Regulation of Notch1 Gene Expression by p53 in Epithelial Cells (original) (raw)
Abstract
The E6 protein of cervical cancer-associated human papillomaviruses (HPVs) is known to suppress keratinocyte differentiation through unidentified mechanisms. Notch1 is a determinant of keratinocyte differentiation and functions as a tumor suppressor in mammalian epidermis. Here, we report that the Notch1 gene is a novel target of p53 and can be down-regulated by E6 through p53 degradation in normal human epithelial cells. Thus, inactivation of p53 by E6 or short-hairpin RNA (shRNA) resulted in reduced Notch1 expression at the transcription level, and a p53-responsive element could be identified in the Notch1 promoter. The expression of E6, p53 shRNA, or Notch1 shRNA suppressed both spontaneous keratinocyte differentiation in culture and its induction upon DNA damage. Furthermore, the induction of Notch1 and differentiation makers as well as thickening of the epidermal layer upon UV irradiation was observed in wild-type but not in p53-deficient mouse skin. Together, our findings not only demonstrate a novel link between p53 and Notch1 in keratinocyte differentiation upon genotoxic stress but also suggest a novel tumor suppressor mechanism of p53 in the development of squamous cell carcinomas, including HPV-induced tumors.
A specific group of so-called high-risk human papillomaviruses (HPVs), such as HPV16 and HPV18, is associated with more than 90% of cervical cancers (60). Infection with these HPVs causes cervical dysplasia or low-grade cervical intraepithelial neoplasia (CIN), and cervical cancers are thought to arise from these lesions after long periods of time (32, 70). The E6 and E7 proteins of HPVs are expressed at relatively low levels in the basal cells of low-grade CIN lesions, where the viral genomes replicate episomally. When high-level expression of E6 and E7 occurs, in most cases with integration of viral genomes into the host genome, neoplastic development is believed to be initiated (59). In fact, E6 and E7 proteins are invariably expressed in HPV-positive cervical cancer cells and inactivate the major tumor suppressors p53 and Rb, respectively, thus contributing to HPV-induced oncogenesis. Sustained expression of E6 and E7 is also required for the maintenance of the transformed phenotype. E6 can inhibit the serum- and calcium-induced differentiation of keratinocytes (49). However, the underlying molecular mechanisms are not fully understood (48).
The Notch gene family encodes evolutionarily conserved cell surface receptors that play a crucial role in cell fate specification and differentiation (22, 29, 42). Upon cell-cell contact, Notch activation is triggered by interaction with its ligands, members of the Delta and Jagged families which are expressed on neighboring cell surfaces. Ligand binding is followed by proteolytic cleavage, release of the Notch intracellular domain (ICD) from the cellular membrane into the cytosol, and translocation of the ICD to the nucleus, where it converts CSL family members {CBF1/RBP-Jκ in mammals, Suppressor of hairless [Su(H)] in Drosophila melanogaster, and Lag1 in Caenorhabditis elegans} from transcriptional repressors into activators. This results in the induction of a number of genes involved in cell growth and differentiation. Among Notch family members, Notch1 has been reported as an oncogene in the development of human T-cell acute lymphoblastic leukemia, where a specific chromosomal translocation generates a constitutively active form of Notch1 that corresponds to the Notch1 ICD (8). In Ras-transformed cells, the activation of Notch1 signaling is reported to be necessary to maintain the neoplastic phenotype (63). Notch1 has also been identified as a key determinant of keratinocyte differentiation, promoting cell cycle arrest through p21 induction and commitment to differentiation (45). In addition, the keratinocyte-specific conditional disruption of Notch1 caused epidermal hyperplasia in mice (38), clearly demonstrating a tumor suppressor function for Notch1 in mammalian epidermis. The immunohistochemical detection of higher levels of Notch1 expression in neoplastic cervical lesions than in normal cervical epithelium suggested a role in carcinogenesis (69), and the activation of Notch1 signaling in cooperation with HPV E6 and E7 was shown to be involved in cellular transformation through a PI3K-Akt-dependent pathway in a spontaneously immortalized keratinocyte cell line, HaCaT (33, 44, 58). In sharp contrast, specific down-modulation of Notch1 was found to be required for sustained HPV E6/E7 expression and malignant conversion in late stages of cervical carcinogenesis (55). However, little is known about the molecular basis for the control of Notch1 expression in the cervix.
Here we present evidence for the positive regulation of Notch1 gene expression by the p53 tumor suppressor in normal human epithelial cells, including keratinocytes, and its down-regulation by E6 through the inactivation of p53. Our results point to a novel molecular mechanism of E6-mediated oncogenesis in the cervix, with implications for p53 mutations in the development of squamous cell carcinomas.
MATERIALS AND METHODS
Cell culture.
The cervical cancer cell lines HeLa, CaSki, SiHa, and C33A, the HaCaT spontaneously immortalized keratinocyte cell line, and the Saos2 osteosarcoma cell line were maintained in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum. Normal human cervical keratinocytes (HCK1s) were obtained with written consent from a patient who underwent abdominal surgery for a gynecological disease other than cervical cancer. Primary human dermal keratinocytes (HDKs) were purchased from Cell Applications Inc. (San Diego, CA). HCK1s were infected with retroviruses transducing the catalytic subunit of human telomerase reverse transcriptase (hTERT) for immortalization. The resultant HCK1 T cells as well as primary HDKs were cultured in serum-free keratinocyte serum-free medium (Invitrogen) supplemented with 5 ng/ml epidermal growth factor (Sigma) and 50 μg/ml of bovine pituitary extract (Hammond CELL TECH). HCK1 T cells in serial cultivation have been subjected to karyotypic analysis, and the cells have maintained normal diploid chromosomes except for trisomy of chromosome 20 at population doublings 10 and 40. In this report, HCK1 T cells after around 30 population doublings were used.
Retroviral vector construction and transduction.
Segments of HPV16 E6E7 (16E6E7), a splice donor site mutant version of E6 (E6SD) (18), a series of E6 mutants, dominant negative forms of p53 (p53C234), ΔNp63α, and hTERT were cloned and recombined into retroviral expression vectors to generate pCLXSN-16E6E7, -16E7, -16E6SD, -16E6 SAT, -16E6 Δ151, -16E6 151V, -18E6SD, -18E6 Δ151, -18E6 158L, -p53C234, and -ΔNp63α and pCLXSH-hTERT, as previously described (21, 53). The construction of the destination vector pDEST-CL-SI-MSCVpuro (designated pSI-CMSCVpuroDEST previously), the p53 short-hairpin RNA (shRNA) retroviral expression vector pCL-SI-MSCVpuro-p53Ri (designated pSI-CMSCVpuro-p53Ri previously), and the entry vector pENTER-H1R-stuffer has been described previously (13, 46). The construction of the shRNA retroviral expression vectors pCL-SI-MSCVpuro-16E6-Ri3 and pCL-SI-MSCVpuro-E6AP-Ri4 was described recently (14, 34). The targeted sequences for 16E6 and E6AP were 5′-GTATGGAACAACATTAGAA-3′ and 5′-GAAATCTAGTGAATGATGA-3′, respectively. To generate the Notch1 shRNA expression vector pCL-SI-MSCVpuro-Notch1Ri, 5′-GGAGCATGTGTAACATCAA-3′ was chosen as the targeted sequence. The production of recombinant retroviruses was as described previously (35). Briefly, the retroviral vector and packaging construct pCL-10A1 were cotransfected into 293FT cells (Invitrogen) using TransIT-293 (Mirus Co., Madison, WI) according to the manufacturer's instructions, and the culture fluid was harvested 48 to 72 h posttransfection. Titers of the recombinant viruses were greater than 2 × 105 drug-resistant CFU/ml with HeLa cells. Following the addition of the recombinant viral fluid to cells in the presence of 4 μg/ml Polybrene, infected cells were selected in the presence of 0.5 μg/ml puromycin or 50 μg/ml G418.
Immunoblotting.
