Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation (original) (raw)
Abstract
The Xa1 gene in rice confers resistance to Japanese race 1 of Xanthomonas oryzae pv. oryzae, the causal pathogen of bacterial blight (BB). We isolated the Xa1 gene by a map-based cloning strategy. The deduced amino acid sequence of the Xa1 gene product contains nucleotide binding sites (NBS) and a new type of leucine-rich repeats (LRR); thus, Xa1 is a member of the NBS-LRR class of plant disease-resistance genes, but quite different from Xa21, another BB-resistance gene isolated from rice. Interestingly, Xa1 gene expression was induced on inoculation with a bacterial pathogen and wound, unlike other isolated resistance genes in plants, which show constitutive expression. The induced expression may be involved in enhancement of resistance against the pathogen.
Bacterial blight (BB), caused by Xanthomonas oryzae pv. oryzae (Xoo), has been one of the most serious diseases in rice, affecting production in irrigated and rain-fed lowland ecosystems throughout Asia, northern Australia, mainland Africa, the southern part of the United States, and Latin America (1). The genetic basis of host resistance to bacterial blight has been studied in depth, and many resistance genes have been identified from cultivated rice and wild rice species (reviewed in ref. 2); some of these resistance genes have been used in rice breeding for BB resistance, and many useful cultivars have been released in Japan and other Asian countries (3).
One of the BB-resistance genes, Xa1, confers a high level of specific resistance to race 1 strains of Xoo in Japan. In 1967, Sakaguchi (4) identified Xa1 and mapped it on rice chromosome 4. Since then, Xa1 has been used in Japanese rice breeding for BB resistance, because race 1 of Xoo has been the most dominant race in Japan. However, as with many plant-resistance genes identified to date, the mechanisms of resistance conferred by expression of Xa1 has remained unknown.
Recently, more than 10 resistance genes in various plants have been cloned by map-based cloning or transposon tagging strategies (reviewed in ref. 5). Surprisingly, the products of these resistance genes share structural similarities, even though these genes confer resistance to diverse pathogens, such as fungi [_Cf-9_, _Cf-2_, _L_6, _M_, _I_2, and _Rpp5_ (6–11)], bacteria [_Pto_ (12), RPS2 (13, 14), RPM1, and Xa21 (15, 16)], virus [_N_ (17)], and a nematode [_Hs1_pro-1 (18)]. This finding suggests that the resistance genes function in common signaling pathways leading defense against pathogen invasion. The presently cloned resistance genes can be grouped into 4 classes: (i) cytoplasmic receptor-like proteins with nucleotide binding sites (NBS) and a leucine-rich repeat (LRR) domain, (ii) a serine-threonine kinase, (iii) transmembrane receptors with a large extracytoplasmic LRR domain, and (iv) a transmembrane receptor with an extracellular LRR domain and an intracellular serine-threonine kinase domain (reviewed in ref. 5).
We report here the map-based cloning and characterization of Xa1. Based on the deduced amino acid sequence, the Xa1 gene is a class 1 resistance gene; a cytoplasmic receptor-like protein with NBS and LRR domains. Thus, the structure is quite different from the first cloned rice resistance gene, Xa21 (16), which is a class 4 gene. The most intriguing finding is that the expression of Xa1, unlike any previously studied resistance genes, is induced by pathogen infection and wound.
MATERIALS AND METHODS
Plant Materials.
Xa1 donor rice cultivars (cv.) IR-BB1 and Kogyoku (Xa1/Xa1) and a susceptible cv. IR24 (xa1/xa1) were used. For high-resolution linkage analysis, F3 populations (4,225 plants) derived from IR24/IR-BB1 and IR24/Kogyoku crosses were tested by bacterial inoculation. For complementation tests of Xa1, susceptible cv. Nipponbare and Kitaake were used for transformation.
cDNA Libraries.
