Isotope Ratio-Based Profiling of Microbial Folates (original) (raw)
. Author manuscript; available in PMC: 2008 Apr 30.
Published in final edited form as: J Am Soc Mass Spectrom. 2007 Mar 23;18(5):898–909. doi: 10.1016/j.jasms.2007.01.017
Abstract
Folate metabolism, which is responsible for one-carbon transfer reactions in critical cellular processes including thymidine biosynthesis, is among the most important targets of antibiotic and anticancer drugs. Analysis of intracellular folates is complicated by three different types of folate modification: oxidation/reduction, methylation, and polyglutamylation. Here we present a method for quantifying the full diversity of intracellular folates by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The method begins with folate extraction using −75°C methanol:water, with ascorbic acid and ammonium acetate added to prevent folate interconversion. The extract is then separated using hydrophilic interaction chromatography with an amino column, ionized by positive mode electrospray, and analyzed on a triple quadrupole instrument using multiple reaction monitoring. The method has been used to profile the folate pools in Escherichia coli and Saccharomyces cerevisiae, with absolute levels of selected folates in E. coli measured by spiking extracts of cells fed uniformly 13C-glucose with purified, unlabeled folate standards. An isotope-ratio-based approach has been applied to study the effects of trimethoprim, a clinically important antibiotic that blocks bacterial dihydrofolate reductase. In addition to causing the expected increase in oxidized and decrease in reduced folates, trimethoprim triggered a dramatic and previously unrecognized shift towards shorter polyglutamate chain lengths. This finding highlights the potential for analysis of the full spectrum of cellular folates by MS/MS to unveil novel biological phenomena.
Introduction
Folates are crucial intermediates in the transfer of one-carbon units [1]. They are involved in the synthesis of compounds including nucleic acids, methionine, and coenzyme A [2]. Structurally, folates consist of pteroic acid (Pte) covalently bound to a (poly)glutamate chain. The pteroic acid can be modified by oxidation-reduction and one-carbon addition-elimination, yielding a diverse family of compounds: folic acid (PteGlun), dihydrofolate (H2PteGlun), tetrahydrofolate (H4PteGlun), 5,10-methenyl-tetrahydrofolate (5,10-CH=H4PteGlun), 5,10-methylene-tetrahydrofolate (5,10-CH2-H4PteGlun), 5-methyl-tetrahydrofolate (5-CH3-H4PteGlun), 5-formyl-tetrahydrofolate (5-CHO-H4PteGlun), and 10-formyl-tetrahydrofolate (10-CHO-H4PteGlun). All of these folate species can contain a glutamate chain of variable length. The polyglutamation is thought to facilitate cellular folate accumulation and to enhance the interaction between folate and certain enzymes, although its role is not necessarily fully known [3].
Analysis of folates can be conducted using either microbiological assays or liquid chromatography (LC) coupled to ultraviolet absorption, fluorescence emission, electrochemical detection, or mass spectrometry; for a comprehensive review, see [3]. Coupling LC with mass spectrometry (LC-MS) has the advantage of readily distinguishing different folate forms and has become a leading analytical tool for folate analysis. A number of LC-MS-based techniques are available in the literature. The chromatography mode can be either reversed phase [4-8] or hydrophilic interaction chromatography (HILIC) [9]. Ionization is generally by electrospray and can be either positive [4,6,8] or negative mode [5,7,9]. The mass spectrometer detection mode can be either MS-only [5,8] or tandem MS [4,6,7,9].
To date, the majority of the LC-MS techniques used in folate analysis only detected those compounds containing one glutamate, in some cases after converting the polyglutamated forms to monoglutamated forms (deconjugation) [4,8]. Fazili et al. recently measured both mono- and diglutamate folates in human erythrocytes, and described the effect of pH, temperature and time on folate deconjugation [10]. Garratt et al. recently developed the first method capable of detecting a large number of mono- and polyglutamated folates and precursors. Their method uses negative ionization after reversed phase LC separation in the presence of the ion-pairing agent _N,N_-dimethylhexylamine [7].
We previously reported the detection of monoglutamated folate and derivatives such as 5-methyl-tetrahydrofolate in positive ionization mode [11]. Here we extend this approach to polyglutamated folates, presenting a systematically validated method using HILIC combined with positive ionization tandem mass spectrometry. A distinguishing feature of the method is its inclusion of multiple reaction monitoring (MRM) events for both unlabeled and uniformly 13C-labeled folates, enabling isotope ratio-based quantitation of cellular folates. The method was applied to study the folate pools in E. coli and yeast, revealing significant amounts of folate species in polyglutamated forms. In addition, the method was used to characterize the response of E. coli folate pools to the dihydrofolate reductase (DHFR) inhibitor trimethoprim. In addition to recapitulating literature findings regarding accumulation of oxidized and depletion of reduced folates in trimethoprim-treated E. coli [12], we demonstrate that trimethoprim treatment leads to profound decreases in folate polyglutamate chain length. This observation, which would not have been feasible using methods focused only on monoglutamated folates, highlights the value of LC-MS/MS methods that examine the full spectrum of cellular folate species.
Experimental
Chemical and Reagents
Standards of 21 different folate species listed in Table 1 were obtained from Schircks Laboratories (Jona, Switzerland). HPLC-grade water and acetonitrile (OmniSolv, EMD Chemical, Gibbstown, NJ) were obtained from VWR International (West Chester, PA); ammonium acetate (99.4%) from Mallinckrodt Chemicals, Phillipsburg, NJ; and ammonium hydroxide solution (29.73%) from Fisher Scientific, Pittsburg, PA. Trimethoprim (≥98%), adenosine-5′-phosphate (AMP, ≥99%), and all the media materials were obtained from Sigma-Aldrich (St. Louis, MO). 13C-glucose was obtained from Cambridge Isotope Laboratories (Andover, MA).
Table 1.
Folate standards used in this study (all purchased from Schircks Laboratories, Jona, Switzerland).