Whole-cell protein extracts were used for immunoblotting as described previously (11). Antibodies against Notch1 (sc-6014; Santa Cruz), activated Notch1 (cleaved Notch1 [Val1744], no. 2421; Cell Signaling Technology), involucrin (clone SY5; Sigma), p53 (Ab6; Oncogene Science), phospho-p53 at Ser15 (9284; Cell Signaling Technology), p63 (clone 4A4; Santa Cruz), p21 (WAF1 Ab1; Oncogene Science), and β-actin (sc-1616; Santa Cruz) were used as probes. An anti-HPV16 E6 monoclonal antibody (clone 47A4) was raised against the 16 N-terminal amino acids and used as a probe. Horseradish peroxidase-conjugated anti-mouse, anti-rabbit (Jackson ImmunoResearch Laboratories), and anti-goat (sc-2033; Santa Cruz) immunoglobulins were used as the secondary antibodies. The LAS3000 charge-coupled-device imaging system (Fujifilm Co. Ltd., Tokyo, Japan) was employed for the detection of proteins visualized by Lumi-light plus Western blotting substrate (Roche).
RNA extraction and Northern blotting.
Total RNA (15 μg) isolated with the RNeasy reagent (QIAGEN) was electrophoresed on 1% agarose-formaldehyde gels, transferred to nylon membranes, and hybridized to 32P-labeled probes. The Notch1 probe was generated by random primer labeling (Amersham) of a Notch1 cDNA corresponding to the ICD. The 36B4 loading control probe was as described previously (11, 17).
Microarray analysis.
Total RNA isolated from wild-type E6, a series of E6 mutants, and control and p53 shRNA-expressing HCK1 T cells was subjected to CodeLink Expression Bioarray analysis using human whole-genome array containing ∼55,000 gene targets according to the manufacturer's instructions (Amersham Biosciences). Transcript levels were obtained as median-normalized values.
Dual-luciferase reporter assay.
The Notch1 promoter reporter N1PR-Luc was constructed by inserting a Notch1 promoter region spanning positions −961 to −1 relative to the translation initiation site (cloned from the bacterial artificial chromosome clone RP11-611D20) into a promoterless luciferase reporter plasmid, PGV-B (Toyo Ink, Japan). For heterologous reporter construction, the downstream candidate of the p53-binding stretch (positions −264 to −228) or the upstream candidate (positions −880 to −783) were inserted into the beta interferon basal-promoter sequence-containing luciferase reporter plasmid (18), generating N1p53cs1-BLuc and N1p53cs2-BLuc, respectively. The mutant reporters were constructed by replacing all core nucleotides, cytosine and guanine, in the putative p53-binding sequences with adenine and thymine, generating N1PRmut-Luc, which contains multiple mutated p53-binding repeats located at nucleotides −880 to −783. N1p53cs1mut-BLuc and N1p53cs2mut-BLuc were constructed in the same manner as N1PRmut-Luc. Cells were cotransfected with the reporters shown in Fig. 5 and the Renilla luciferase construct for normalization, with or without p53, using Lipofectamine 2000 (Invitrogen). Cell lysates were harvested at 48 h posttransfection and subjected to a dual-luciferase reporter assay according to the manufacturer's instructions (Promega).
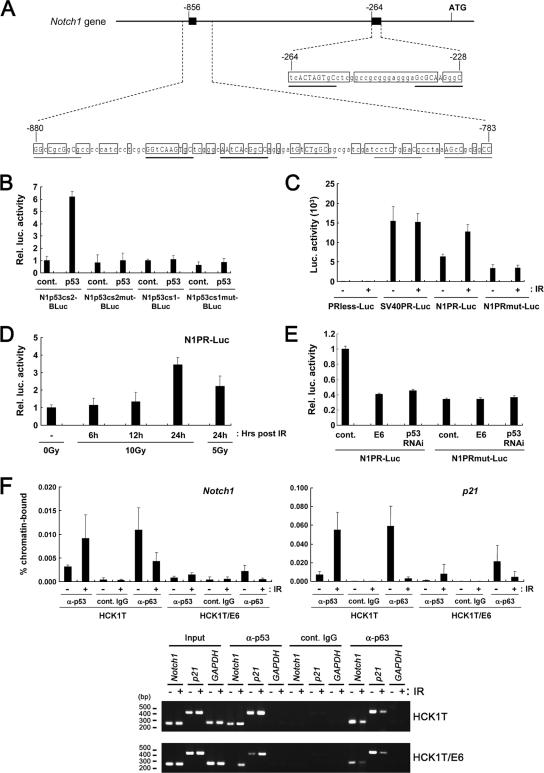
FIG. 5.
Transcriptional activation of the Notch1 promoter by p53. (A) Schematic representation of the two potential p53-responsive sites in the Notch1 promoter. The two p53-responsive candidates in the Notch1 promoter region are depicted, with position +1 representing the translation initiation site. Shown are the bases conserved among human, mouse, and rat sequences (open boxes) and the putative p53-binding consensus sequence (the thick line indicates where more than 70% of bases match among human, mouse, and rat sequences; the narrow line indicates adjacent sequences containing the conserved core nucleotides, cytosine and guanine, among human, mouse, and rat sequences). Uppercase letters indicate identity with the consensus sequence in the putative p53-binding elements. (B) Saos2 cells were transfected with the indicated heterologous luciferase reporters carrying the distal p53-binding candidate (N1p53cs2-BLuc, the wild-type reporter; N1p53cs2mut-BLuc, the p53-binding site-mutated reporter) or the proximal p53-binding candidate (N1p53cs1-BLuc, the wild-type reporter; N1p53cs1mut-BLuc, the p53-binding site-mutated reporter), with or without a p53 expression plasmid. The cell lysates at 48 h posttransfection were subjected to dual-luciferase reporter assays. Rel. Luc., relative luciferase; cont., control. (C) HCK1 T cells were transfected with the promoterless construct (PRless-Luc), the SV40 minimum enhancer promoter (SV40PR-Luc), the 1-kb Notch1 promoter (N1PR-Luc), or the 1-kb Notch1 promoter having the distal p53 binding site mutation (N1PRmut-Luc). Twenty-four hours after transfection, cells were exposed to 10 Gy gamma irradiation (IR) or left untreated. Cell lysates were prepared after another 24-h incubation. (D) HCK1 T cells were transfected with the N1PR-Luc reporter, and samples were collected at 6, 12, and 24 h after 5 or 10 Gy gamma irradiation (48 h after transfection). (E) HCK1 T cells stably expressing the N1PR-Luc or N1PRmut-Luc reporters were transduced with retroviral vectors encoding 16E6 or p53 shRNA. (F) The binding of endogenous p53 to the Notch1 promoter was assessed by ChIP assay. Either HCK1 T cells (HCK1T) or HCK1 T-cell-expressing 16E6 (HCK1T/E6) cells were exposed to 10 Gy gamma irradiation or left untreated, and 24 h later, cells were processed for preparation of the soluble chromatin fraction. ChIP was performed using specific antibodies against p53, p63, or control IgGs. α, anti. Input chromatin represents the portion of the sonicated chromatin prior to immunoprecipitation. The immunoprecipitates were analyzed by PCR amplification with primers specific for the distal p53-binding candidate in the Notch1 promoter shown to be responsive to p53 in Fig. 4A, for the p21 promoter-containing p53-responsive elements, and for the GAPDH gene as a control. The amounts of immunoprecipitated DNA fragments and input DNA were determined by quantitative real-time PCR. The percentages of immunoprecipitated promoter fragments relative to the total DNA input are shown in the top panel. Amplified PCR products in the linear range are shown in the bottom panel.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were carried out using an acetyl-histone H3 ChIP assay kit (Upstate Biotechnology). Briefly, 1 × 107 keratinocytes were fixed with 1% formaldehyde, neutralized by the addition of 125 mM glycine. Cells were washed twice in ice-cold phosphate-buffered saline and lysed in sodium dodecyl sulfate lysis buffer (1% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]) containing protease inhibitors, and DNA in the cross-linked chromatin preparations was sonicated to an average fragment size of 0.6 kb. The insoluble material was removed by centrifugation, and soluble chromatin samples were precleared with a 50% slurry of protein G-Sepharose-salmon sperm DNA. Each sample was incubated overnight at 4°C with 2 μg of monoclonal antibodies against p53 (clone DO-7; Oncogene Science), p63 (clone 4A4; Santa Cruz), or control immunoglobulin G (IgG) (Southern Biotechnology). Immune complexes were collected with protein G-Sepharose and eluted. Input templates were purified from 10% of the original lysates in parallel with the eluted immunoprecipitated samples. Cross-linking was reversed by incubation at 65°C for 6 h. After phenol-chloroform extraction and ethanol precipitation, the recovered DNA (4 μl from 25-μl immunoprecipitated chromatin DNA samples or 1 μl from the 100-μl input DNA control) was subjected to PCR amplification using a SYBR green PCR core reagent kit (Applied Biosystems) with a iCycler iQ real-time PCR detection system (Bio-Rad) or PCR amplification in the linear range. The specific primers for this analysis were as follows: 5′-GTGACCGAGGAGCGTGTC-3′ and 5′-CTAGCCCAGCGGCTTCACT-3′ for the Notch1 promoter, 5′-CCAGCCCTTTGGATGGTTT-3′ and 5′-GCCTCCTTTCTGTGCCTGA-3′ for the p21waf1 promoter, and 5′-AAAAGCGGGGAGAAAGTAGG-3′ and 5′-CTAGCCTCCCGGGTTTCTCT-3′ for the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene.
p53-deficient mice and UV irradiation.