To obtain pathogen-induced as well as constitutive mRNA, 2-month-old IR-BB1 was inoculated with T7174, a representative strain of Japanese Xoo race 1 (19). One gram of 5-cm leaf tips were harvested at 1, 2, 3, 4, and 5 days after inoculation (DAI) and pooled for RNA isolation. Poly(A)+ RNA was isolated by using oligotex-dT30 (TaKaRa). cDNA was synthesized from 5 μg of poly(A)+ RNA and cloned into vector lambda gt10 (Amersham). A total of 2.5 × 105 plaque-forming units (pfu) were screened with Y5212 DNA. To separate the YAC DNA from yeast chromosomes, DNAs in the YAC clone Y5212 were electrophoresed with CHEF (contour-clamped homogeneous electric field) in 0.5× TBE (89 mM Tris base/89 mM boric acid/2 mM EDTA, pH 8.0) at 14°C by using 1% gel for 60 hr at 4 V/cm by switching from 20 to 60 sec. YAC DNA was extracted from the gel with the Prep-A-gene DNA purification kit (Bio-Rad). For plaque hybridization, 30 ng of 32P-labeled YAC DNA was used as a probe.
Another cDNA library was constructed with the same cDNA described above in vector Lambda ZAPII. A total of 3.9 × 105 pfu were screened to obtain the full-length cDNA corresponding to Xa1 gene. Labeling of the probes and signal detection were done with the ECL system (Amersham) following the manufacturer’s instructions.
Similarity Search.
Computer searches for similar sequences were carried out with the data registered in nonredundant protein sequence databases GenPept, PDB, SwissProt, SPupdate, and PIR by using the blast algorithm (20).
Rapid Amplification of cDNA Ends (RACE)-PCR.
RACE-PCR was performed with the Marathon cDNA amplification kit (CLONTECH) according to the manufacturer’s instructions. Template mRNA was extracted as described above. RACE products were cloned into the PCRII vector (Invitrogen) and sequenced.
Cosmid Library.
_Sau_3AI partially digested DNA from the cv. IR-BB1 was ligated with _Bam_HI-digested cosmid vector SuperCosI (Stratagene). The clones were packaged in vitro with GigapackIII Gold (Stratagene) and transfected into competent Escherichia coli XL1-BlueMR. Colony filters including 5 × 104 colony-forming units were screened with restriction fragment length polymorphism markers with the ECL system (Amersham).
Transformation.
Protoplasts were isolated from suspension culture cells of rice (cv. Nipponbare or Kitaake) and purified according to ref. 21. Protoplast density was adjusted to about 2 × 106 with transformation buffer (22). Cosmid 3-2 and pCH (23), the plasmid harboring the 35S promoter-hygromicin resistance gene, or CLD04541XA and pCH were added to the protoplast suspension and mixed gently and well. The polyethylene glycol (PEG) treatment was done according to the method of Datta (24) with modification to transform large DNA; PEG solution was added to tubes containing the protoplast–DNA mixture and mixed by shaking. Tubes were incubated at room temperature for 10 min. Washing solution (20 ml of 0.4 M mannitol and 0.01 M CaCl2) was added and the tubes were incubated for another 15 min. After incubation, the layer of protoplasts and that of washing solution were mixed well by gently inverting the tube. Protoplasts were collected by centrifugation and washed 4 times by repeating the above steps. After PEG treatment, protoplasts were cultured according to ref. 25.
Southern Hybridization.
Genomic DNA was isolated from green leaves by the CTAB method (26). Genomic DNA was digested with the restriction enzymes _Bam_HI, _Bgl_II, _Dra_I, _Eco_RI, _Eco_RV, or _Hin_dIII; separated by electrophoresis on 0.6% agarose gel; and transferred onto positively charged nylon membrane (Boehringer Mannheim). Labeling of probes and signal detection were done with the ECL system (Amersham).
Northern Analysis.
The poly(A)+ RNAs (2 μg) were separated in 1% agarose gel and blotted onto a positively charged nylon membrane (Boehringer Mannheim). The blot was hybridized with 32P-labeled c42-1 and the 1.5-kb insert of cDNA clone (DDBJ accession number D40094) that completely matches rice actin 1 (GenBank accession no. X16280). Hybridization was done in Rapid hybridization buffer (Amersham) at 65°C overnight. The blot was washed twice in 1× SSC and 1% SDS at room temperature for 10 min, then twice in 0.1× SSC and 1% SDS at 65°C for 30 min.
Reverse Transcriptase–PCR (RT-PCR).