| Standard | Abbreviated name | Formula |
|---|---|---|
| Pteroic acid | Pteroic acid | C14H12N6O3 |
| _p_-aminobenzoyl-L-glutamate | pABGlu | C12H14N2O5 |
| Folic acid | PteGlu | C19H19N7O6 |
| 7,8-dihydrofolic acid | H2PteGlu (DHF) | C19H21N7O6 |
| (6R,S)-5,6,7,8-tetrahydrofolic acid | H4PteGlu (THF) | C19H23N7O6 |
| (6R,S)-5,10-methenyl-5,6,7,8-tetrahydrofolic acid | 5,10-CH=H4PteGlu | C20H21N7O6 |
| (6R,S)-5,10-methylene-5,6,7,8-tetrahydrofolic acid | 5,10-CH2-H4PteGlu | C20H23N7O6 |
| (6S)-5-methyl-5,6,7,8-tetrahydrofolic acid | 5-CH3-H4PteGlu (MTHF) | C20H25N7O6 |
| (6S)-5-formyl-5,6,7,8-tetradrofolic acid | 5-CHO-H4PteGlu | C20H23N7O7 |
| (6R,S)-5-methyl-5,6,7,8-tetrahydropteroyldi-γ-L-glutamate | 5-CH3-H4PteGlu2 | C25H32N8O9 |
| (6R,S)-5-formyl-5,6,7,8-tetrahydropteroyldi-γ-L-glutamate | 5-CHO-H4PteGlu2 | C25H30N8O10 |
| _p_-aminobenzyltri-γ-L-glutamate | pABGlu3 | C22H28N4O11 |
| folic acid triglutamate | PteGlu3 | C29H33N9O12 |
| (6R,S)-5-methyl-5,6,7,8-tetrahydropteroyltri-γ-L-glutamate | 5-CH3-H4PteGlu3 | C30H39N9O12 |
| (6R,S)-5-formyl-5,6,7,8-tetrahydropteroyltri-γ-L-glutamate | 5-CHO-H4PteGlu3 | C30H37N9O12 |
| (6R,S)-5-methyl-5,6,7,8-tetrahydropteroyltetra-γ-L-glutamate | 5-CH3-H4PteGlu4 | C35H46N10O15 |
| (6R,S)-5-formyl-5,6,7,8-tetrahydropteroyltetra-γ-L-glutamate | 5-CHO-H4PteGlu4 | C35H44N10O16 |
| _p_-aminobenzylpenta-γ-L-glutamate | pABGlu5 | C32H42N6O17 |
| (6R,S)-5-methyl-5,6,7,8-tetrahydropteroylpenta-γ-L-glutamate | 5-CH3-H4PteGlu5 | C40H53N11O18 |
| (6R,S)-5-formyl-5,6,7,8-tetrahydropteroylpenta-γ-L-glutamate | 5-CHO-H4PteGlu5 | C40H51N11O19 |
| _p_-aminobenzylhepta-γ-L-glutamate | pABGlu7 | C42H56N8O23 |
Instrumentation
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses were performed on a Finnigan TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Electron Corporation, San Jose, CA) equipped with electrospray ionization (ESI) source, coupled to a LC-10A HPLC system (Shimadzu, Columbia, MD). The MS instrumental parameters were: spray voltage 3200 V for positive ESI, 3000 V for negative ESI, nitrogen as sheath gas at 30 psi and as auxiliary gas at 10 psi, argon as the collision gas at 1.5 mTorr, and capillary temperature 325°C. Scan time for each MRM was 0.1 s with a scan width of 1 m/z. General LC parameters were: autosampler temperature 4°C, column temperature 15°C, and injection volume 20 μl.
LC-MS/MS method development
The fragmentation pattern of each parent ion was determined by infusing a 1 μg/ml solution of each folate species listed in Table 1 into the mass spectrometer at a flow rate of 50 μl/min. The instrument performed an automatic “compound optimization” to determine the optimal collision energy (CE) for the four most abundant product ions in both positive and negative ESI. The infusion solvent was 50:50 methanol:water (unbuffered) or 50:50 acetonitrile:water (with 10 mM ammonium acetate + 10 mM ammonium hydroxide, pH 9.4); both gave very similar results.
The LC approach was HILIC using an aminopropyl column (Luna NH2, 5 μm particle size, Phenomenex, Torrance, CA) at basic pH [11]. Solvent A was 20 mM ammonium acetate + 20 mM ammonium hydroxide in 95:5 water:acetonitrile, pH 9.4. Solvent B was acetonitrile. Balancing of separation, sensitivity, and running time resulted in a selection of column dimensions of 50 × 2 mm, flow rate of 200 μL/min, with a gradient of t = 0, 85% B; t = 3 min, 0% B; t = 14.5 min, 0% B; t = 15.5 min, 85% B; t = 20 min, 85% B. The final flow rate was 200 μl/min.
MS/MS parameters were finalized by running the available folate standards on LC-MS/MS and monitoring all of their possible MRMs. The best MRM for each standard was selected based on the signal-to-noise ratio. Based on the best MRMs for compounds with standards available, MRMs were inferred for those compounds where standards were not available. The final LC-MS/MS method contained MRMs for ∼ 60 folate compounds containing up to 8 glutamates.
Compound stability and sample handling
Reduced folates are unstable with respect to oxidation [13-17]. To evaluate folate stability, the available folate standards were added at a concentration of 1 μg/ml to a variety of different solvent mixtures, with systematic data collected for three solvent systems: 50:50 methanol:water, 50:50 methanol:water + 0.1% ascorbic acid (pH 3.8), and 50:50 methanol:water + 0.1% ascorbic acid + 20mM ammonium acetate (NH4OAc) (pH 6.2). The stability samples were analyzed using the above-developed LC-MS/MS method repeatedly over a period of 48 hours. All samples included 500 μg/ml AMP as internal standard. The selection of AMP was based on its being a stable compound with a similar RT to many folate compounds. The observed signal for each compound, after normalization to AMP signal, was plotted as a function of time and fit to a single exponential decay function to determine the compound half-life.
To evaluate the stability of the folates during LC separation step, additional experiments were performed for seven monoglutamated folate species (folic acid, DHF, THF, 5,10-CH=H4PteGlu, 5,10-CH2-H4PteGlu, 5-CH3-H4PteGlu and 5-CHO-H4PteGlu) under the precise chromatographic running conditions (15°C, pH 9.4, no ascorbic acid added). Samples were analyzed through direct infusion into the mass spectrometer without column separation repeatedly over a period of 20 min immediately following their dissolution into the chromatographic running buffer (50:50 Solvent A: Solvent B).
LC-MS/MS method validation
The reproducibility, limit of detection (LOD) and linearity of the LC-MS/MS method was assessed using the 21 available folate standards (Table 1) dissolved in 50:50 methanol:water + 0.1% ascorbic acid + 20mM ammonium acetate (NH4OAc) (pH 6.2). Fresh stock solutions were prepared minimally every three months and were stored at −80°C. For reproducibility studies, mixed solutions of the 21 folate standards at 1 μg/ml with 500 ng/ml AMP as internal standard were prepared and divided into three parts. The first aliquot was analyzed four times on Day 1. The remaining aliquots were stored at −80°C and analyzed on Days 2 and 3, four times in each case. The resulting data, normalized to the AMP signal, were used to evaluate the intra- and interday reproducibility.
To determine the linearity and LOD, mixtures of the standards at various concentrations (1, 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000 ng/ml) were analyzed. The observed signal was normalized to AMP (500 ng/mL) signal. Linearity was assessed by regression. The LOD was defined as the lowest concentration at which the signal-to-noise was at least 5.
Microbial culture conditions and extraction
Escherichia coli K-12 strain NCM3722 was grown in liquid culture in a shaking flask at 37°C in a minimal salt media [18] with 10 mM ammonium chloride as the nitrogen source and 0.4% glucose (either unlabelled or uniformly 13C-labeled) as the carbon source. Exponential-phase cultures were quenched and extracted when optical density at 650 nm (A650) reached 0.5. The bacteria were pelleted by centrifugation for 4 min at 5000 g at room temperature, the supernatant immediately aspirated, and the pellet resuspended and mixed in 300 μl 80:20 methanol:water + 0.1% ascorbic acid + 20 mM ammonium acetate (−75°C; prepared fresh daily) and transferred to a 1.5 ml conical tube and allowed to sit on dry ice for 15 min. The sample was then spun in a micro-centrifuge at maximal speed for 5 min at 4°C and the supernatant taken as the first extract. The pellet was then resuspended in 200 μl 80:20 methanol:water + 0.1% ascorbic acid + 20 mM ammonium acetate and sonicated in an ice-bath for 10 min. The sample was again spun in a micro-centrifuge at maximal speed for 5 min at 4 °C and the supernatant taken as the second extract. The second extract was combined with the first extract to give a total of 500 μl extract.
