8.5 is not known, but TUNEL analyses indicated that the smaller, less developed _Gcn5_hat/hat embryos had elevated levels of apoptotic cells at E8.5 (Fig. 6A, right bottom panel), similar to that we reported previously for Gcn5 null embryos (Fig. 6A, left bottom panel) (38). No such increase in apoptosis (relative to _Gcn5_hat/+ or wild-type littermates) was observed in the larger, more developed _Gcn5_hat/hat embryos as assayed by TUNEL analyses (Fig. 6A, top panel) or immunostaining for activated caspase 3 (Fig. 6B), indicating that the loss of Gcn5 HAT activity is not sufficient to induce this phenotype.
FIG. 5.
_Gcn5_hat/hat embryos exhibit defects in neural tube closure and exencephaly. Lateral views of embryos at the indicated time points. (A) Wild-type embryo; (B and B') _Gcn5_hat/hat embryos. In all other panels, the embryo on the left is a wild-type or _Gcn5_hat/+ embryo, and the embryo on the right (and in the middle in panel F) is a _Gcn5_hat/hat embryo.
FIG. 6.
Apoptosis in _Gcn5_hat/hat embryos. (A) TUNEL analyses of lateral sections of E8.5 Gcn5+/+, _Gcn5_hat/hat, and _Gcn5_−/− embryos, as indicated. Boxed regions are shown at higher magnification in the insets. Both larger (top right panel) and smaller (bottom right panel) _Gcn5_hat/hat embryos are shown. Few TUNEL-positive cells are observed in the wild-type or larger _Gcn5_hat/hat embryos, whereas increased numbers of apoptotic cells are observed in the _Gcn5_−/− (bottom left panel) and smaller _Gcn5_hat/hat embryos. hd, head fold; hrt, heart; so, somite. (B) Immunohistochemistry to detect activated caspase 3 in sections of Gcn5+/+ or larger _Gcn5_hat/hat E8.5 embryos. Boxed regions are shown at higher magnifications in the bottom panels to show the few positive cells observed. As described above, only low levels of apoptotic cells are observed in either type of embryo.
Similar levels of immunostaining for H3 serine 10 phosphorylation, which marks mitotic cells, were also observed in both wild-type and _Gcn5_hat/hat embryos (Fig. 7A), suggesting cell proliferation was not grossly affected by loss of Gcn5 catalytic activity. To more accurately gauge the effects of the Gcn5 mutation on cell proliferation, we analyzed growth of MEFs isolated from E13.5 embryos. Proliferation of third passage MEFs isolated from _Gcn5_hat/hat embryos was initially similar to that of third passage wild-type or _Gcn5_hat/+ MEFs (Fig. 7B), but doubling of the _Gcn5_hat/hat mutant cells was somewhat slower 4 to 6 days after plating (Fig. 7B, days 4 to 6). Flow cytometry did not reveal any gross abnormalities in the cell cycle of _Gcn5_hat/hat MEFs. Similar distributions of G1, S, and G2 cells were detected in MEFs of all three genotypes from passages 2 to 10 (Fig. 7C). No change in apoptosis, as detected by annexin V immunostaining, was observed in the mutant MEFs at any time (Fig. 7D), consistent with normal (i.e., not increased) levels of apoptosis in the more developed _Gcn5_hat/hat embryos from which the MEFs were isolated (Fig. 6A and B). These results indicate that Gcn5 catalytic activity is not required for overall cell survival or cell cycle progression. The reason for the slower growth of _Gcn5_hat/hat MEFs is unclear.
Neural tube defects (NTDs) and exencephaly in _Gcn5_hat/hat embryos.