RT-PCR was carried out by using the Thermostable r_Tth_ Reverse Transcriptase RNA PCR Kit (Perkin–Elmer) according to the manufacturer’s instruction: 250 ng of total RNA was applied and 35 PCR cycles were used with denaturing at 95°C for 1 min, and annealing and extension at 60°C for 1 min. Primers were synthesized for amplification of Xa1 and actin genes based on the sequences of c42 and RAc1 (GenBank accession no. X16280), respectively: Xa1 gene, 5′-ACTGCCCTCTTGCACACGCCTTTGG-3′ (sense) and 5′-CCGGTACATCAGTATTGTCCATCGG-3′ (antisense); actin, 5′-TATGGTCAAGGCTGGGTTCG-3′ (sense) and 5′-CCATGCTCGATGGGGTACTT-3′ (antisense).
RESULTS
Identification of the Xa1 Candidate cDNA Clone by High-Resolution Mapping and Sequence Analysis.
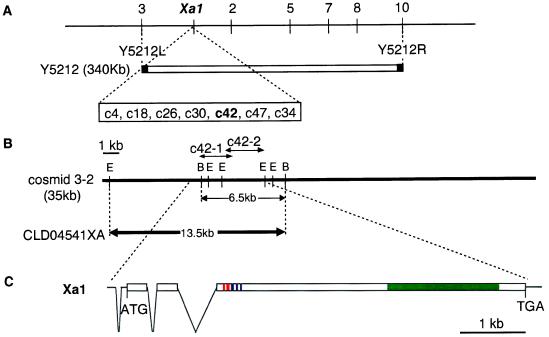
Previously, we isolated a 340-kb YAC clone, Y5212, that spans the Xa1 locus (Fig. 1A) (27). To identify expressed sequences in the region of Y5212, a cDNA library of IR-BB1 (Xa1/Xa1) was constructed with vector lambda gt10 and screened with Y5212 clone DNA. Thirty-three positive clones were obtained; they were classified into 21 classes based on their 3′ sequences.
Figure 1.
(A) Genetic map of the Xa1 region on chromosome 4. Numbers above the vertical lines indicate recombinations between Xa1 and cDNAs derived from the Y5212 region. Y5212L and Y5212R are YAC end clones (27). Seven cDNAs in the open rectangle linked to Xa1 without recombination. (B) Partial restriction enzyme map of the cosmid 3-2 and CLD04541XA: B, _Bam_HI; E, _Eco_RI. The 13.5-kb _Eco_RI-_Bam_HI fragment was subcloned into binary vector CLD04541 (kindly provided by J. D. G. Jones) to generate CLD04541XA. Arrows indicate the positions and lengths of the cDNAs c42-1 and c42-2. (C) Physical structure of the Xa1 gene. The initiation (ATG) and termination (TGA) codons are indicated. Exons are indicated by horizontal lines and open squares, and introns are indicated by lines angled downward. Deduced motifs of the Xa1 gene product are indicated: red, NBS; blue, conserved domains (15); and green, LRR.
We used these cDNAs for high-resolution linkage mapping to select cDNAs that cosegregated with Xa1. Based on responses to inoculation with a race 1 strain T7174 of Xoo, 965 susceptible plants were selected from F3 populations, each of which segregated for Xa1. The susceptible segregants were used for pooled-sampling linkage analysis (28, 29). Seven out of the 21 cDNAs were linked to Xa1 without recombination (Fig. 1A). For the other 14 cDNAs, we identified more than one plant with a recombination event between the cDNA and Xa1.
We determined the full nucleotide sequences of the seven candidate cDNAs. From among these sequenced cDNAs, a homology search with the corresponding deduced amino acid sequences revealed one, c42-1, with significant homology to NBS-LRR type-resistance genes, such as RPS2 (13, 14), RPM1 (15), N (17), and _L_6 (8). Therefore, c42-1 was further investigated as an Xa1 candidate.