For yeast, Saccharomyces cerevisiae strain CEN-PK was grown in standard Yeast Nitrogen Base media containing glucose and ammonia but no amino acids. This media notably contains, among other micronutrients, 2 mg/L of folic acid. The cells were grown in a shaking flask at 30°C to A600 = 0.6 and extracted. The extraction procedure was identical to that used for E. coli except for the centrifuge speed (3000 g instead of 5000 g).
Assessment of ion suppression due to the biological matrix
Twenty unlabeled folate standards (the 21 listed in Table 1, except pteroic acid) were spiked into either (i) extract from 13C-glucose grown E. coli cell culture or (ii) 80:20 methanol:water + 0.1% ascorbic acid + 20 mM ammonium acetate to a final concentration of 1 μg/ml. Ion signals from the two samples were compared to evaluate the extent of ion suppression from the biological matrix [19].
Profiling of folate pools in E. coli and yeast
Profiling of the folate pools in E. coli and S. cerevisiae was based on duplicate cultures using either unlabeled glucose or 13C-labeled glucose as the carbon source. Each sample was analyzed a total of four times, with runs spaced 80 min apart, which enabled estimation of intra-sample reproducibility and also rough assessment of the stability of the detected folate species (based on changes in signal intensity over the duration of the four runs). For 13C-glucose labeling, cells were grown at least 10 generations in uniformly 13C-glucose prior to sample collection. The MRM parameters for the uniformly 13C-folates were based on the number of carbon atoms in the parent and product ions (see Table 2).
Table 2.
LC-MS/MS parameters for 64 folate compounds.
| Compoundsa | Parent formula | 12C-parent m/z | 13C-parent m/z | Collision energy (eV) | 12C-product m/z | 13C-product m/z | Product formula | RT (minute)b |
|---|---|---|---|---|---|---|---|---|
| Pteroic acid | C14H13N6O3+ | 313.14 | 327.15 | 19 | 295.09 | 309.14 | C14H11N6O2+ | 6.5 |
| pABGlu | C12H15N2O5+ | 267.1 | 279.14 | 18 | 120.02 | 127.07 | C7H4O2+ | 6.1 |
| PteGlu | C19H20N7O6+ | 442.15 | 461.21 | 27 | 295.09 | 309.14 | C14H11N6O2+ | 6.6 |
| H2PteGlu | C19H22N7O6+ | 444.16 | 463.23 | 27 | 178.07 | 185.1 | C7H8N5O1+ | 6.1 |
| H4PteGlu | C19H24N7O6+ | 446.18 | 465.24 | 20 | 299.13 | 313.17 | C14H15N6O2+ | 6.2 |
| 5,10-CH=H4PteGlu | C20H22N7O6+ | 456.16 | 476.23 | 31 | 412.17 | 431.24 | C19H22N7O4+ | 6.2 |
| 5,10-CH2-H4PteGlu | C20H24N7O6+ | 458.18 | 478.25 | 23 | 311.13 | 326.18 | C15H15N6O2+ | 6.2 |
| 5-CH3-H4PteGlu | C20H26N7O6+ | 460.19 | 480.26 | 25 | 313.14 | 328.19 | C15H17N6O2+ | 6.2 |
| 5/10-CHO-H4PteGlu | C20H24N7O7+ | 474.17 | 494.24 | 20 | 327.12 | 342.17 | C15H15N6O3+ | 6.1 |
| pABGlu2 | C17H22N3O8+ | 396.14 | 413.20 | 25 | 120.02 | 127.07 | C7H4O2+ | N/A |
| PteGlu2 | C24H27N8O9+ | 571.19 | 595.27 | 30 | 295.09 | 309.14 | C14H11N6O2+ | N/A |
| H2PteGlu2 | C24H29N8O9+ | 573.21 | 597.29 | 30 | 178.07 | 185.1 | C7H8N5O1+ | N/A |
| H4PteGlu2 | C24H31N8O9+ | 575.22 | 599.3 | 30 | 299.13 | 313.17 | C14H15N6O2+ | N/A |
| 5,10-CH=H4PteGlu2 | C25H29N8O9+ | 585.22 | 610.29 | 30 | 412.17 | 431.24 | C19H22N7O4+ | N/A |
| 5,10-CH2-H4PteGlu2 | C25H31N8O9+ | 587.22 | 612.31 | 30 | 311.13 | 326.18 | C15H15N6O2+ | N/A |
| 5-CH3-H4PteGlu2 | C25H33N8O9+ | 589.24 | 614.32 | 25 | 313.14 | 328.19 | C15H17N6O2+ | 6.3 |
| 5/10-CHO-H4PteGlu2 | C25H31N8O10+ | 603.22 | 628.3 | 24 | 327.12 | 342.17 | C15H15N6O3+ | 6.2 |
| pABGlu3 | C22H29N4O11+ | 525.18 | 547.26 | 35 | 120.02 | 127.07 | C7H4O2+ | 6.3 |
| PteGlu3 | C29H34N9O12+ | 700.23 | 729.33 | 32 | 295.09 | 309.14 | C14H11N6O2+ | 7.0 |
| H2PteGlu3 | C29H36N9O12+ | 702.25 | 731.35 | 35 | 178.07 | 185.1 | C7H8N5O1+ | 6.7 |
| H4PteGlu3 | C29H38N9O12+ | 704.26 | 733.36 | 35 | 299.13 | 313.17 | C14H15N6O2+ | 6.7 |
| 5,10-CH=H4PteGlu3 | C30H36N9O12+ | 714.25 | 744.35 | 35 | 412.17 | 431.24 | C19H22N7O4+ | N/A |
| 5,10-CH2-H4PteGlu3 | C30H38N9O12+ | 716.26 | 746.36 | 35 | 311.13 | 326.18 | C15H15N6O2+ | 6.8 |
| 5-CH3-H4PteGlu3 | C30H40N9O12+ | 718.28 | 748.38 | 32 | 313.14 | 328.19 | C15H17N6O2+ | 6.7 |
| 5/10-CHO-H4PteGlu3 | C30H38N9O12+ | 732.26 | 762.36 | 36 | 327.12 | 342.17 | C15H15N6O3+ | 6.6 |
| pABGlu4 | C27H36N5O14+ | 654.23 | 681.32 | 43 | 120.02 | 127.07 | C7H4O2+ | N/A |
| PteGlu4 | C34H41N10O15+ | 829.28 | 863.39 | 34 | 295.09 | 309.14 | C14H11N6O2+ | N/A |
| H2PteGlu4 | C34H43N10O15+ | 831.29 | 865.41 | 40 | 178.07 | 185.1 | C7H8N5O1+ | 7.1 |
| H4PteGlu4 | C34H45N10O15+ | 833.31 | 867.42 | 40 | 299.13 | 313.17 | C14H15N6O2+ | N/A |
| 5,10-CH=H4PteGlu4 | C35H43N10O15+ | 843.29 | 878.41 | 40 | 412.17 | 431.24 | C19H22N7O4+ | N/A |
| 5,10-CH2-H4PteGlu4 | C35H45N10O15+ | 845.31 | 880.42 | 40 | 311.13 | 326.18 | C15H15N6O2+ | 7.2 |
| 5-CH3-H4PteGlu4 | C35H47N10O15+ | 847.32 | 882.44 | 38 | 313.14 | 328.19 | C15H17N6O2+ | 7.1 |
| 5/10-CHO-H4PteGlu4 | C35H45N10O16+ | 861.3 | 896.42 | 45 | 327.12 | 342.17 | C15H15N6O3+ | 7.1 |