_Gcn5_hat/hat embryos exhibited defects in neural tube closure early in development and exencephaly at later times (Fig. 5C to G). Neural tube closure normally begins at three sites at E8.5. The neural folds lift and fuse first at the hindbrain/cervical boundary (site 1), followed by fusions at the midbrain/forebrain boundary (site 2), and finally, fusion at the rostral end of the neural tube (site 3) (10). Fusion then continues from these sites, leading to complete neural tube closure by E9.5. The neural folds of E9.5 _Gcn5_hat/hat embryos remained open, despite the extensive posterior development of these embryos (Fig. 5C). Exencephaly was seen in _Gcn5_hat/hat embryos from E11.5 onwards (Fig. 5D to G) and was quite dramatic in some embryos (e.g., the rightmost embryo in Fig. 5F).
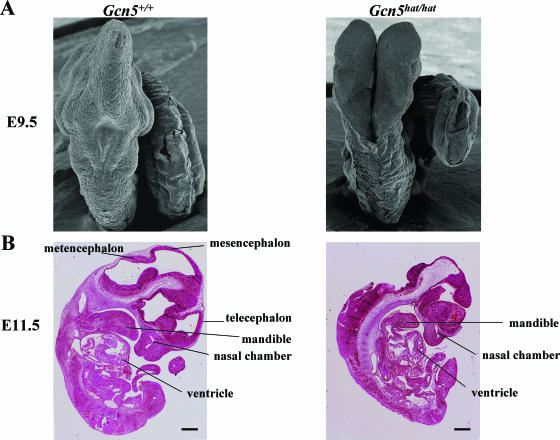
To further define the nature of the NTDs in _Gcn5_hat/hat embryos, we performed scanning electron microscopy on E9.5 embryos. As expected, neural tube closure was completed in wild-type littermates at this time point (Fig. 8A, left panel), but the neural tube remained open from the hindbrain-cervical boundary (Fig. 8A, right panel) rostrally towards the forebrain in _Gcn5_hat/hat embryos (Fig. 5C to G; also data not shown). Normal neural tube fusion was achieved along the spinal region of the _Gcn5_hat/hat embryos.
FIG. 8.
Neural tube closure defects are limited to the cranial region of _Gcn5_hat/hat embryos. (A) Scanning electron micrographs showing dorsal views of E9.5 wild-type or _Gcn5_hat/hat embryos. The neural tube is completely closed in the wild-type embryo, but it is open from the hindbrain-cervical boundary forward towards the forebrain in the _Gcn5_hat/hat embryo. (B) Hematoxylin-and-eosin-stained lateral sections of wild-type and _Gcn5_hat/hat embryos at E11.5 reveal severe morphological changes in the regions of the metencephalon, mesencephalon, and telecephalon in the mutant embryo. Bar, 430 μm.
Comparison of hematoxylin-and-eosin-stained lateral sections of wild-type and _Gcn5_hat/hat E11.5 embryos revealed that the lack of the cranial vault in the mutants was associated with severe morphological changes in several areas of the brain, including the metencephalon, mesencephalon, and telecephalon (Fig. 8B). These analyses also confirmed that development of posterior structures was not grossly affected in the mutant embryos. These data further establish the severity of the NTDs in the _Gcn5_hat/hat embryos and indicate these defects are limited to the cranial region.
Expression of mesodermal and neural markers in _Gcn5_hat/hat embryos.
The cranial NTDs in the _Gcn5_hat/hat embryos may reflect a failure of the neural folds to elevate in the midbrain region, thereby preventing fusion (10). Alternatively, cranial NTDs might arise from defective development of head mesenchyme. Others have shown that decreased or increased numbers of head mesenchyme cells around the cranial neural tube lead to neural tube defects (8, 44, 45). In addition, sonic hedge hog (Shh) signaling is required for proper bending of the neural folds to form the neural tube (10, 11). To determine whether any of these steps is affected by loss of Gcn5 HAT activity, we compared the expression of key marker genes for these processes in _Gcn5_hat/hat embryos to that in their wild-type or heterozygous littermates.