Northern analysis of mRNA corresponding to c42-1 revealed that the length of this mRNA was approximately 6 kb (data not shown), suggesting that c42-1, which was 2.3 kb in length, was only a partial cDNA. To obtain the full-length cDNA corresponding to the gene including c42-1, another cDNA library was constructed in vector Lambda ZAPII and screened with c42-1 as the probe. A positive clone, c42-2, containing a 3.0-kb insert cDNA was obtained; the 5′ region of c42-2 overlapped the 3′ end of c42-1 and differed from the latter by the presence of a 2,754-bp extension (Fig. 1B). A 797-bp 5′ fragment flanking c42-1 was obtained by RACE-PCR. The length of the combined sequence of these cDNAs was 5,910 bp, consistent with the length of mRNA detected in the Northern analysis. We designate the combined cDNA as c42.
Complementation Test Reveals that the Genomic Clone Including the Candidate cDNA Confers Race-Specific Resistance.
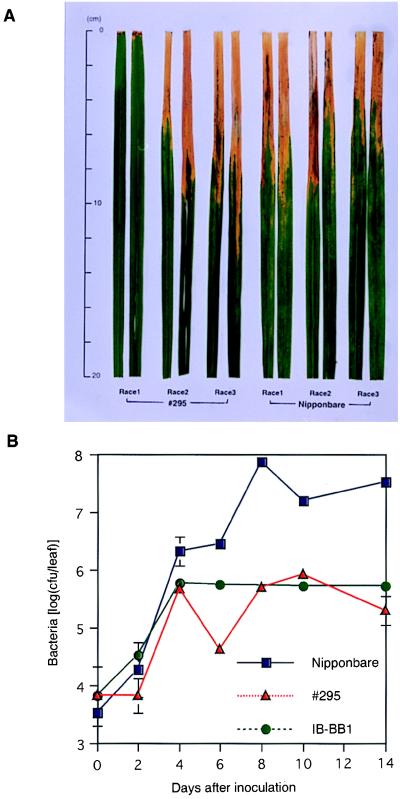
To examine whether the gene corresponding to c42 functions as Xa1, we conducted genetic complementation tests. We screened a cosmid genomic library of IR-BB1 by using c42 as a probe. Three positive clones were obtained. Physical mapping of positive cosmid clones indicated that the cosmid clone 3-2 included the entire c42-coding region and 7 kb of the 5′ flanking region (Fig. 1B). This cosmid was introduced into a susceptible rice cv., Nipponbare, by the PEG-mediated method. By this transformation, a total of 540 hygromycin-resistant plants from 314 lines were obtained. Three months after transformation, the plants were inoculated with T7174, a representative strain of Japanese Xoo race 1. One plant, #295, showed the same resistance reaction as did IR-BB1 (Fig. 2A). A 6.5-kb _Bam_HI-_Bam_HI fragment including c42 derived from cosmid 3-2 (see Fig. 1B) was revealed to exist in the plant #295 (data not shown). The bacterial growth rate on plant #295 was similar to that on IR-BB1, which was 100-fold lower than that on Nipponbare (Fig. 2B). To confirm that the resistance to Xoo was because of the introduction of cosmid 3-2, plant #295 was crossed with Nipponbare and 20 resulting BF1 plants were analyzed by Southern hybridization and bacterial inoculation. The 11 BF1 plants that inherited the introduced 6.5 kb _Bam_HI-_Bam_HI fragment at a single locus were resistant to T7174. The nine other plants, lacking the introduced fragments, were susceptible. Thus, Xa1 was confined to a 35-kb region of cosmid 3-2.
Figure 2.
(A) Race-specific resistance to Xoo conferred by the cloned Xa1 gene. #295 is a transformant made by introduction of a cosmid-containing Xa1 gene into a susceptible line, Nipponbare. The leaves were inoculated with the Japanese Xoo race 1 strain T7174, race 2 strain T7147, and race 3 strain T7133 by the clipping method (19). Photograph taken 14 days after inoculation. (B) Growth of Xoo strain T7174R, a spontaneous, rifampicin-resistant mutant from T7174 (Japanese race 1), in leaves of transgenic rice containing Xa1 (#295), IR-BB1 with Xa1, and the control line Nipponbare. The bacterial populations were determined from two to three leaf samples at each time point. Leaf tips were ground, suspended in sterilized water, and plated on peptone-sucrose media containing 20 mg/ml rifampicin and 100 mg/ml cycloheximide. Data points are the means. The standard deviations of the means are shown by vertical bars (not shown if smaller than the symbols).