| pABGlu5 | C32H43N6O17+ | 783.27 | 815.38 | 45 | 120.04 | 127.07 | C7H4O2+ | 7.0 |
| PteGlu5 | C39H48N11O18+ | 958.32 | 997.45 | 20 | 295.09 | 309.14 | C14H11N6O2+ | N/A |
| H2PteGlu5 | C39H50N11O18+ | 960.33 | 999.46 | 20 | 178.07 | 185.1 | C7H8N5O1+ | 8.0 |
| H4PteGlu5 | C39H52N11O18+ | 962.35 | 1001.5 | 20 | 299.13 | 313.17 | C14H15N6O2+ | N/A |
| 5,10-CH=H4PteGlu5 | C40H50N11O18+ | 972.35 | 1012.5 | 20 | 412.17 | 431.24 | C19H22N7O4+ | N/A |
| 5,10-CH2-H4PteGlu5 | C40H52N11O18+ | 974.35 | 1014.5 | 20 | 311.13 | 326.18 | C15H15N6O2+ | 7.8 |
| 5-CH3-H4PteGlu5 | C40H54N11O18+ | 976.36 | 1016.5 | 43 | 313.14 | 328.19 | C15H17N6O2+ | 7.6 |
| 5/10-CHO-H4PteGlu5 | C40H52N11O19+ | 990.34 | 1030.48 | 53 | 327.12 | 342.17 | C15H15N6O3+ | 7.6 |
| pABGlu6 | C37H50N7O20+ | 912.31 | 949.44 | 48 | 120.02 | 127.07 | C7H4O2+ | N/A |
| PteGlu6 | C44H55N12O21+ | 1087.36 | 1131.51 | 21 | 295.09 | 309.14 | C14H11N6O2+ | N/A |
| H2PteGlu6 | C44H57N12O21+ | 1089.38 | 1133.52 | 21 | 178.07 | 185.1 | C7H8N5O1+ | N/A |
| H4PteGlu6 | C44H59N12O21+ | 1091.39 | 1135.54 | 21 | 299.13 | 313.17 | C14H15N6O2+ | N/A |
| 5,10-CH=H4PteGlu6 | C45H57N12O21+ | 1101.38 | 1146.53 | 21 | 412.17 | 431.24 | C19H22N7O4+ | N/A |
| 5,10-CH2-H4PteGlu6 | C45H59N12O21+ | 1103.39 | 1148.54 | 21 | 311.13 | 326.18 | C15H15N6O2+ | N/A |
| 5-CH3-H4PteGlu6 | C45H61N12O21+ | 1105.41 | 1150.56 | 21 | 313.14 | 328.19 | C15H17N6O2+ | 8.3 |
| 5/10-CHO-H4PteGlu6 | C45H59N12O22+ | 1119.39 | 1164.54 | 25 | 327.12 | 342.17 | C15H15N6O3+ | N/A |
| pABGlu7 | C42H58N8O232+ | 521.17 | 542.25 | 30 | 120.02 | 127.07 | C7H4O2+ | 9.0 |
| PteGlu7 | C49H63N13O242+ | 608.7 | 633.29 | 22 | 295.09 | 309.14 | C14H11N6O2+ | N/A |
| H2PteGlu7 | C49H65N13O242+ | 609.71 | 634.29 | 22 | 178.07 | 185.1 | C7H8N5O1+ | N/A |
| H4PteGlu7 | C49H67N13O242+ | 610.71 | 635.3 | 22 | 299.13 | 313.17 | C14H15N6O2+ | N/A |
| 5,10-CH=H4PteGlu7 | C50H65N13O242+ | 615.71 | 640.80 | 22 | 412.17 | 431.24 | C19H22N7O4+ | N/A |
| 5,10-CH2-H4PteGlu7 | C50H67N13O242+ | 616.71 | 641.8 | 22 | 311.13 | 326.18 | C15H15N6O2+ | N/A |
| 5-CH3-H4PteGlu7 | C50H69N13O242+ | 617.72 | 642.81 | 22 | 313.14 | 328.19 | C15H17N6O2+ | 9.6 |
| 5/10-CHO-H4PteGlu7 | C50H67N13O252+ | 624.71 | 649.8 | 26 | 327.12 | 342.17 | C15H15N6O3+ | N/A |
| PteGlu8 | C54H70N14O272+ | 673.22 | 700.32 | 23 | 295.09 | 309.14 | C14H11N6O2+ | N/A |
| H2PteGlu8 | C54H72N14O272+ | 674.23 | 701.32 | 23 | 178.07 | 185.1 | C7H8N5O1+ | N/A |
| H4PteGlu8 | C54H74N14O272+ | 675.23 | 702.33 | 23 | 299.13 | 313.17 | C14H15N6O2+ | N/A |
| 5,10-CH=H4PteGlu8 | C55H72N14O272+ | 680.23 | 707.83 | 23 | 412.17 | 431.24 | C19H22N7O4+ | N/A |
| 5,10-CH2-H4PteGlu8 | C55H74N14O272+ | 681.23 | 708.84 | 23 | 311.13 | 326.18 | C15H15N6O2+ | N/A |
| 5-CH3-H4PteGlu8 | C55H76N14O272+ | 682.24 | 709.84 | 23 | 313.14 | 328.19 | C15H17N6O2+ | N/A |
| 5/10-CHO-H4PteGlu8 | C55H74N14O282+ | 689.23 | 716.83 | 27 | 327.12 | 342.17 | C15H15N6O3+ | N/A |
Absolute quantitation of selected folates in E. coli was based on cells grown in 13C-glucose. These cells were extracted in duplicate using solvent spiked with ten folate standards (5-CH3-H4PteGlu1-5 and 5-CHO-H4PteGlu1-5) at a concentration of 1 μg/ml (only the first extraction step used solvent spiked with the unlabeled folates). Using the resulting ratios of labeled to unlabeled peak heights, absolute concentrations of folates were determined as described previously [20], using the cell dry weight (CDW) of 13.7 mg for a 50 ml culture at A650 = 0.6.
Effect of trimethoprim on folate pools in E. coli
An isotope ratio-based scheme was used to probe the effect of antimicrobial drug trimethoprim on the folate pools in E. coli. Extracts of untreated cells grown in unlabeled glucose were pooled with extracts of drug-treated cells grown in uniformly 13C-glucose, and vice versa. Trimethoprim was added to exponentially growing cells at a final concentration of 4 μg/ml when A650 reached 0.45. The addition of trimethoprim dramatically slowed the cell growth. The drug-treated 13C-cells were extracted after 1 hour (A650 = 0.5). The untreated 12C-cells were extracted at A650 = 0.5. The two extracts were combined and analyzed. Experiments were performed in triplicate, yielding a total of six independent measurements for the drug-treated and six for the untreated cells (three each with and without 13C labeling).
Results and discussion
LC-MS/MS method
The method developed here involves LC separation coupled with positive ESI tandem mass spectrometry. Positive mode was selected based on superior sensitivity for most folates (consistent with Freisleben et al [4]). In Table 2 we list LC-MS/MS parameters, including the parent ion, collision energy, product ion, and retention time (where available) for ∼ 60 folate species. For folate species containing more than 6 glutamates, the best signal was obtained using doubly charged parent ions, i.e., (M+2H)2+.