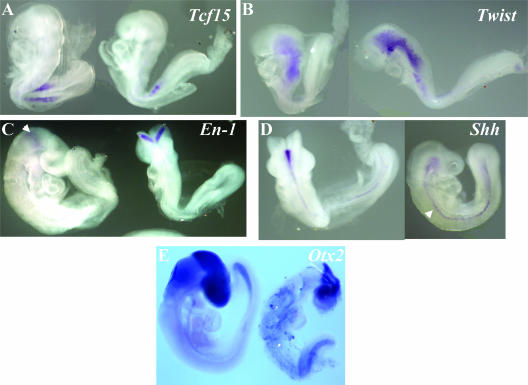
Whole-mount in situ hybridizations indicate that expression of Tcf15 occurs at the right time and place in _Gcn5_hat/hat embryos (Fig. 9A). Together with our data above demonstrating normal expression of Fgf8 and Dll in _Gcn5_−/− _p53_−/− embryos (Fig. 2A and B), these findings indicate that neither Gcn5 expression nor HAT activity is required for presomitic mesoderm formation. Levels and location of Twist expression also appeared normal in E9.0 _Gcn5_hat/hat embryos (Fig. 9B), indicating that head mesenchyme is formed properly in the absence of Gcn5 HAT activity. _En_-1 expression was evident in the midbrain region of _Gcn5_hat/hat embryos around E9.0 (Fig. 9C). Expression of Shh occurred along the midline of the mutant animals, as is observed in wild-type littermates (Fig. 9D). Otx2 was also expressed normally in the anterior and midbrain regions of E9.5 _Gcn5_hat/hat embryos (Fig. 9E). These data, then, indicate that the NTDs observed in the _Gcn5_hat/hat embryos are not due to a failure in development of head mesenchyme or Shh expression. These data also indicate that Gcn5 HAT activity is not required for anterior, midbrain, or hindbrain specification. The NTDs in the _Gcn5_hat/hat embryos must reflect deficiency in another process required for neural fold elevation and neural tube fusion.
FIG. 9.
Normal expression of developmental markers in _Gcn5_hat/hat embryos. Whole-mount in situ hybridizations were performed on E8.5 to E9.0 embryos using probes for the indicated genes. In each panel, the embryo on the right is _Gcn5_hat/hat, and the embryo on the left is a wild-type or heterozygous littermate. The arrow in panel C points to _En_-1 expression in the midbrain. The arrow in panel D points to Shh expression in the midline of the embryo.
DISCUSSION
Our previous work revealed that paraxial mesoderm and chordamesoderm are specified but do not survive in _Gcn5_−/− embryos due to increased apoptosis relative to wild-type or heterozygous littermates (38). We show here that elimination of the p53 tumor suppressor allows _Gcn5_−/− embryos to survive longer and to develop somites. These results confirm our earlier conclusion that Gcn5 is not required for the specification of presomitic mesoderm but is required for the survival of this tissue lineage (in the presence of p53). We show here that loss of Gcn5 HAT activity is not sufficient to induce early embryo lethality but causes cranial NTDs, which culminate in exencephaly in mid- to late gestation. These findings demonstrate that Gcn5 HAT activity is required for proper neural tube closure in the mouse.
The phenotype of our _Gcn5_−/− _p53_−/− embryos was somewhat reminiscent of that reported by Yamauchi et al. for mouse mutants lacking Gcn5 C-terminal sequences, which encode the HAT domain and bromodomain (40). Mice homozygous for that Gcn5 allele were smaller than their wild-type or heterozygous littermates and appeared delayed in development, just as we see in the _Gcn5_−/− _p53_−/− double null mutants.