Dorsal areas of wild-type (p53+/+), heterozygous (p53+/−), and null (p53−/−) mice (56) were shaved 2 days prior to irradiation with UVB at a single dose of 50 mJ/cm2 with a peak wavelength of 312 nm. Within 15 min after irradiation, mice were treated with cyclosporine (3.0 mg) intraperitoneally and dorsal-skin biopsies were taken at 0 or 48 h postirradiation.
Immunostaining.
HCK1 T cells expressing various constructs were seeded on chamber slides and fixed in 4% paraformaldehyde. Mouse skin samples were embedded in Tissue-Tek optimal cutting temperature compound (Sakura), and frozen sections (5 μm) were fixed in 4% paraformaldehyde. For immunofluorescence analysis, the following antibodies were used: anti-Notch1 (sc-6014; Santa Cruz), anti-involucrin (clone SY5; Sigma), anti-K10 (clone DE-K10; Covance), anti-mouse involucrin (PRB-140C; Covance), anti-mouse K10 (PRB-159P; Covance), and anti-Loricrin (PRB-145P; Covance). Alexa Flour 488- or Alexa 594-conjugated donkey anti-goat or -rabbit or goat anti-rabbit or -mouse IgGs (Molecular Probes) were used as the secondary antibodies. DAPI was applied to the sections with the secondary antibodies.
RESULTS
E6-mediated inactivation of p53 correlates with the repression of the Notch1 tumor suppressor.
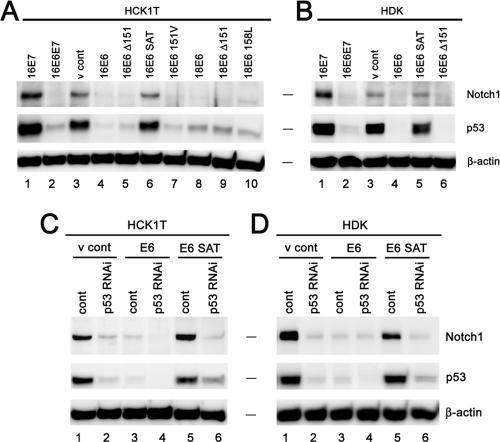
Previous reports have shown that Notch1 is down-regulated in cervical cancer cell lines in comparison to its level in primary keratinocytes and that the exogenously introduced Notch1 ICD induces the growth arrest of HPV-positive cervical cancer cells through the suppression of either E6/E7 expression (55) or E47 activity (54). We here confirmed the striking down-regulation of Notch1 in cervical cancer cell lines, relative to levels in normal counterparts (see Fig. S1, left panel, in the supplemental material). Since our data are consistent with a potential tumor-suppressive function for Notch1, we assessed its expression in other cancer-derived cell lines. For this purpose, we examined a range of lung cancer cell lines and found that Notch1 is indeed down-regulated in all lines we tested (see Fig. S1, right panel, in the supplemental material). In order to further verify the growth-suppressive activity of Notch1, we examined the effects of the exogenously expressed Notch1 ICD on cellular proliferation in a panel of cervical cancer cell lines, a spontaneously immortalized keratinocyte cell line, HaCaT, different types of normal epithelial cells, including normal HCK1s, primary HDKs, normal human mammary epithelial cells (HMECs), normal human bronchial epithelial cells, normal human small-airway epithelial cells, and normal human foreskin fibroblasts (HFFs), all (except HDKs) immortalized with hTERT. With the exception of the HPV-negative cervical cancer cell line C33A, HMECs, and HFFs, all the other cervical cancer cell lines and normal epithelial cells underwent massive growth inhibition triggered by the Notch1 ICD (see Fig. S2A and B in the supplemental material). Notch1 ICD-induced growth suppression was also seen in HCK1 T cells or HDKs expressing E6 and E7 driven by a heterologous promoter, suggesting that Nocth1 activation can override the proliferative signals generated by E6 and E7, independently of the transcriptional down-modulation of these viral genes. Interestingly, the Notch1 ICD showed rather a growth-stimulatory effect in HMECs, indicating that the converse activity can be elicited in this cell type. Having established tumor-suppressive effects and a marked reduction in Notch1 levels in cervical cancer cells, we hypothesized that E6 and/or E7 might have the ability to down-regulate Notch1 expression. To clarify the role of HPV oncoproteins, we introduced wild-type or mutant HPV16 E6 or wild-type E7 by retroviral-gene transfer into HCK1 T cells and examined endogenous Notch1 protein levels (Fig. 1A). Down-regulation of Notch1 expression was induced by wild-type E6 or the mutant E6 whose C terminus was deleted (E6 Δ151), which is incapable of binding to PDZ domain (14)-containing proteins (Fig. 1A, lanes 4 and 5), whereas Notch1 expression levels were sustained in cells expressing the E6 mutant that is defective in p53 inactivation (E6 SAT) (Fig. 1A, lane 6). The expression of E7 resulted in an up-regulation of Notch1 levels (Fig. 1A, lane 1), in line with the increase in p53 levels known to occur due to previously defined mechanisms (3, 61). Essentially the same results were obtained with primary keratinocytes, HDKs which have not been genetically manipulated for immortalization (Fig. 1B), indicating a common regulatory mechanism for Notch1 expression. The correlation between E6's ability to inactivate p53 and its repression of Notch1 further indicates that the enhanced p53 degradation by E6 and the ubiquitin ligase E6AP (47) is responsible for the observed Notch1 down-regulation. In accordance with this notion, E6 or E6AP silencing in cervical cancer cell lines resulted in an increase in Notch1 levels as well as in the restoration of p53 (Fig. 2A, lanes 2 and 4, and B, lanes 2, 4, and 6), suggesting the involvement of E6's activity in the down-modulation of Notch1 even in the transformed cells and its contribution to the progression of cervical cancer.
FIG. 1.
Down-regulation of Notch1 expression in normal keratinocytes by HPV E6 or p53 silencing. (A and B) Down-regulation of Notch1 by HPV E6. HCK1 T cells (HCK1T) (A) and HDKs (B) were transduced with the indicated genes by retroviral gene transfer. 16E6, 16E7, 16E6E7, and 18E6, high-risk-HPV16- or -HPV18-derived E6 and E7; E6 Δ151, the E6 mutant with 1 amino acid deleted at the C-terminal end, which lacks the ability to bind to PDZ domain (14)-containing proteins; E6 SAT, the E6 mutant with a 3-amino-acid substitution at the N terminus, which is defective in p53 inactivation; 16E6 151V, 16E6 in which the C-terminal leucine was replaced with valine; 18E6 158L, 18E6 in which the C-terminal valine was replaced with leucine; v cont, vector control. Expression levels of Notch1 and p53 proteins were analyzed by immunoblotting. The band corresponding to the furin-processed transmembrane subunit of Notch1 with a molecular mass of 120 kDa is shown as Notch1 hereinafter. β-Actin was used as the loading control. Note that the down-regulation of endogenous Notch1 expression was induced by p53-inactivating E6 proteins, including the wild-type, the C terminus deletion mutant, and the C terminus substitution mutant E6 proteins, but not by wild-type 16E7 or the 16E6 SAT mutant. (C and D) Down-regulation of Notch1 by knock-down of p53 by shRNA. HCK1 T cells (C) and HDKs (D) expressing wild-type 16E6, 16E6 SAT, or the vector control (v cont) were transduced with either a control (cont) or a p53 shRNA-bearing retroviral vector. Expression levels of Notch1 and p53 proteins were analyzed by immunoblotting. β-Actin was used as the loading control.