Separation in the current method uses a HILIC approach on an aminopropyl column in which folates are strongly retained in acetonitrile and elute in the aqueous phase, with increasing numbers of glutamates leading to increased retention (consistent with the positively charged column binding strongly to polyanionic analytes). Folate elution is very sensitive to the buffer pH, with higher pH giving faster elution (consistent with the column becoming less cationic with increasing pH). While the current method detected folates containing ≤ 8 glutamates, it can be modified to detect compounds containing even more glutamates by either increasing the elution time in aqueous buffer or increasing the buffer pH.
Folate stability
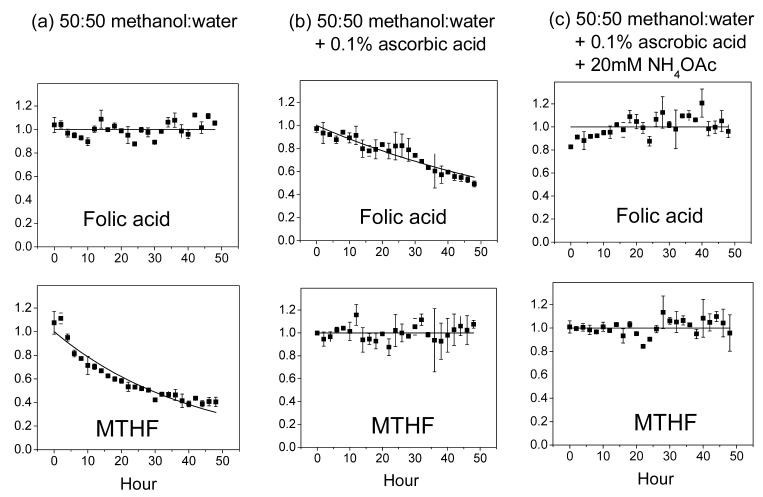
We systematically evaluated folate stability under three different test conditions (which were selected based on literature analysis and preliminary laboratory experiments). Results are listed in Table 3. Without antioxidant, most reduced folate species are highly unstable. For example, dihydrofolate and tetrahydrofolate both decay with t1/2 < 1 h. As shown previously, ascorbic acid helps prevent folate oxidation. However, folic acid (PteGlu) is unstable in methanol:water + 0.1% ascorbic acid as seen in Fig.1; this may be due to trace metal contamination or dissolved gas in the solvent. Folic acid instability is slightly exacerbated by higher ascorbic acid levels, such as those previously used in the literature, e.g., 1% [3,16] (data not shown). Increasing solution pH by addition of 20 mM ammonium acetate helps restore the folic acid stability as seen in Fig. 1. Thus, we recommend methanol:water + 0.1% ascorbic acid + 20 mM ammonium acetate as a preferred buffer for folate extraction and storage.
Table 3.
Method validation results for the 21 folate compound standards listed in Table 1.
| Compound | LLOQ (ng/ml) | Linearity (R2) | Intraday reproducibility (%RSD) | Interday reproducibility (%RSD) | Stability in different storage conditions: t1/2 (hour) | ||
|---|---|---|---|---|---|---|---|
| 50:50 methanol: water | 50:50 methanol: water + 0.1% ascorbic acid | 50:50 methanol: water + 0.1% ascorbic acid + 20mM NH4OAc | |||||
| Pteroic acid | 5 | 0.970 | 14 | 26 | 24 | 24 | 16 |
| pABGlu | 1 | 0.998 | 9 | 10 | > 96 | > 96 | > 96 |
| PteGlu | 5 | 0.998 | 10 | 9 | > 96 | 78 | > 96 |
| H2PteGlu | 5 | 0.978 | 8 | 15 | <0.5 | 10 | > 96 |
| H4PteGlu | 5 | 0.994 | 15 | 21 | <0.5 | 93 | 46 |
| 5,10-CH=H4PteGlu | 100 | 0.998 | 18 | 7 | 12 | > 96 | > 96 |
| 5,10-CH2-H4PteGlu | 5 | 0.989 | 10 | 13 | 4.5 | 14 | > 96 |
| 5-CH3-H4PteGlu | 1 | 0.998 | 11 | 11 | 42 | > 96 | > 96 |
| 5-CHO-H4PteGlu | 5 | 0.993 | 9 | 5 | > 96 | > 96 | > 96 |
| 5-CH3-H4PteGlu2 | 5 | 0.997 | 13 | 13 | 7.8 | > 96 | > 96 |
| 5-CHO-H4PteGlu2 | 5 | 0.997 | 9 | 8 | 85 | > 96 | > 96 |
| pABGlu3 | 5 | 0.987 | 10 | 3 | > 96 | > 96 | > 96 |
| PteGlu3 | 10 | 0.998 | 12 | 9 | > 96 | > 96 | > 96 |
| 5-CH3-H4PteGlu3 | 5 | 0.989 | 9 | 12 | 9.7 | > 96 | > 96 |
| 5-CHO-H4PteGlu3 | 5 | 0.999 | 10 | 2 | > 96 | > 96 | > 96 |
| 5-CH3-H4PteGlu4 | 10 | 0.978 | 12 | 14 | 18 | > 96 | > 96 |
| 5-CHO-H4PteGlu4 | 10 | 0.997 | 11 | 17 | 70 | > 96 | > 96 |
| pABGlu5 | 50 | 0.995 | 16 | 21 | > 96 | > 96 | > 96 |
| 5-CH3-H4PteGlu5 | 50 | 0.976 | 13 | 28 | 42 | > 96 | > 96 |
| 5-CHO-H4PteGlu5 | 50 | 0.994 | 18 | 18 | 39 | > 96 | > 96 |
| pABGlu7 | 200 | 0.982 | 6 | 4 | > 96 | > 96 | > 96 |
Fig. 1.
Representative compound stability traces. The top panel shows PteGlu (folic acid) and the bottom 5-CH3-H4PteGlu (MTHF), whose stabilities were measured in the indicated solvent systems. Each solid square is the average of two independent experiments, with the error bar being ± one standard error of the mean. The data is fit to single-exponential decay function, except when there is no obvious decay (in which case the line is for illustration purposes only).
Two additional analyses of folate stability were conducted. The first examined the short term stability of seven monoglutamate folate standards (folic acid, DHF, THF, 5,10-CH=THF, 5,10-CH2-THF, 5-CH3-THF, 5-CHO-THF ) under the LC running conditions (pH 9.4, 15°C) in the absence of ascorbic acid. The only folates to decay detectably over 20 min were 5,10-CH=THF (t1/2 ∼ 5 min) and THF (t1/2 ∼ 30 min). The second analysis involved data collected during profiling of cellular folates. Biological extracts stored at 4°C were run repeatedly over ∼ 4 h. Of the folates which were detected from E. coli or yeast extracts (see Tables 4 and 5), all were stable for the 4 h period, except for H4PteGlu3, which decayed with t1/2 ∼ 2 h.
Table 4.