The fact that those mutant embryos and our original Gcn5 null embryos die much earlier than our _Gcn5_hat/hat embryos indicates that Gcn5 has important developmental functions that are independent of its acetyltransferase activity. Otherwise, the catalytic site mutations would render a phenotype just as severe as that of Gcn5 deletion. Gcn5-containing complexes in mice likely contain multiple functional modules, as they do in yeast (2, 19, 37). Nonenzymatic proteins likely contribute essential functions to the complexes as well. In flies, for example, two Ada2 variants are assembled into separate Gcn5-containing complexes that have distinct roles during development (20). The complete absence of the Gcn5 protein in our _Gcn5_−/− mice (38) and the presence of (at most) a truncated protein in mice expressing the Gcn5 allele of Yamauchi et al. (40) might affect the integrity of all Gcn5-containing complexes in these mutant embryos. In contrast, complex integrity is predicted to be intact in our _Gcn5_hat/hat mice on the basis of the precedent of catalytically inactive Gcn5 proteins in yeast (6, 35). We have not yet tested this prediction, as we do not have immortalized cell lines expressing this allele, and biochemical purification would be difficult, if not impossible, from the limited amounts of embryonic material or primary MEFs that we could obtain. The fact that the _Gcn5_hat/hat embryos survive until E13.5 to E16.5, though, supports the idea that this allele preserves important Gcn5 complex functions. Interestingly, overexpression of catalytically inactive forms of Gcn5 in yeast caused dominant-negative effects, likely due to competition of the mutant and wild-type forms of the proteins for assembly into SAGA or other Gcn5-containing complexes. We have not observed any dominant-negative effects of the mutant allele in our _Gcn5_hat/+ mice, which likely reflects that normal expression (i.e., not overexpression) of the mutant allele in these mutant mice.
Gcn5 is expressed ubiquitously in early mouse embryos, with the exception of the developing heart (38). It is somewhat surprising, then, that developmental defects in the _Gcn5_hat/hat mutants appear to be limited to neural tissues. The normal development of other tissues may reflect Gcn5 redundancy with PCAF or overlapping functions with p300. PCAF null mice do not exhibit any abnormal phenotypes, so PCAF functions are not normally essential during development (38, 40). However, _Gcn5_−/− _PCAF_−/− double mutant embryos die much earlier than _Gcn5_−/− single mutant embryos do, indicating PCAF shares some redundant functions with Gcn5 during development (38). Similarly, the phenotype of mice heterozygous for null alleles of both Gcn5 and p300 exhibit decreased viability relative to mice heterozygous for either single mutation, indicating that Gcn5 and p300 share some functions during embryogenesis (31). Mouse embryos with only one functional p300 allele exhibit an increased frequency of NTDs, indicating this HAT is also important for neural tube closure (42). Interestingly, mutations in the acetyltransferase (AT) domain of p300 are dominant, unlike our Gcn5 AT domain mutations. The presence of a single p300 AT mutant allele results in defective development of the lungs, cardiovascular system, and small intestine in mice (33). None of these organs are affected by loss of Gcn5 AT activity. Although these HATs physically interact and share some functions during development, they clearly have a number of distinct functions as well.
Neural tube closure is a complex and multistep process that involves lifting, bending, and fusion of the neural folds (10). Alterations in tissue proliferation or in cell survival affect this process, but our data indicate that _Gcn5_hat/hat fibroblasts divide somewhat slowly in vitro and are not subject to increased apoptosis. It seems likely, then, that Gcn5 affects neural tube closure through effects on cell growth and/or gene transcription. Gcn5 functions as a transcriptional coactivator, and although we found that Gcn5 HAT activity is not required for expression of Twist, _En_-1, Shh, or Otx2, it may regulate expression of one or more of the ∼100 other genes required for proper neural tube closure (10, 45). This possibility will be addressed in future studies.
Interestingly, another subunit of Gcn5-containing complexes, Ataxin-7, is also implicated in neural functions (17, 18, 25). The Sca7 gene that encodes Ataxin-7 is subject to polyglutamine (poly-Q) expansions in humans, giving rise to spinocerebellar ataxia (15). Mouse models of poly-Q expansions of Sca7 exhibit many phenotypes similar to those found in the human disease, including ataxia and retinal defects (15, 43). Current models suggest that the mutant proteins have adverse effects on transcriptional regulation of downstream target genes. Recent work indicates that such defects are associated with increased histone acetylation (17), but other studies indicate that poly-Q expanded Ataxin-7 proteins inhibit Gcn5 functions (25, 30). Our current findings support the importance of Gcn5 acetyltransferase activity in normal neural development. The mutant Gcn5 alleles that we have generated will allow us to determine directly whether decreased Gcn5 protein levels or catalytic activity modulates the Sca7 poly-Q mutant phenotype in mice.