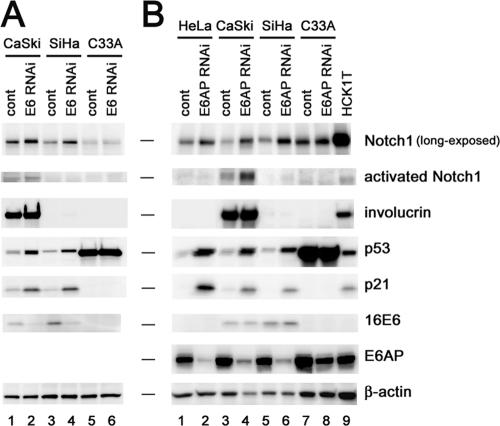
FIG. 2.
Effects of E6 or E6AP silencing on Notch1 expression in cervical cancer cells. HeLa, CaSki, SiHa, and C33A cells were transduced with either a control (cont) or an shRNA-bearing retroviral vector for 16E6 (A) or E6AP (B). Cell extracts were analyzed by immunoblotting with the indicated antibodies. HCK1T, HCK1 T cells.
The knock-down of p53 down-regulates Notch1 expression, while p53 activation leads to up-regulation.
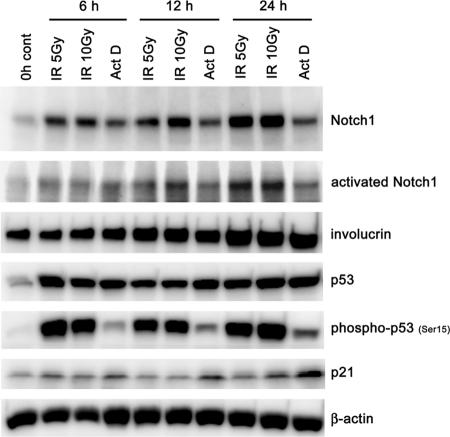
In order to more specifically investigate the consequence of p53's loss of function upon Notch1 expression, we employed the RNA interference (RNAi) approach. This resulted in the reduction of endogenous Notch1 protein in both HCK1 T cells and HDKs expressing the control vector or the E6 SAT mutant (Fig. 1C, lanes 2 and 6, and D, lanes 2 and 6). In E6-expressing cells, p53 protein levels were further reduced by p53 shRNA, leading to a more intensive down-regulation of Notch1 (Fig. 1C, lane 4, and D, lane 4). Conversely, when HCK1 T cells were exposed to ionizing radiation or treated with actinomycin D, the up-regulation of Notch1 was induced in accordance with the accumulation and activation of p53 (Fig. 3), further suggesting that p53 is directly involved in Notch1 gene expression in normal human epithelial cells upon genotoxic stress. In addition, to examine whether a dominant negative mutant of p53 might similarly function in terms of Notch1 regulation, a temperature-sensitive mutant of p53, p53Val138, was introduced into normal epithelial cells. Not only in keratinocytes (HCK1 T cells and HDKs) but also in human small-airway epithelial T cells and HMEC T cells, the inactivation of p53's function by a temperature shift to 37°C from the permissive temperature resulted in a reduction of Notch1, while Notch1 expression was scarcely detected under either condition in HFF T cells (see Fig. S3 in the supplemental material). The results suggest that the regulation of Notch1 expression by p53 is a mechanism conserved in different epithelial cell types, although different mechanisms to suppress Notch1 expression might exist in the fibroblasts.
FIG. 3.
Induction of Notch1 expression induced by ionizing radiation (IR) or actinomycin D (ActD) treatment in parallel with p53 activation. HCK1 T cells were exposed to 5 or 10 Gy gamma irradiation, treated with 5 nM actinomycin D, or left untreated (control [cont]), and cell lysates were prepared at 6, 12, and 24 h posttreatment. Cell extracts were analyzed by immunoblotting with the indicated antibodies.
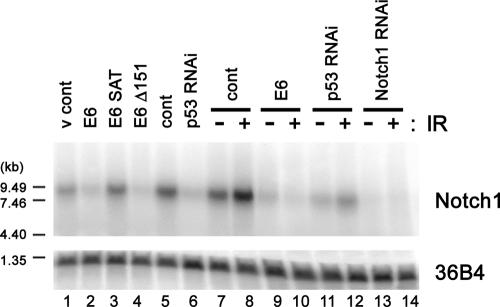
To determine whether Notch1 expression is regulated by p53 at the transcriptional level and to examine gene expression profiles comprehensively, we performed microarray analysis. The expression of E6, E6 Δ151, or p53 shRNA reduced the Notch1 transcript levels compared to levels in the control vector or E6 SAT mutant-expressing cases (Table 1). In addition, among the four Notch family members, Notch1 was specifically down-regulated by E6 and p53 silencing. Northern blot analysis confirmed the microarray result that Notch1 was regulated by p53 at the level of transcription (Fig. 4, lanes 1 to 6). The down-regulated Notch1 transcript levels before and after ionizing radiation in E6-, p53 shRNA-, and Notch1 shRNA-expressing cells were also verified (Fig. 4, lanes 7 to 14).
TABLE 1.
Notch1 expression is down-regulated by both E6 and p53 RNAi in normal cervical keratinocytes at the transcriptional level
| Line | Relative gene expressiona | ||||
|---|---|---|---|---|---|
| p53 | Notch1 | Notch2 | Notch3 | Notch4 | |
| 16E6 | 1.2 | 0.3 | 1.3 | 0.9 | 1.1 |
| 16E6 SAT | 1.1 | 1.0 | 1.7 | 1.2 | 1.1 |
| 16E6 Δ151 | 1.1 | 0.4 | 0.9 | 0.8 | 1.0 |
| 16E6 SATΔ151 | 1.0 | 0.9 | 1.5 | 1.2 | 0.8 |
| 16E6 151V | 1.5 | 0.3 | 1.1 | 1.0 | 1.2 |
| 16E6E7 | 0.9 | 0.3 | 1.0 | 1.4 | 1.1 |
| p53 RNAi | 0.4 | 0.4 | 0.8 | 1.1 | 1.1 |
FIG. 4.
Notch1 mRNA levels in HCK1 T cells expressing HPV E6, p53 shRNA, or Notch1 shRNA. Total RNA (15 μg) extracted from subconfluent cultures of HCK1 T cells expressing wild-type or mutant HPV16 E6, p53 shRNA, Notch1 shRNA, or control vectors (v cont) were subjected to Northern blotting. Cells were harvested 24 h postexposure to 10 Gy gamma irradiation (IR) (+) or no irradiation (−). 36B4 was used as the loading control.
The Notch1 promoter is transactivated by p53.
Next, we examined the ability of p53 to transactivate the Notch1 promoter. There are several putative p53-responsive sequences in a promoter region of the human Notch1 gene. Critical p53-binding motifs in two candidates are conserved among human, mouse, and rat. One is located 856 bp upstream from the translation initiation site, consisting of multiple tandem repeats of a putative p53-binding motif with imperfect matches. The other is located 264 bp upstream, featuring two tandem repeats (Fig. 5A). In Saos2 cells, which are deficient in p53, ectopically expressed p53 caused a significant induction of luciferase expression from the heterologous reporter containing the distal candidate sequence, while the reporter containing the proximal candidate as well as examples with mutations in the p53-binding motif failed to elicit a response to p53 (Fig. 5B). We then assessed the activity of the 1-kb Notch1 promoter region, including the two candidate elements in HCK1 T cells. The 1-kb Notch1 promoter showed considerable activity that was about half as strong as simian virus 40 (SV40) early enhancer promoter activity, which was further activated by ionizing radiation in a dose-dependent manner (Fig. 5C and D), while mutations in the distal putative p53-binding motifs resulted in a reduction of the basal activity and the lack of a response to ionizing radiation (Fig. 5C). In addition, the activity of the 1-kb Notch1 promoter was found to be down-regulated by the expression of E6 or p53 shRNA (Fig. 5E).
To explore whether p53 binds to the region containing the distal element in vivo, we carried out ChIP assays. While p53 binding was detected in HCK1 T cells at steady state and was significantly enhanced upon ionizing radiation, E6 expression resulted in a significant reduction even after such exposure to radiation (Fig. 5F). Therefore, we conclude that the distal putative p53-binding stretch in Notch1 promoter functions as a p53-responsive element.
Recent findings of cross-regulation between Notch1 and the p53 family member p63 (36) prompted us to investigate the relevance of p63 in Notch1 gene expression. Intriguingly, the ChIP analysis revealed p63 binding to the Notch1 promoter and its dissociation upon genotoxic stress (Fig. 5F). Thus, Notch1 gene expression may be controlled by a functional interplay between p63 and p53.
The expression of keratinocyte differentiation markers is repressed by E6 or the silencing of p53 or Notch1.