Folate pools in exponentially growing E. coli. Data are from the average of two duplicate biological extracts each analyzed four times by LC-MS/MS. Reported values are the mean peak heights. Intra-sample reproducibility (determined from the four analytical runs of the first biological replicate) and inter-sample reproducibility (determined by comparing the two biological replicates) are shown in brackets as relative standard deviation (RSD). “0” indicates that no signal was detected; “+” indicates trace signal. Note that direct comparison of the signals between different compounds is complicated by different compounds having different response factors, i.e., different species give different ion counts when present in identical concentration.
| n = | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| pAB-Glun | 0 | + | 4541 (16%, 1%) | 0 | 0 | 0 | 0 | 0 |
| PteGlun | 0 | 0 | + | 0 | 0 | 0 | 0 | 0 |
| H2PteGlun | 0 | + | 40540 (9%, 13%) | 3250 (14%, 8%) | 1208 (41%, 7%) | 0 | 0 | 0 |
| H4PteGlun | 0 | + | 18893 (N/Aa, 3%) | + | 0 | 0 | 0 | 0 |
| 5,10-CH=H4PteGlun | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5,10-CH2-H4PteGlun | 0 | + | 18300 (15%, 20%) | 2172 (17%, 20%) | 1312 (39%, 40%) | 0 | 0 | 0 |
| 5-CH3-H4PteGlun | 5947 (10%, 12%) | 11205 (27%, 23%) | 270889 (19%, 18%) | 40747 (8%, 4%) | 11119 (18%, 9%) | 1180 (42%, 14%) | 0 | 0 |
| 5/10-CHO-H4PteGlun | + | 696 (45%, 5%) | 35301 (19%, 12%) | 2106 (27%, 11%) | + | 0 | 0 | 0 |
Table 5.
Folate pools in exponentially growing S. cerevisiae. Methods and nomenclature are as in Table 4.
| n = | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| pAB-Glun | 0 | 6686 (18%, 16%) | + | 0 | 0 | 0 | 0 | 0 |
| PteGlun | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H2PteGlun | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H4PteGlun | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5,10-CH=H4PteGlun | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5,10-CH2-H4PteGlun | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5-CH3-H4PteGlun | 1623 (40%, 61%) | 5911 (13%, 2%) | 5485 (34%, 6%) | 3181 (35%, 15%) | 439 (17%, 4%) | 5374 (25%, 19%) | 2501 (16%, 8%) | 0 |
| 5/10-CHO-H4PteGlun | + | + | 0 | 0 | 0 | 0 | 0 | 0 |
Method validation and performance
Mixtures of the 21 compound standards listed in Table 1 were used for method validation. The majority of compound standards have a LOD of 5 - 10 ng/ml (Table 3). The observed signal is linear with respect to the concentration from the respective LOD to 2 μg/ml, with a median R2= 0.994. The reproducibility for the standards is characterized by a median relative standard deviation of 11% for intra-day reproducibility and 12% for inter-day reproducibility.
In comparing our method to the previously published LC-MS/MS method for quantitation of folate polyglutamates from Garratt et al. [7], three major differences are the ionization mode (positive vs. negative), the chromatographic resin (aminopropyl vs. C18), and ion pairing agent (none vs _N,N_-dimethylhexylamine). The selection of ion pairing agent and ionization mode are intertwined: use of a cationic ion pairing agent precludes positive ESI detection. Despite their substantial differences, the methods perform similarly in terms of sensitivity, specificity, and reproducibility. One advantage of Garratt et al.'s approach is the good separation and sharp peaks afforded by the ion-pairing reversed phase chromatography. One advantage of the present approach is that it avoids ion pairing, thereby facilitating implementation of the present method on a multiuser instrument that is sometimes operated in both positive and negative ion mode.
Profiling of polyglutamated folates in E. coli and yeast
To examine the biological applicability of our method, we generated folate extracts from pelleted E. coli and S. cerevisiae. Because cells were pelleted before metabolome quenching, all biological results presented herein apply directly only to cells after centrifugation. Accordingly, it would be useful to cross-validate our results with other quenching procedures (which have their own pros and cons) in future biological studies [21,22].
Extracts from both organisms were prepared in duplicate, with each sample analyzed four times to evaluate analytical versus biological reproducibility (Tables 4 and 5). Example chromatograms are provided in Fig. 2. For E. coli, the median analytical (intra-sample) RSD was 19% and the median biological (inter-sample) RSD 12%; corresponding values for S. cerevisiae were 30% and 12%, respectively. The suboptimal analytical reproducibility reflects the challenges of accurate quantitation of many low abundance compounds in parallel—when signals are small, splitting of scan time over many compounds can compromise reproducibility and repeated runs are required to obtain precise results.
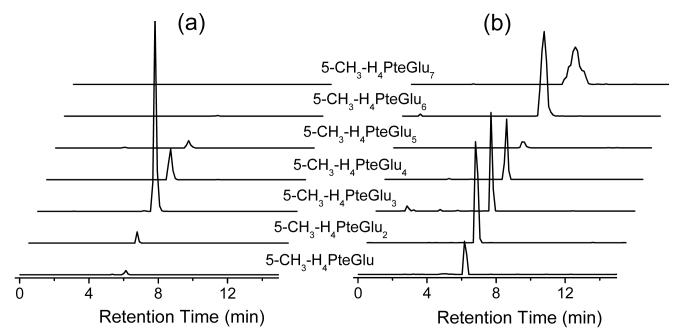
Fig. 2.
Representative chromatography traces for 5-CH3-H4PteGlun (MTHF-Glun, n from 1 to 7) from (a) E. coli and (b) yeast extract.
One potential contributor to analytical variability is ion suppression. To assess the extent of ion suppression occurring in biological extracts, 13C-glucose grown E. coli extract was spiked with 20 unlabeled folate standards (those listed in Table 1, except pteroic acid). The median signal of the standards in the biological matrix was reduced by 48%, indicating non-trivial (but also not severe) signal loss due to folate absorption and/or ion suppression.
Examining the overall trends in Tables 4 and 5, for E. coli a wide variety of folate oxidation-reduction and one carbon modified states are detectable, with polyglutamate chains of length 3 and 4 dominating across all oxidation and one carbon modification states. In S. cerevisiae a very large number of polyglutamate states (N = 1 – 7) are present in non-trivial amounts, but almost all of the pteroic acid is in the 5-methyl-tetrahydrofolate state. Thus, in both organisms, the folate polyglutamate, oxidation, and one carbon modification states follow logical patterns consistent with simple reaction pathways, in which polyglutamate labeling does not markedly alter oxidation-reduction or one carbon modification states, or vice versa. In contrast, the only previous paper to examine the full range of folate species, which studied spinach and mouse liver, found that different glutamate chain lengths predominant for different folate oxidation and one-carbon states [7]. While this literature finding might reflect more complex interplay between polyglutamate labeling and other folate reactions in these higher organisms, it may also reflect failure of the prior method to accurately capture the true cell folate profiles (e.g., because of stability problems).
Isotope ratio-based folate profiling
To enable isotope ratio-based folate absolute quantitation, we grew E. coli in uniformly 13C-glucose. To examine the efficacy of this procedure for labeling of folates, we compared signal heights of uniformly 12C-folates from E. coli grown in unlabeled glucose to those of uniformly 13C-folates from E. coli grown in uniformly 13C-glucose (Figure 3). As would be expected if our 13C-MRMs were correct and full folate labeling with 13C had occurred, signals were similar in both cases for each given folate species.
Fig. 3.
Comparing the 12C-signal from 12C-glucose-grown E. coli extract with the 13C-signal from 13C-glucose-grown E. coli extract for 17 folate compounds. No 12C-signals were seen in the 13C-glucose grown cells, or vice versa. The line of unity is drawn as a reference. When the data were fit by linear regression, the slope was 1.00 ± 0.09, R2 = 0.90. Specific folate compounds were those showing quantitative signal in Table 4: pAB-Glu3, H2PteGlu3-5, H4PteGlu3, 5,10-CH2-H4PteGlu3-5, 5-CH3-H4PteGlu1-6, 5/10-CHO-H4PteGlu2-4.