Acknowledgments
We thank R. Behringer and members of the Dent lab for helpful discussions. We thank John Latham for reading of the manuscript. We thank Kenneth Dunner for help with scanning electron microscopy of embryos.
Y.A.E. was supported by a Sowell-Huggins Fellowship. This work was supported by a grant from the NIH (GM067718) to S.Y.R.D. DNA sequencing, flow cytometry, and scanning electron microscopy were performed at UTMDACC core facilities supported by the UTMDACC Cancer Center Support Grant (CA16672) from the NCI.
Footnotes
▿
Published ahead of print on 26 February 2007.
REFERENCES
- 1.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8**:**1243-1254. [DOI] [PubMed] [Google Scholar]
- 2.Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286**:**2151-2153. [DOI] [PubMed] [Google Scholar]
- 3.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20**:**3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274**:**18285-18289. [DOI] [PubMed] [Google Scholar]
- 5.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84**:**843-851. [DOI] [PubMed] [Google Scholar]
- 6.Candau, R., J. Zhou, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16**:**555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavusoglu, N., M. Brand, L. Tora, and A. Van Dorsselaer. 2003. Novel subunits of the TATA binding protein free TAFII-containing transcription complex identified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry following one-dimensional gel electrophoresis. Proteomics 3**:**217-223. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z. F., and R. R. Behringer. 1995. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9**:**686-699. [DOI] [PubMed] [Google Scholar]
- 9.Ciurciu, A., O. Komonyi, T. Pankotai, and I. M. Boros. 2006. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol. Cell. Biol. 26**:**9413-9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copp, A. J., N. D. Greene, and J. N. Murdoch. 2003. The genetic basis of mammalian neurulation. Nat. Rev. Genet. 4**:**784-793. [DOI] [PubMed] [Google Scholar]
- 11.Echelard, Y., D. J. Epstein, B. St-Jacques, L. Shen, J. Mohler, J. A. McMahon, and A. P. McMahon. 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75**:**1417-1430. [DOI] [PubMed] [Google Scholar]
- 12.Edmondson, D. G., J. K. Davie, J. Zhou, B. Mirnikjoo, K. Tatchell, and S. Y. Dent. 2002. Site-specific loss of acetylation upon phosphorylation of histone H3. J. Biol. Chem. 277**:**29496-29502. [DOI] [PubMed] [Google Scholar]
- 13.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11**:**1640-1650. [DOI] [PubMed] [Google Scholar]
- 14.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates III, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2**:**863-867. [DOI] [PubMed] [Google Scholar]
- 15.Grote, S. K., and A. R. La Spada. 2003. Insights into the molecular basis of polyglutamine neurodegeneration from studies of a spinocerebellar ataxia type 7 mouse model. Cytogenet. Genome Res. 100**:**164-174. [DOI] [PubMed] [Google Scholar]
- 16.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90**:**595-606. [DOI] [PubMed] [Google Scholar]
- 17.Helmlinger, D., S. Hardy, G. Abou-Sleymane, A. Eberlin, A. B. Bowman, A. Gansmuller, S. Picaud, H. Y. Zoghbi, Y. Trottier, L. Tora, and D. Devys. 2006. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 4**:**e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmlinger, D., S. Hardy, S. Sasorith, F. Klein, F. Robert, C. Weber, L. Miguet, N. Potier, A. Van-Dorsselaer, J. M. Wurtz, J. L. Mandel, L. Tora, and D. Devys. 2004. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 13**:**1257-1265. [DOI] [PubMed] [Google Scholar]
- 19.Ingvarsdottir, K., N. J. Krogan, N. C. Emre, A. Wyce, N. J. Thompson, A. Emili, T. R. Hughes, J. F. Greenblatt, and S. L. Berger. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25**:**1162-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23**:**3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19**:**1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marmorstein, R., and S. Y. Roth. 2001. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 11**:**155-161. [DOI] [PubMed] [Google Scholar]