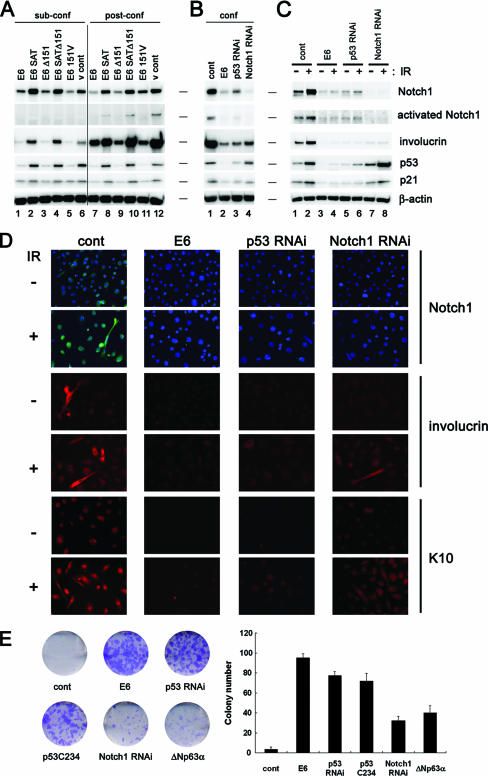
The E6 oncoprotein is known to suppress the differentiation of keratinocytes (48, 49), although the underlying molecular mechanism is largely unexplored. To assess the possibility that the repression of Notch1 by E6 could be involved, the expression of involucrin, an established keratinocyte-specific early differentiation marker, was analyzed. Since a report emphasized the importance of confluence-triggered cell-cell contact for the onset of keratinocyte differentiation (19), we examined involucrin levels in HCK1 T cells expressing wild-type E6, a series of E6 mutants, p53 shRNA, or Notch1 shRNA in both sub- and postconfluent states. Unlike in the control case, in which involucrin expression was markedly up-regulated as cells became confluent (Fig. 6A, lane 12), E6-expressing cells showed reduced involucrin levels, suppressive effects correlating with E6's targeting of p53 (Fig. 6A, lanes 7, 9, and 11). Consistent with this observation, p53 silencing also caused the repression of involucrin induction in accordance with reduced levels of Notch1 expression (Fig. 6B, lane 3). More importantly, E6, p53 shRNA, and Notch1 shRNA all repressed involucrin expression induced by either cell-cell contact or gamma irradiation (Fig. 6B, lanes 2 to 4, and C, lanes 4, 6, and 8), indicating that endogenous levels of p53 support keratinocyte differentiation through Notch1, and induced p53 enhances this process. Morphological and immunocytochemical analyses with antibodies against other differentiation markers, including K10, provided support for this notion (Fig. 6D). To further assess the suppressive effects of E6, p53 shRNA, a dominant negative mutant of p53 (p53C234), or Notch1 shRNA on differentiation and the consequential advantages to the proliferative potential, we carried out clonogenic growth assays after genotoxic stimulation or culture in serum-containing medium to induce differentiation. We also analyzed the effect of ΔNp63α overexpression in the assay since this isoform has been shown to be down-regulated in response to UV radiation (27, 66) or keratinocyte differentiation (36, 39) (see Fig. S4 in the supplemental material), and thus a counteracting function is expected. Colony formation of the control HCK1 T cells was almost completely inhibited by irradiation with 1 Gy (Fig. 6E) or more (data not shown). However, the expression of E6, p53 shRNA, or p53C234 resulted in the formation of numbers of colonies, and Notch1 silencing also conferred clonogenic ability, albeit to a lesser extent. The effect of ΔNp63α expression was equivalent to that of Notch1 silencing in this setting (Fig. 6E). Similarly, the clonogenicity of HCK1 T cells in serum-containing medium was restored by the expression of E6, p53 shRNA, or Notch1 shRNA to some extent (see Fig. S5 in the supplemental material). These results strongly suggest that Notch1 is a mediator of keratinocyte differentiation, which can be regulated by p53.
FIG. 6.
Inhibition of keratinocyte differentiation by expression of E6, p53 shRNA, or Notch1 shRNA. HCK1 T cells expressing the indicated series of E6 mutants (A) and shRNAs for p53 or Notch1 (B) were harvested in the sub- and postconfluent states (sub-conf and post-conf, respectively), and cell extracts were analyzed by immunoblotting with the indicated antibodies. cont, control. (C) HCK1 T cells expressing the control vector, wild-type E6, p53 shRNA, or Notch1 shRNA were exposed to 10 Gy gamma irradiation (IR) (+) or left untreated (−), and cell lysates were prepared at 24 h posttreatment. Cell extracts were analyzed by immunoblotting with the indicated antibodies. (D) Cells seeded on the chamber slide were treated similarly to those shown in panel C and subjected to immunocytochemical analysis with the indicated antibodies. DAPI (4′,6′-diamidino-2-phenylindole) staining is shown in samples for Notch1 detection. Magnification, ×30. (E) Aliquots of 500 HCK1 T cells expressing the control vector, wild-type E6, p53 shRNA, a dominant negative mutant of p53 (p53C234), Notch1 shRNA, or ΔNp63α were seeded on 35-mm dishes under sparse conditions and then exposed to 1 Gy gamma irradiation (+) or left untreated (−). After being cultivated for 2 weeks, the cells were stained with Giemsa's dye, and numbers of colonies were counted. The photographs are of representative dishes, and the graph illustrates means ± standard deviations.
The expression of Notch1 and differentiation markers is induced upon UV irradiation in vivo.
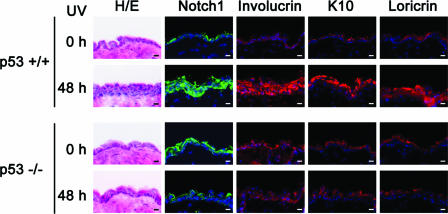
Though we have shown that the silencing of endogenous p53 represses the differentiation of keratinocytes in vitro, no skin phenotype has been reported for p53-deficient mice. To explore the physiological relevance of the p53-dependent regulation of Notch1, p53-deficient mouse skin was irradiated with UVB. As reported previously, the epidermal layer was thickened (4, 28) and the differentiation markers involucrin, loricrin, and K10 were all up-regulated (26), which correlates well with increased Notch1 staining 48 h after UVB irradiation in p53+/+ and p53+/− mouse skin, while such changes were far less evident, if present at all, in p53−/− mouse skin (Fig. 7 and data not shown for p53+/− mouse skin). These results suggest an involvement of p53 in keratinocyte differentiation also in vivo upon DNA damage.
FIG. 7.
p53-dependent differentiation of mouse epidermis induced by UVB irradiation. Wild-type (p53+/+) and p53-null (p53−/−) mice were irradiated with UVB, and dorsal skin biopsies were taken at 48 h postexposure. Frozen skin sections were subjected to hematoxylin and eosin staining (H/E) or immunohistochemical analysis with the indicated antibodies and DAPI. Scale bars, 10 μm.
DISCUSSION
Notch1 gene as a novel direct target of p53 in normal human epithelial cells.
In the present study, using normal human epithelial cells from uterine cervix (HCK1 T cells) and skin (HDKs), we uncovered a functional relationship between Notch1 and p53. Our results revealed that the Notch1 gene is a novel target of the transcriptional factor p53 in epithelial cells (Fig. 5) but not in fibroblasts (data not shown). Recently, the Notch1 gene was suggested to be a p53 target on the basis of ChIP analysis with the paired-end, ditag sequencing strategy in a cancer cell line, HCT116 (62). In stark contrast, p53 has been suggested to negatively regulate Notch1 expression and activation at the posttranscriptional level in thymocytes and to play a role in T-cell development (25). Therefore, the regulatory mechanism of Notch1 expression in thymocytes could be different from that in epithelial cells and remained to be explored in more detail.
Implications of p53-mediated Notch1 gene expression.