The 13.5-kb _Eco_RI-_Bam_HI fragment of cosmid 3-2 contains the entire c42-coding region and no other possible ORFs. To confirm that the c42 ORF was responsible for Xa1 activity, the 13.5-kb fragment was cloned into the vector CLD04541 (Fig. 1B), and the resulting clone (CLD04541XA) was introduced into susceptible rice cv., Kitaake, by the PEG-mediated method. Approximately 1,200 hygromycin-resistant plants were grown and tested by inoculation at 3 months after transformation. Among them, 14 transformants were resistant to T7174 and carried the introduced 13.5-kb _Eco_RI-_Bam_HI fragment. Thus, we concluded that c42 corresponds to the functional Xa1 gene.
The specificity of the transformants containing Xa1 to confer resistant interactions with race 1 strains of Xoo was examined by inoculation #295 and 14 CLD04541XA-transformants with strains of two races that should be virulent to Xa1 (races 2 and 3). The transformants were resistant to race 1 strain T7174 but were susceptible to strains T7147 (race 2) and T7133 (race 3) (Fig. 2A), indicating that the race specificity of the transformants was consistent with that of IR-BB1.
Structure of the Xa1 Gene: Xa1 Has NBS Motifs and a Unique LRR Domain.
The composite nucleotide sequence of the Xa1 cDNA encoded a 5,406-bp ORF that was flanked by 5′ and 3′ untranslated regions of 112 and 392 bp, respectively. The nucleotide sequence of the 13.5-kb _Eco_RI-_Bam_HI genomic fragment (Fig. 1B) was also determined and compared with that of the cDNA sequence. The Xa1 gene was composed of four exons separated by three introns (Fig. 1C). The derived 1,802-aa sequence for the Xa1 gene harbored several regions with similarity to deduced polypeptide domains of other R genes, RPS2 (13, 14), RPM1 (15), N (17), and _L_6 (8): two motifs of NBS [amino acids 326–334 (P-loop) and 399–408 (kinase 2a)], three “conserved domains” indicated in (15) [amino acids 438–449 (kinase 3a?), 501–513, and 557–564], and an LRR from amino acids 1,093–1,650 (Fig. 3A). The Xa1 LRR is composed of six almost perfect repeats, each 93 aa long. In each repeat unit, six occurrences of a consensus sequence (LXXLXL/IXXN/CXX) were found (Fig. 3B). Of six direct-repeat units, the first to fifth units are almost identical not only at the amino acid level (92–99% simple homology), but also at the nucleotide sequence level (97–99%). The sixth unit is less similar to the other five units: 62–67% similarity to the amino acid sequence and 73–75% at the nucleotide sequence level. In addition, at amino acid positions 3 to 8 of each LRR unit, there is a sequence, GHGEDG, matching the corresponding consensus NBS sequence (GXGXXG) (Fig. 3B). Beside these structures, the putative XA1 protein contains 22 potential N-linked glycosylation sites [NX(S/T)] (Fig. 3A). Six of these sites are at amino acid position 36 of each LRR repeat (Fig. 3B). These structural characteristics suggest that the Xa1 product interacts with other proteins in a defense reaction–signal transduction pathway (5).
Figure 3.
Deduced amino acid sequence of the Xa1 gene product. (A) NBS domains with significant homology to other resistance genes (15) are bold and underlined; “conserved domains” indicated in (15) are underlined; LRR are in italics; asparagines (potential N-linked glycosylation sites) are in bold. (B) LRR with consensus sequences listed at bottom. Leucines, asparagines, and cysteines matching with the consensus sequences are in bold; the NBS domains are bold and underlined. Numbers on the left indicate the positions of residues in the Xa1 amino acid sequence. The GenBank accession number for Xa1 genomic and amino acid sequences is AB002266.
Xa1 Is a Single-Copy Gene.
Southern hybridization analysis was carried out by using the genomic DNAs from resistance line IR-BB1 and susceptible isogenic line IR24. The NBS region of Xa1 hybridized to a single band of DNA from IR-BB1 and IR24, suggesting that Xa1 is a single-copy gene and IR24 also has a sequence homologous to Xa1 at the same locus.