Armed with this evidence that the 13C-labeling was effective and did not disrupt the cellular folate pool sizes, we then extracted uniformly 13C-labeled E. coli with solvent spiked with selected unlabeled folate standards. This enabled absolute quantitation of these folate species. Results are shown in Figure 4, and confirm the major trends seen in Table 4 and discussed above: the most prevalent species were those with three glutamates, with both the 5-methyl-tetrahydrofolate and the somewhat less abundant 5- or 10-formyl-tetrahydrofolate species (which are isomeric and thus may not be distinguished by MS) showing a similar pattern of folate abundance as a function of glutamate chain length. One caveat regarding these results is that our standards have gamma-linked glutamates. While the first three glutamates of folates in E. coli are gamma-linked, subsequent glutamates are alpha-linked [23,24]. This discrepancy might impact the quantitation of longer chain folates from E. coli.
Fig. 4.
Absolute quantitation of 5-CH3-H4PteGlun and 5/10-CHO-H4PteGlun from E. coli. Experiments were performed in duplicate, with each sample run three times. Error bars show the biological (inter-sample) standard deviation.
Effect of trimethoprim on folate pools in E. coli
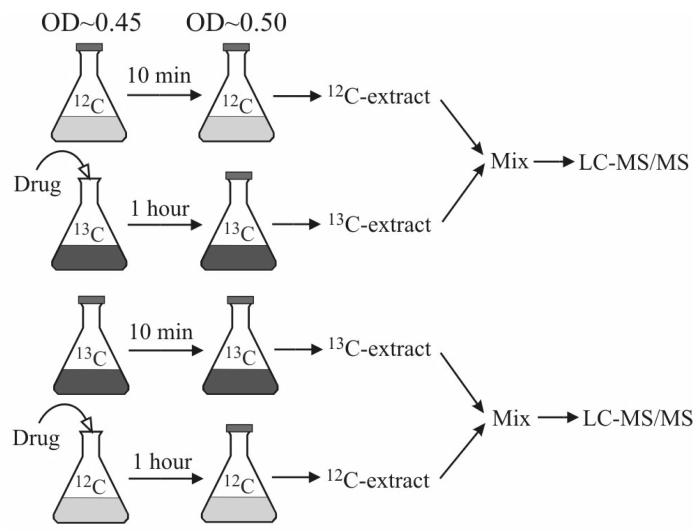
The antimicrobial drug trimethoprim is an inhibitor of DHFR [12,25]. Combined with sulfonamide, it is used clinically to treat diseases caused by enteric bacteria such as urinary tract infections. We employed an isotope ratio-based profiling scheme (Figure 5) to obtain a snapshot of the full response of the cellular folate pools to trimethoprim. This scheme eliminated the risk of reaching erroneous biological conclusions due to certain analytical problems such as ion suppression artifacts—the effect of ion suppression on the co-eluting 12C- and 13C-folates will be similar, thus “correcting” for the degree of suppression [19]. It also significantly reduced analysis time, which was especially valuable given the limited stability of folate compounds. Results are shown in Table 6.
Fig. 5.
Isotope ratio-based experimental scheme for studying the effect of the antibiotic drug trimethoprim on the folate pools in E. coli.
Table 6.
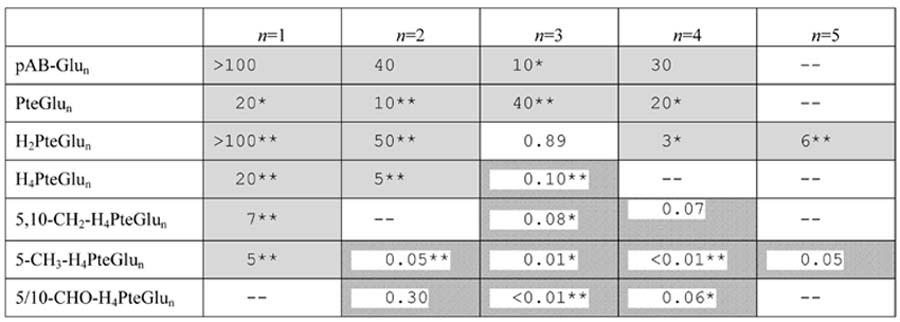
Effect of the DHFR inhibitor trimethoprim on the folate pools of E. coli. The overall experimental design is shown schematically in Fig. 5. Values in the table indicate fold-change in the level of the indicated folate species upon exposure of E. coli to 4 μg/mL trimethoprim for 1 h. Fold change = (peak height in drug-treated sample) / (peak height in the untreated sample). In cases where the signal in one condition is below the LOD, this signal was considered to be 100 ion counts, the typical LOD. Shaded boxes indicate significantly increased signal (p < 0.05) while hatched boxes indicate significantly decreased signal. “--” indicates that the compound was not quantifiable in either the drug-treated or untreated sample. Highly statistically significant results are highlighted by * (p < 0.01) or ** (p < 0.001).
All but one of the 27 folate-related species that we quantified successfully changed substantially (> 2-fold and p < 0.05) in concentration in response to the drug. Certain of these responses were expected based on literature knowledge of the drug's mechanism of action and effect on cellular folate pools [12]: in general, tetrahydrofolate species decreased and dihydrofolate (or yet more oxidized) species increased (in Table 6, this is reflected as values > 1 predominating in the top half of the table and < 1 predominating in the lower half of the table). These results confirm the ability of our LC-MS/MS method to recapitulate a well characterized pharmacological response.
Less expected, however, was the finding that the shift towards more oxidized folate species is matched by an equally pronounced shift towards folate species with shorter glutamate chain lengths (in Table 6, this is reflected as values > 1 predominating in the left half of the table and < 1 predominating in the right half of the table). Such a shift has, to our knowledge, never been previously reported. As evidence of the profundity of the shift to shorter glutamate chain length species, tetrahydrofolate species bearing a single glutamate actually increased during trimethoprim treatment, despite blockade of DHFR. The propensity for glutamate chain lengths to decrease when DHFR is blocked may be explained by ability to the enzyme folylpolyglutamate synthetase to add glutamate to tetrahydrofolate (and its formyl and methylene derivatives) but not more oxidized folate species [26]. Further experiments are required to test this possible mechanism, as well as to map the detailed dynamics of cellular folate pool responses to DHFR blockade. Nevertheless, the present data highlight the potential of applying the unique analytical power of LC-MS/MS to make discoveries regarding folates of pharmacological significance.
Conclusion
We present a comprehensive LC-MS/MS method for profiling cellular levels of folates. The method starts with folate extraction using 50:50 methanol:water + 0.1% ascorbic acid + 20 mM ammonium acetate at cold temperature, followed by HILIC separation and analysis on a triple quadrupole mass spectrometer in positive ESI MRM mode. Contributions of the present method relative to a closely related method that was recently published [7] include optimization of the extraction conditions for folate stability and of the MRM scans for sensitivity. In addition, the complications of ion pairing are avoided. Finally, the present method is readily compatible with one convenient metabolomic method [11], facilitating analysis of folates and other water soluble cellular metabolites in rapid succession.
The distinguishing features of the present work lie, however, not primarily in these technical advantages, but in the ways we have applied the method. The present manuscript is the first to observe logical, structured patterns in intracellular folate pools, where the most abundant glutamation state is conserved across different oxidation-reduction and one-carbon modification states, and vice versa. The method has been applied to conduct isotope ratio-based absolute quantitation and concentration profiling. This is, to our knowledge, the first example of global analysis of intracellular folates using an isotope ratio-based approach. Finally, the method has been used to confirm one previously described response of E. coli to a DHFR inhibitor antibiotic drug (that the drug leads to net folate oxidation), while also discovering a novel and equally pronounced cellular biochemical response (net shortening of the folate glutamate chain length). We believe that continued refinement and application of methods for global folate profiling is poised to substantially advance understanding of folate biology and the pharmacology of anti-folate drugs.