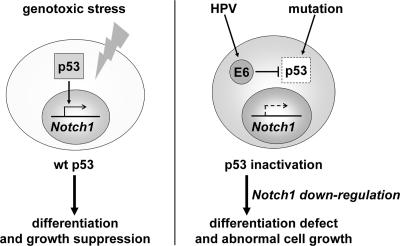
The repression of a keratinocyte differentiation marker by E6 or p53 silencing (Fig. 6) indicates that the down-regulation of Notch1 is a critical mechanism by which E6 suppresses keratinocyte differentiation (48, 49). Considering the established roles of Notch1 in keratinocyte growth control and differentiation (6, 19, 30, 37, 45), it is conceivable that p53 governs genomic integrity in normal epithelial cells by inducing growth suppression and/or differentiation through the up-regulation of Notch1. Supporting this, a role of p53 in keratinocyte differentiation has been suggested by analyses of Mdm2 transgenic mice (10). As p53 is mutated in many cancers, including squamous cell carcinomas, Notch1 might be down-regulated in such cases. Indeed, Notch1 expression was also repressed in a range of cervical cancer cell lines as well as lung cancer cell lines, compared to levels in normal counterparts (see Fig. S1 in the supplemental material). In addition, the regulation of Notch1 expression by p53 was suggested to be a mechanism conserved in different epithelial cell types (see Fig. S3 in the supplemental material). Thus, we propose the hypothesis that p53 mutations may also promote the development of squamous cell carcinomas through Notch1 down-regulation (Fig. 8).
FIG. 8.
Proposed model for the significance of p53-driven Notch1 expression. In normal epithelial cells, p53 might have an alternative mechanism to govern genomic integrity in response to genotoxic stress, featuring growth suppression or differentiation through up-regulation of the Notch1 gene. Inactivation of p53 by HPV E6 expression or mutation would bring about the down-regulation of Notch1, thereby promoting carcinogenesis in a broad spectrum of epithelial tissues.
Here we have described evidence indicating that endogenous levels of p53 are involved in Notch1 expression and the differentiation of keratinocytes in culture. However, we cannot conclude that p53 is involved in the normal differentiation of keratinocytes. It is more likely that the endogenous levels of p53 observed under monolayer culture conditions were induced by relatively weak environmental stress (e.g., reactive oxygen species). In accordance with this idea, p53-deficient mice are developmentally normal (7), and thus p53 is not essential for the differentiation of keratinocytes in the developmental stage, though it remains possible that the regulation of Notch1 might be compensated by the other p53 family members in p53-deficient mice. From a comparison of skin responses in wild-type and p53-deficient mice (Fig. 7), we speculate that the regulation of Notch1 by p53 could have evolved as a defensive mechanism against the DNA damage induced by UVB in close relation to its possible regulation by another p53 family member, p63.
It is well known that the biological consequences of p53 activation are dependent on the degree of DNA damage. Since higher doses of ionizing radiation in clonogenic growth assays resulted in no colony formation and since wild-type-p53 mice receiving higher doses of UV showed apoptotic figures rather than an up-regulation of epidermal differentiation (data not shown), the p53-Notch1 pathway could function when cells are exposed to rather mild genotoxic stress in the environment.
Possible involvement of other p53 family members in Notch1 regulation.
The pivotal role of p63 in epithelial development has been addressed previously (31, 68). The p63 gene encodes two major isoforms with or without the N-terminal transactivation domain, TAp63 or ΔNp63, respectively, through two different promoters, and each of them has three transcription variants, α, β, and γ, produced by alternative splicing (67). TAp63 isoforms are required for the initiation of epithelial stratification during early development (20), and a recent report has also suggested a tumor suppressor function (9). On the other hand, ΔNp63 isoforms are predominantly expressed in basal cells of mature epidermis and have functional links with the proliferative potential of squamous epithelial cells (1, 15, 40). More importantly, the overexpression of ΔNp63 isoforms has been suggested to counteract Notch1's ability to restrict growth and promote the differentiation of keratinocytes (36). Recently, it was reported that no Notch1 expression was detected in p63-deficient embryonic epidermis, suggesting the involvement of p63 in Notch1 expression (24). Intriguingly, we detected p63 binding to the p53-responsive elements in the Notch1 promoter and its replacement by p53 upon gamma irradiation (Fig. 5F), raising the noteworthy possibility that the predominant form of p63, ΔNp63, might function as a negative regulator of Notch1 gene expression. In this regard, we tested the effect of the exogenous expression of ΔNp63α, a predominant isoform in mature epidermis (2), on Notch1 expression and found it to be marginal (data not shown). This is in agreement with a recent report showing that ΔNp63α overexpression by itself did not significantly enhance proliferation (39). The results suggest that the level of endogenous ΔNp63 proteins suffices to antagonize Notch1 expression in the steady state. However, this does not explain the recent finding that p63 is required for Notch1 expression in embryonic epidermis (24). Clearly, further studies are required to clarify the molecular mechanisms in more detail.
Notch1: oncogene or tumor suppressor?
The oncogenic potential of activated Notch proteins has already been addressed with regard to the development of human T-cell leukemia and mouse mammary carcinomas (8, 16, 65). Notch1 expression is reported to be up-regulated in human breast carcinomas (63), though its significance has yet to be determined. Conversely, activated Notch signaling has been shown to inhibit the growth of hepatocellular carcinoma, small-cell lung cancer, and prostate cancer cells (41, 50, 51). In the development of cervical cancer, contradictory actions of Notch1 have been described; it has been called both an oncogene product (33, 43, 44, 52, 58, 63) and a tumor suppressor (54, 55). Based on the crucial role of Notch1 in keratinocyte differentiation (19, 30, 37, 45) and tumor suppression (38), it is conceivable that the biological consequence of Notch1 signaling is determined by a balance of many potential functions which require combinations of ubiquitous and cell-type-specific factors. Indeed, we found that the Notch1 ICD did not induce growth suppression in mammary epithelial cells (see Fig. S2 in the supplemental material). Therefore, the tumor-suppressive function of Notch1 might prevail over its oncogenic actions in normal epidermis and stratified squamous epithelia. However, it is possible that, even in keratinocytes, Notch1 can act as an oncogene product through the disruption of specific pathways, leading to tumor suppression under selective pressure for a growth advantage. In line with this notion, the growth of an HPV-negative cervical cancer cell line, C33A, was merely suppressed by the Notch1 ICD (see Fig. S2 in the supplemental material). The elevated Notch1 protein levels observed in squamous metaplasia, CIN lesions, and well-differentiated superficial carcinomas of the cervix (5, 12, 69), but not in invasive cervical cancers (55), could simply be a consequence of an increased number of cells with differentiation capacity, not the cause. In fact, we detected Notch1 in cervical cancer cell lines, although the levels were considerably lower than in normal keratinocytes (see Fig. S1 in the supplemental material). A previous report described considerable levels of Notch1 expression in cervical cancer cell lines (23). However, the conclusion was based on the observation that the Notch1 levels were comparable with those in HaCaT cells, which express mutant p53 and a much lower level of Notch1 than that in normal keratinocytes (data not shown). As previously shown (57), we also detected activated Notch1 proteins in CaSki cells, which appeared to be at a relatively high level compared to that in normal keratinocytes (Fig. 2B, lane 3 versus lane 9). Interestingly, however, we observed high involucrin levels in the CaSki cells, different from the levels in other cervical cancer cell lines, namely, HeLa, SiHa, and C33A , which have negligible levels. The knock-down of E6 or E6AP in CaSki cells further increased both activated Notch1 and involucrin (Fig. 2A, lane 2, and B, lane 4). In addition, the exogenous expression of the Notch1 ICD induced the up-regulation of involucrin, concomitant with the down-regulation of ΔNp63α in the CaSki cells as well as the normal HCK1 T cells (data not shown). These results imply that the CaSki cells preserve the Notch1-mediated differentiation program, as do normal keratinocytes. By these features, CaSki cells might be exceptional among cervical cancer cell lines with regard to the regulation of Notch1. However, we consider it possible that Notch1 activation is required for the proliferation or generation of the neoplastic phenotype of some fraction of cervical cancers, which has been implied by other investigators using CaSki cells (57, 63, 64). However, further study is required to determine the underlying molecular mechanisms by which abnormal proliferation associated with increased Notch1, especially in the early stages of cervical carcinogenesis, is induced.
In conclusion, our data provide the first mechanistic link between p53 and Notch1 as two factors which are important for tumor suppression in keratinocytes and insight into the potentially novel significance of p53 inactivation in squamous cell carcinomas, including cervical cancers. We now need to focus on specific interactions with ligands and signaling effectors or modifiers to determine how Notch1 might function as a tumor suppressor in one cellular environment and an oncogene product in another.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank S. Aizawa, H. Arakawa, and M. Futamura for p53-deficient mice and J. Yokota and T. Kohno for lung cancer cell lines. We also are grateful to M. Saito and M. Enari for technical assistance and useful discussions. We furthermore express our sincere thanks to T. Ishiyama, M. Umeki, Y. Hanada, and A. Noguchi for providing expert technical assistance.