The Expression Pattern of the Xa1 Gene Was Investigated by Northern Blot Analysis.
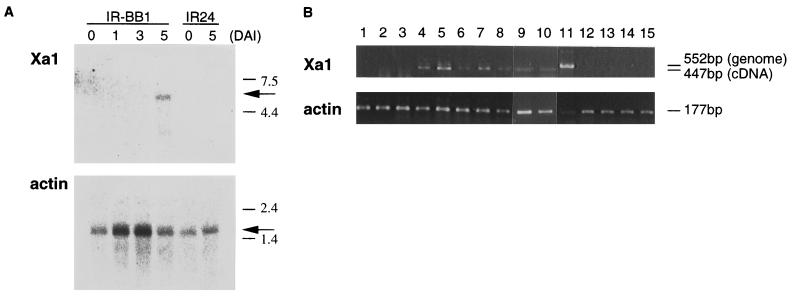
We investigated the expression pattern of Xa1 by Northern blot analysis. IR-BB1 and IR24 were inoculated with the incompatible strain, T7174. Xa1 mRNA was detected only in the IR-BB1 sample taken 5 days after inoculation (DAI). No signal could be detected immediately after inoculation (IAI), 1 and 3 DAI of IR-BB1 and IAI, and 5 DAI of IR24 (Fig. 4A). With RT-PCR, a Xa1 cDNA-specific, 447-bp fragment was detected in IR-BB1 from 3 DAI and continuously thereafter up to 15 DAI, although no such fragment was detected in IR24 (Fig. 4B). Further, the expression of Xa1 in IR-BB1 inoculated with water and compatible strain T7133 was also analyzed by RT-PCR. At 5 DAI, Xa1 mRNA was detected in both water and T7133-inoculated plants. Thus, Xa1 expression was induced by wounding and pathogen inoculation in resistant plant IR-BB1. As shown in Fig. 2B, significant differences in bacterial growth rates between resistant and susceptible plants were detected from 4 DAI. The timing of this bacterial growth limitation is consistent with the pattern of accumulation of Xa1 mRNA, causing us to speculate that the induction of expression is involved in enhancement of resistance to the pathogen, although the mechanism inducing Xa1 expression remains unknown.
Figure 4.
Expression of the Xa1 gene. (A) Northern blot analysis of Xa1 mRNA. Four microgram aliquots of poly(A)+ RNA were extracted from leaf tip tissue of IR-BB1 and IR24 at 0 (IAI), 1, 3, and 5 DAI, and 0 (IAI) and 5 DAI, respectively. The blot was hybridized with 32P-labeled c42-1 and actin probes, and positive signals were analyzed by using a BS2000 Bio-imaging Analyzer (Fuji). Size markers (RNA ladder, GIBCO/BRL) are shown in kilobases. (B) Xa1 cDNA detection by RT-PCR. Total RNA was extracted from leaf tip tissue of IR-BB1 and IR24 inoculated by incompatible strain T7174 at 0 (IAI), 1, 2, 3, 5, 7, 10, and 15 (lanes 1–8), and 0 (IAI), 3, 5, and 10 DAI (lanes 12–15), respectively. Total RNA from IR-BB1 of 5 DAI with water (lane 9) and compatible strain T7133 (lane 10) was also analyzed. Genomic DNA (lane 11) was extracted from IR-BB1 (26). To prevent amplification of contaminant genomic DNA in total RNA samples, _Xa1_-specific primers were designed for both sides of the second intron (105 bp). Thus, the expected lengths of amplified products from total RNA and genomic DNA were 447 and 552 bp, respectively.
DISCUSSION
In this study, we used a map-based cloning strategy to isolate Xa1, one of the BB-resistance genes in rice. In the complementation test of the Xa1 candidate genomic sequences, we used the PEG-mediated method. Although 1,700 hygromycin-resistance plants were regenerated, only 15 BB-resistance plants were obtained. Because most of BB-susceptible plants with hygromycin resistance were not analyzed by Southern hybridization, whether low efficiency of regeneration of BB resistant plants is because of low transformation efficiency is unclear. However, among 10 BB-susceptible regenerated plants analyzed, one plant showed multiple introduced fragments but the others had no introduced fragment. We supposed that cotransformation could lead to a lower number of cosmid 3-2 and CLD04541XA than the plasmid harboring the hygromycin-resistance gene.