- 23.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273**:**23781-23785. (Erratum, **273:**27755.) [DOI] [PubMed] [Google Scholar]
- 24.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21**:**6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon, S. J., M. G. Pray-Grant, D. Schieltz, J. R. Yates IIII, and P. A. Grant. 2005. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc. Natl. Acad. Sci. USA 102**:**8478-8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizzen, C. A., J. E. Brownell, R. G. Cook, and C. D. Allis. 1999. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 304**:**675-696. [DOI] [PubMed] [Google Scholar]
- 27.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in _mdm2_-deficient mice by deletion of p53. Nature 378**:**203-206. [DOI] [PubMed] [Google Scholar]
- 28.Nagy, A., M. Gertsenstein, K. Vintersten, and R. R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94**:**35-44. [DOI] [PubMed] [Google Scholar]
- 30.Palhan, V. B., S. Chen, G. H. Peng, A. Tjernberg, A. M. Gamper, Y. Fan, B. T. Chait, A. R. La Spada, and R. G. Roeder. 2005. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl. Acad. Sci. USA 102**:**8472-8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan, H. M., A. W. Xu, C. Coco, G. Srajer, S. Wyszomierski, Y. A. Evrard, R. Eckner, and S. Y. Dent. 2005. GCN5 and p300 share essential functions during early embryogenesis. Dev. Dyn. 233**:**1337-1347. [DOI] [PubMed] [Google Scholar]
- 32.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70**:**81-120. [DOI] [PubMed] [Google Scholar]
- 33.Shikama, N., W. Lutz, R. Kretzschmar, N. Sauter, J. F. Roth, S. Marino, J. Wittwer, A. Scheidweiler, and R. Eckner. 2003. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 22**:**5175-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19**:**86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, L., C. Mizzen, C. Ying, R. Candau, N. Barlev, J. Brownell, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol. Cell. Biol. 17**:**519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6**:**601-604. [DOI] [PubMed] [Google Scholar]
- 37.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15**:**199-208. [DOI] [PubMed] [Google Scholar]
- 38.Xu, W., D. G. Edmondson, Y. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of GCN5 leads to increased apoptosis and mesodermal defects during mouse development. Nature Genet. 26**:**229-232. [DOI] [PubMed] [Google Scholar]
- 39.Xu, W., D. G. Edmondson, and S. Y. Roth. 1998. Mammalian GCN5 and P/CAF acetyltransferases share homologous amino-terminal domains important for the recognition of nucleosomal substrates. Mol. Cell. Biol. 18**:**5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of the histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97**:**11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, X.-J., V. V. Ogryzko, J.-I. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP associated factor that competes with the adenoviral oncoprotein E1A. Nature 382**:**319-324. [DOI] [PubMed] [Google Scholar]
- 42.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93**:**361-372. [DOI] [PubMed] [Google Scholar]
- 43.Yoo, S. Y., M. E. Pennesi, E. J. Weeber, B. Xu, R. Atkinson, S. Chen, D. L. Armstrong, S. M. Wu, J. D. Sweatt, and H. Y. Zoghbi. 2003. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron 37**:**383-401. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, Q., R. R. Behringer, and B. de Crombrugghe. 1996. Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat. Genet. 13**:**275-283. [DOI] [PubMed] [Google Scholar]
- 45.Zohn, I. E., K. V. Anderson, and L. Niswander. 2005. Using genomewide mutagenesis screens to identify the genes required for neural tube closure in the mouse. Birth Defects Res. Part A 73**:**583-590. [DOI] [PubMed] [Google Scholar]