This work was supported in part by a grant-in-aid for cancer research from the Ministry of Health, Labor, and Welfare, a grant-in-aid for scientific research in the cancer priority area from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and a grant-in-aid from the Takeda Science Foundation to T.K.
Footnotes
▿
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Barbieri, C. E., C. E. Barton, and J. A. Pietenpol. 2003. Delta Np63 alpha expression is regulated by the phosphoinositide 3-kinase pathway. J. Biol. Chem. 278**:**51408-51414. [DOI] [PubMed] [Google Scholar]
- 2.Barbieri, C. E., and J. A. Pietenpol. 2006. p63 and epithelial biology. Exp. Cell Res. 312**:**695-706. [DOI] [PubMed] [Google Scholar]
- 3.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395**:**124-125. [DOI] [PubMed] [Google Scholar]
- 4.Berton, T. R., A. Pavone, and S. M. Fischer. 2001. Ultraviolet-B irradiation alters the cell cycle machinery in murine epidermis in vivo. J. Investig. Dermatol. 117**:**1171-1178. [DOI] [PubMed] [Google Scholar]
- 5.Daniel, B., A. Rangarajan, G. Mukherjee, E. Vallikad, and S. Krishna. 1997. The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J. Gen. Virol. 78**:**1095-1101. [DOI] [PubMed] [Google Scholar]
- 6.Devgan, V., C. Mammucari, S. E. Millar, C. Brisken, and G. P. Dotto. 2005. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 19**:**1485-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356**:**215-221. [DOI] [PubMed] [Google Scholar]
- 8.Ellisen, L. W., J. Bird, D. C. West, A. L. Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66**:**649-661. [DOI] [PubMed] [Google Scholar]
- 9.Flores, E. R., S. Sengupta, J. B. Miller, J. J. Newman, R. Bronson, D. Crowley, A. Yang, F. McKeon, and T. Jacks. 2005. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7**:**363-373. [DOI] [PubMed] [Google Scholar]
- 10.Ganguli, G., J. Abecassis, and B. Wasylyk. 2000. MDM2 induces hyperplasia and premalignant lesions when expressed in the basal layer of the epidermis. EMBO J. 19**:**5135-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gewin, L., H. Myers, T. Kiyono, and D. A. Galloway. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 18**:**2269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, G. E., R. S. Mann, E. Mitsiadis, D. Henrique, M. L. Carcangiu, A. Banks, J. Leiman, D. Ward, D. Ish-Horowitz, and S. Artavanis-Tsakonas. 1999. Human ligands of the Notch receptor. Am. J. Pathol. 154**:**785-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haga, K., S. Ohno, T. Yugawa, M. Narisawa-Saito, M. Fujita, M. Sakamoto, D. A. Galloway, and T. Kiyono. 2007. Efficient immortalization of primary human cells by p16INK4a-specific short hairpin RNA or Bmi-1, combined with introduction of hTERT. Cancer Sci. 98**:**147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handa, K., T. Yugawa, M. Narisawa-Saito, S. Ohno, M. Fujita, and T. Kiyono. 2007. E6AP-dependent degradation of DLG4/PSD95 by high-risk human papillomavirus type 18 E6 protein. J. Virol. 81**:**1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hibi, K., B. Trink, M. Patturajan, W. H. Westra, O. L. Caballero, D. E. Hill, E. A. Ratovitski, J. Jen, and D. Sidransky. 2000. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 97**:**5462-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jhappan, C., D. Gallahan, C. Stahle, E. Chu, G. H. Smith, G. Merlino, and R. Callahan. 1992. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 6**:**345-355. [DOI] [PubMed] [Google Scholar]
- 17.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396**:**84-88. [DOI] [PubMed] [Google Scholar]
- 18.Kiyono, T., A. Hiraiwa, S. Ishii, T. Takahashi, and M. Ishibashi. 1994. Inhibition of p53-mediated transactivation by E6 of type 1, but not type 5, 8, or 47, human papillomavirus of cutaneous origin. J. Virol. 68**:**4656-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolly, C., M. M. Suter, and E. J. Muller. 2005. Proliferation, cell cycle exit, and onset of terminal differentiation in cultured keratinocytes: pre-programmed pathways in control of C-Myc and Notch1 prevail over extracellular calcium signals. J. Investig. Dermatol. 124**:**1014-1025. [DOI] [PubMed] [Google Scholar]
- 20.Koster, M. I., S. Kim, A. A. Mills, F. J. DeMayo, and D. R. Roop. 2004. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 18**:**126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyo, S., M. Nakamura, T. Kiyono, Y. Maida, T. Kanaya, M. Tanaka, N. Yatabe, and M. Inoue. 2003. Successful immortalization of endometrial glandular cells with normal structural and functional characteristics. Am. J. Pathol. 163**:**2259-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, E. C. 2004. Notch signaling: control of cell communication and cell fate. Development 131**:**965-973. [DOI] [PubMed] [Google Scholar]
- 23.Lathion, S., J. Schaper, P. Beard, and K. Raj. 2003. Notch1 can contribute to viral-induced transformation of primary human keratinocytes. Cancer Res. 63**:**8687-8694. [PubMed] [Google Scholar]
- 24.Laurikkala, J., M. L. Mikkola, M. James, M. Tummers, A. A. Mills, and I. Thesleff. 2006. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development 133**:**1553-1563. [DOI] [PubMed] [Google Scholar]
- 25.Laws, A. M., and B. A. Osborne. 2004. p53 regulates thymic Notch1 activation. Eur. J. Immunol. 34**:**726-734. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. H., H. T. An, J. H. Chung, K. H. Kim, H. C. Eun, and K. H. Cho. 2002. Acute effects of UVB radiation on the proliferation and differentiation of keratinocytes. Photodermatol. Photoimmunol. Photomed. 18**:**253-261. [DOI] [PubMed] [Google Scholar]
- 27.Liefer, K. M., M. I. Koster, X. J. Wang, A. Yang, F. McKeon, and D. R. Roop. 2000. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res. 60**:**4016-4020. [PubMed] [Google Scholar]
- 28.Lu, Y. P., Y. R. Lou, P. Yen, D. Mitchell, M. T. Huang, and A. H. Conney. 1999. Time course for early adaptive responses to ultraviolet B light in the epidermis of SKH-1 mice. Cancer Res. 59**:**4591-4602. [PubMed] [Google Scholar]
- 29.Lubman, O. Y., S. V. Korolev, and R. Kopan. 2004. Anchoring notch genetics and biochemistry; structural analysis of the ankyrin domain sheds light on existing data. Mol. Cell 13**:**619-626. [DOI] [PubMed] [Google Scholar]
- 30.Mammucari, C., A. Tommassi di Vignano, A. A. Sharov, J. Neilson, M. C. Havrda, D. R. Roop, V. A. Botchkarev, G. R. Crabtree, and G. P. Dotto. 2005. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev. Cell 8**:**665-676. [DOI] [PubMed] [Google Scholar]
- 31.Mills, A. A., B. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398**:**708-713. [DOI] [PubMed] [Google Scholar]
- 32.Munger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78**:**11451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair, P., K. Somasundaram, and S. Krishna. 2003. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J. Virol. 77**:**7106-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narisawa-Saito, M., K. Handa, T. Yugawa, S. Ohno, M. Fujita, and T. Kiyono. 4 December 2006. HPV16 E6-mediated stabilization of ErbB2 in neoplastic transformation of human cervical keratinocytes. Oncogene. doi: 10.1038/sj.onc.1210118. [DOI] [PubMed]
- 35.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70**:**5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen, B. C., K. Lefort, A. Mandinova, D. Antonini, V. Devgan, G. Della Gatta, M. I. Koster, Z. Zhang, J. Wang, A. Tommasi di Vignano, J. Kitajewski, G. Chiorino, D. R. Roop, C. Missero, and G. P. Dotto. 2006. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 20**:**1028-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nickoloff, B. J., J. Z. Qin, V. Chaturvedi, M. F. Denning, B. Bonish, and L. Miele. 2002. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 9**:**842-855. [DOI] [PubMed] [Google Scholar]