Acknowledgements
We thank Robert Moder for contributing to initial method development efforts and Andrew L. Bognar for providing valuable insight into the properties of folylpolyglutamate synthetase. This research was supported by the NIGMS/NIH Center for Quantitative Biology at Princeton University (P50GM071508) and grants from the Beckman Foundation and American Heart Association to J.D.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanson AD, Roje S. One-carbon metabolism in higher plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:119–137. doi: 10.1146/annurev.arplant.52.1.119. [DOI] [PubMed] [Google Scholar]
- 2.Rebeille F, Ravanel S, Jabrin S, Douce R, Storozhenko S, Van Der Straeten D. Folates in plants: biosynthesis, distribution, and enhancement. Physiologia Plantarum. 2006;126:330–342. [Google Scholar]
- 3.Quinlivan EP, Hanson AD, Gregory JF. The analysis of folate and its metabolic precursors in biological samples. Anal. Biochem. 2006;348:163–184. doi: 10.1016/j.ab.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Freisleben A, Schieberle P, Rychlik M. Specific and sensitive quantification of folate vitamers in foods by stable isotope dilution assays using high-performance liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2003;376:149–156. doi: 10.1007/s00216-003-1844-y. [DOI] [PubMed] [Google Scholar]
- 5.Hart DJ, Finglas PM, Wolfe CA, Mellon F, Wright AJA, Southon S. Determination of 5-methyltetrahydrofolate (C-13-labeled and unlabeled) in human plasma and urine by combined liquid chromatography mass spectrometry. Anal. Biochem. 2002;305:206–213. doi: 10.1006/abio.2002.5662. [DOI] [PubMed] [Google Scholar]
- 6.Kok RM, Smith DEC, Dainty JR, van den Akker JT, Finglas PM, Smulders YM, Jakobs C, de Meer K. 5-methyltetrahydrofolic acid and folic acid measured in plasma with liquid chromatography tandem mass spectrometry: applications to folate absorption and metabolism. Anal. Biochem. 2004;326:129–138. doi: 10.1016/j.ab.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Garratt LC, Ortori CA, Tucker GA, Sablitzky F, Bennett MJ, Barrett DA. Comprehensive metabolic profiling of mono- and polyglutamated folates and their precursors in plant and animal tissue using liquid chromatography/negative ion electrospray ionisation tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:2390–2398. doi: 10.1002/rcm.2074. [DOI] [PubMed] [Google Scholar]
- 8.Patring JDM, Jastrebova JA. Application of liquid chromatography-electrospray ionization mass spectrometry for determination of dietary folates: Effects of buffer nature and mobile phase composition on sensitivity and selectivity. J. Chromatogr. A. 2007 doi: 10.1016/j.chroma.2006.12.079. in press. [DOI] [PubMed] [Google Scholar]
- 9.Garbis SD, Melse-Boonstra A, West CE, van Breemen RB. Determination of folates in human plasma using hydrophilic interaction chromatography-tandem mass spectrometry. Anal. Chem. 2001;73:5358–5364. doi: 10.1021/ac010741y. [DOI] [PubMed] [Google Scholar]
- 10.Fazili Z, Pfeiffer CM, Zhang M, Jain R. Erythrocyte folate extraction and quantitative determination by liquid chromatography-tandem mass spectrometry: comparison of results with microbiologic assay. Clin. Chem. 2005;51:2318–2325. doi: 10.1373/clinchem.2005.053801. [DOI] [PubMed] [Google Scholar]
- 11.Bajad SU, Lu WY, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Quinlivan EP, McPartlin J, Weir DG, Scott J. Mechanism of the antimicrobial drug trimethoprim revisited. Faseb J. 2000;14:2519–2524. doi: 10.1096/fj.99-1037com. [DOI] [PubMed] [Google Scholar]
- 13.Indrawati, Van Loey A, Hendrickx M. Pressure and temperature stability of 5-methyltetrahydrofolic acid: A kinetic study. J. Agric. Food Chem. 2005;53:3081–3087. doi: 10.1021/jf048370k. [DOI] [PubMed] [Google Scholar]
- 14.Wilson SD, Horne DW. Evaluation of Ascorbic-Acid in Protecting Labile Folic-Acid Derivatives. Proc. Natl. Acad. Sci. USA. 1983;80:6500–6504. doi: 10.1073/pnas.80.21.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeiffer CM, Rogers LM, Gregory JF. Determination of folate in cereal-grain food products using trienzyme extraction and combined affinity and reversed-phase liquid chromatography. J. Agric. Food Chem. 1997;45:407–413. [Google Scholar]
- 16.Nilsson C, Johansson M, Yazynina E, Stralsjo L, Jastrebova J. Solid-phase extraction for HPLC analysis of dietary folates. Eur. Food Res. Technol. 2004;219:199–204. [Google Scholar]
- 17.Baggott JE. Hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate at pH 2.5 to 4.5. Biochem. 2000;39:14647–14653. doi: 10.1021/bi001362m. [DOI] [PubMed] [Google Scholar]
- 18.Gutnick DC,JM, Klopotowski T, Ames BN. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacterio. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Annesley TM. Ion suppression in mass spectrometry. Clin. Chem. 2003;49:1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 20.Lu WY, Kimball E, Rabinowitz JD. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. J. Am. Soc. Mass Spectrom. 2006;17:37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Wittmann C, Krömer JO, Kiefer P, Binz T, Heinzle E. Impact of the cold shock phenomenon on quantitation of intracellular metabolism in bacteria. Anal. Biochem. 2004;327:135–139. doi: 10.1016/j.ab.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Rabinowitz JD. Cellular metabolomics in Escherichia coli. Exp. Rev. Proteomics. 2007 doi: 10.1586/14789450.4.2.187. in press. [DOI] [PubMed] [Google Scholar]
- 23.Ferone R, Hanlon MH, Singer SC, Hunt DF. Alpha-Carboxyl-Linked Glutamates in the Folylpolyglutamates of Escherichia-Coli. J. Biol. Chem. 1986;261:6356–6362. [PubMed] [Google Scholar]
- 24.Ferone R, Singer SC, Hunt DF. Invitro Synthesis of Alpha-Carboxyl-Linked Folylpolyglutamates by an Enzyme Preparation from Escherichia-Coli. J. Biol. Chem. 1986;261:6363–6371. [PubMed] [Google Scholar]
- 25.Finberg RW, Moellering RC, Tally FP, Craig WA, Pankey GA, Dellinger EP, West MA, Joshi M, Linden PK, Rolston KV, Rotschafer JC, Rybak MJ. The importance of bactericidal drugs: Future directions in infectious disease. Clin. Infect. Diseas. 2004;39:1314–1320. doi: 10.1086/425009. [DOI] [PubMed] [Google Scholar]
- 26.Masurekar M, Brown GM. Partial-Purification and Properties of an Enzyme from Escherichia-Coli That Catalyzes Conversion of Glutamic-Acid and 10-Formyltetrahydropteroylglutamic Acid to 10-Formyltetrahydropteroyl-Gamma-Glutamylglutamic Acid. Biochem. 1975;14:2424–2430. doi: 10.1021/bi00682a024. [DOI] [PubMed] [Google Scholar]