- 38.Nicolas, M., A. Wolfer, K. Raj, J. A. Kummer, P. Mill, M. van Noort, C. C. Hui, H. Clevers, G. P. Dotto, and F. Radtke. 2003. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33**:**416-421. [DOI] [PubMed] [Google Scholar]
- 39.Okuyama, R., E. Ogawa, H. Nagoshi, M. Yabuki, A. Kurihara, T. Terui, S. Aiba, M. Obinata, H. Tagami, and S. Ikawa. 22 January 2007. p53 homologue, p51/p63, maintains the immaturity of keratinocyte stem cells by inhibiting Notch1 activity. Oncogene. doi: 10.1038/sj.onc.1210235. [DOI] [PubMed]
- 40.Patturajan, M., S. Nomoto, M. Sommer, A. Fomenkov, K. Hibi, R. Zangen, N. Poliak, J. Califano, B. Trink, E. Ratovitski, and D. Sidransky. 2002. DeltaNp63 induces beta-catenin nuclear accumulation and signaling. Cancer Cell 1**:**369-379. [DOI] [PubMed] [Google Scholar]
- 41.Qi, R., H. An, Y. Yu, M. Zhang, S. Liu, H. Xu, Z. Guo, T. Cheng, and X. Cao. 2003. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 63**:**8323-8329. [PubMed] [Google Scholar]
- 42.Radtke, F., and K. Raj. 2003. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3**:**756-767. [DOI] [PubMed] [Google Scholar]
- 43.Ramdass, B., T. T. Maliekal, S. Lakshmi, M. Rehman, P. Rema, P. Nair, G. Mukherjee, B. K. Reddy, S. Krishna, and M. Radhakrishna Pillai. 2007. Coexpression of Notch1 and NF-kappaB signaling pathway components in human cervical cancer progression. Gynecol. Oncol. 104**:**352-361. [DOI] [PubMed] [Google Scholar]
- 44.Rangarajan, A., R. Syal, S. Selvarajah, O. Chakrabarti, A. Sarin, and S. Krishna. 2001. Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt. Virology 286**:**23-30. [DOI] [PubMed] [Google Scholar]
- 45.Rangarajan, A., C. Talora, R. Okuyama, M. Nicolas, C. Mammucari, H. Oh, J. C. Aster, S. Krishna, D. Metzger, P. Chambon, L. Miele, M. Aguet, F. Radtke, and G. P. Dotto. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20**:**3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawada, M., T. Kiyono, S. Nakashima, J. Shinoda, T. Naganawa, S. Hara, T. Iwama, and N. Sakai. 2004. Molecular mechanisms of TNF-alpha-induced ceramide formation in human glioma cells: P53-mediated oxidant stress-dependent and -independent pathways. Cell Death Differ. 11**:**997-1008. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75**:**495-505. [DOI] [PubMed] [Google Scholar]
- 48.Sherman, L., H. Itzhaki, A. Jackman, J. J. Chen, D. Koval, and R. Schlegel. 2002. Inhibition of serum- and calcium-induced terminal differentiation of human keratinocytes by HPV 16 E6: study of the association with p53 degradation, inhibition of p53 transactivation, and binding to E6BP. Virology 292**:**309-320. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, L., A. Jackman, H. Itzhaki, M. C. Stoppler, D. Koval, and R. Schlegel. 1997. Inhibition of serum- and calcium-induced differentiation of human keratinocytes by HPV16 E6 oncoprotein: role of p53 inactivation. Virology 237**:**296-306. [DOI] [PubMed] [Google Scholar]
- 50.Shou, J., S. Ross, H. Koeppen, F. J. de Sauvage, and W. Q. Gao. 2001. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 61**:**7291-7297. [PubMed] [Google Scholar]
- 51.Sriuranpong, V., M. W. Borges, R. K. Ravi, D. R. Arnold, B. D. Nelkin, S. B. Baylin, and D. W. Ball. 2001. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 61**:**3200-3205. [PubMed] [Google Scholar]
- 52.Subramanyam, D., and S. Krishna. 2006. c-Myc substitutes for Notch1-CBF1 functions in cooperative transformation with papillomavirus oncogenes. Virology 347**:**191-198. [DOI] [PubMed] [Google Scholar]
- 53.Takeda, Y., T. Mori, H. Imabayashi, T. Kiyono, S. Gojo, S. Miyoshi, N. Hida, M. Ita, K. Segawa, S. Ogawa, M. Sakamoto, S. Nakamura, and A. Umezawa. 2004. Can the life span of human marrow stromal cells be prolonged by bmi-1, E6, E7, and/or telomerase without affecting cardiomyogenic differentiation? J. Gene Med. 6**:**833-845. [DOI] [PubMed] [Google Scholar]
- 54.Talora, C., S. Cialfi, O. Segatto, S. Morrone, J. K. Choi, L. Frati, G. P. Dotto, A. Gulino, and I. Screpanti. 2005. Constitutively active Notch1 induces growth arrest of HPV-positive cervical cancer cells via separate signaling pathways. Exp. Cell Res. 305**:**343-354. [DOI] [PubMed] [Google Scholar]
- 55.Talora, C., D. C. Sgroi, C. P. Crum, and G. P. Dotto. 2002. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 16**:**2252-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukada, T., Y. Tomooka, S. Takai, Y. Ueda, S. Nishikawa, T. Yagi, T. Tokunaga, N. Takeda, Y. Suda, S. Abe, et al. 1993. Enhanced proliferative potential in culture of cells from p53-deficient mice. Oncogene 8**:**3313-3322. [PubMed] [Google Scholar]
- 57.Veeraraghavalu, K., M. Pett, R. V. Kumar, P. Nair, A. Rangarajan, M. A. Stanley, and S. Krishna. 2004. Papillomavirus-mediated neoplastic progression is associated with reciprocal changes in JAGGED1 and manic fringe expression linked to notch activation. J. Virol. 78**:**8687-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veeraraghavalu, K., V. K. Subbaiah, S. Srivastava, O. Chakrabarti, R. Syal, and S. Krishna. 2005. Complementation of human papillomavirus type 16 E6 and E7 by Jagged1-specific Notch1-phosphatidylinositol 3-kinase signaling involves pleiotropic oncogenic functions independent of CBF1;Su(H);Lag-1 activation. J. Virol. 79**:**7889-7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Knebel Doeberitz, M. 2002. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur. J. Cancer 38**:**2229-2242. [DOI] [PubMed] [Google Scholar]
- 60.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189**:**12-19. [DOI] [PubMed] [Google Scholar]
- 61.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1**:**20-26. [DOI] [PubMed] [Google Scholar]
- 62.Wei, C. L., Q. Wu, V. B. Vega, K. P. Chiu, P. Ng, T. Zhang, A. Shahab, H. C. Yong, Y. Fu, Z. Weng, J. Liu, X. D. Zhao, J. L. Chew, Y. L. Lee, V. A. Kuznetsov, W. K. Sung, L. D. Miller, B. Lim, E. T. Liu, Q. Yu, H. H. Ng, and Y. Ruan. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124**:**207-219. [DOI] [PubMed] [Google Scholar]
- 63.Weijzen, S., P. Rizzo, M. Braid, R. Vaishnav, S. M. Jonkheer, A. Zlobin, B. A. Osborne, S. Gottipati, J. C. Aster, W. C. Hahn, M. Rudolf, K. Siziopikou, W. M. Kast, and L. Miele. 2002. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8**:**979-986. [DOI] [PubMed] [Google Scholar]
- 64.Weijzen, S., A. Zlobin, M. Braid, L. Miele, and W. M. Kast. 2003. HPV16 E6 and E7 oncoproteins regulate Notch-1 expression and cooperate to induce transformation. J. Cell. Physiol. 194**:**356-362. [DOI] [PubMed] [Google Scholar]
- 65.Weng, A. P., A. A. Ferrando, W. Lee, J. P. T. Morris, L. B. Silverman, C. Sanchez-Irizarry, S. C. Blacklow, A. T. Look, and J. C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306**:**269-271. [DOI] [PubMed] [Google Scholar]
- 66.Westfall, M. D., A. S. Joyner, C. E. Barbieri, M. Livingstone, and J. A. Pietenpol. 2005. Ultraviolet radiation induces phosphorylation and ubiquitin-mediated degradation of DeltaNp63alpha. Cell Cycle 4**:**710-716. [DOI] [PubMed] [Google Scholar]
- 67.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dotsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2**:**305-316. [DOI] [PubMed] [Google Scholar]
- 68.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398**:**714-718. [DOI] [PubMed] [Google Scholar]
- 69.Zagouras, P., S. Stifani, C. M. Blaumueller, M. L. Carcangiu, and S. Artavanis-Tsakonas. 1995. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc. Natl. Acad. Sci. USA 92**:**6414-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2**:**342-350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]