So far, one other BB-resistance gene, Xa21, has been isolated from rice (16). XA21 was predicted to be a transmembrane protein containing extracellular LRR and a cytoplasmic kinase domain (16, 30). It is assumed that the extracellular LRR of XA21 constitute the most probable domain that could participate in the protein–protein interactions that might affect pathogen recognition (16). However, the predicted protein structure of the Xa1 gene is quite different from that of the Xa21 gene. The Xa1 gene product consists of NBS regions at the amino-terminal side and LRR at the carboxyl-terminal side, but no distinct transmembrane domain. Thus, the Xa1 gene belongs to the NBS-LRR class of plant disease-resistance genes and is believed to encode a cytoplasmic protein. We speculate that the interaction between the Xa1 gene product and a ligand that may be specified by the avr genes for Xoo races may occur in the cytoplasm at sites like RPS2 and RPM1 (31), although the Xa21 gene product has an extracellular domain. The significant difference between the two gene products implies that there are at least two different ligand-recognition systems for specific Xoo races in rice. On the contrary, the tomato-resistance genes Cf-9 and Cf-2 (6, 7) confer resistance to different races of the same pathogen, Cladosporium fuluvum. These genes encode similar structures that are putative transmembrane receptors with a large extracytoplasmic LRR domain. The remarkable homology in their C-terminal portions and in LRR implies that the extracellular LRR may interact with extracellular elicitors. At the interaction between tomato and C. fuluvum, it is suggested that the genes Cf-9 and Cf-2 recognize avr gene products by a common mechanism (7, 30).
The C-terminal half of Xa1 gene is composed of LRR, the common motif of the several classes of resistance genes. The LRR of other R-genes, such as RPS2 (13, 14), RPM1 (15), N (17), Cf-9, Cf-2 (6, 7), and Xa21 (16), are composed of imperfect repeats, 20–26 aa in length. The leucine-rich region in _L_6 (8) contains two direct repeats of 146 and 148 aa with 74% identity. In contrast, the Xa1 LRR is unique in that it is composed of six almost perfect repeats, each 93 aa long, with 62–99% simple homology at the amino acid level, to each other. Specially, the first to fifth units showed 97–99% simple homology at the nucleotide sequence level. The conservation of Xa1 LRR suggests that the duplication of the 93-aa unit occurred recently during the evolution of this gene. Recently, Ellis et al. (32) suggested that the 3′ half of the rust-resistance genes in L locus of Flax, containing LRR, is important in specificity control by the analysis of in vivo intragenic recombinant alleles. The duplication of the LRR units in Xa1 may be involved in the evolution of the race-specific pathogen recognition by Xa1 gene product.
So far, there is no report of induced expressed R gene. Interestingly, Xa1 mRNA was detected from rice leaves at 5 days after cutting with water and inoculation of both the compatible and incompatible strains of Xoo, but was not detected in intact leaves. These findings suggested that the Xa1 gene expression may be induced by stimulus of wounding involved in pathogen infection, and accumulation of Xa1 gene product may lead to high efficiency of interaction with avr gene product. This interaction may activate the signal transductions involved in _Xa1_-madiated BB resistance. Thus, it is likely that Xa1 plays an important role in pathogen recognition, which is supported by the analysis of race-specific resistance of the Xa1 transformants. Which factors regulate the expression of Xa1 remains an interesting question.
We believe that accumulation of information about resistance gene-mediated host defense mechanism will facilitate engineering of genes conferring durable resistance to a broad spectrum of pathogens.
Acknowledgments
We thank Yosuke Umehara, Hiroshi Tanoue, Yoshihide Kuboki, and Ayahiko Shomura for technical assistance, Jonathan D. G. Jones for providing CLD04541, Chuanyin Wu and Fumio Takaiwa for transformation, Norio Matsushima for sequence analysis, and Yuzo Minobe, Koichi Hasegawa, and Kaetsu Kobayashi for establishing collaborations. This research was supported by the Japanese Ministry of Agriculture, Forestry and Fisheries, and the Japan Racing Association. S.Y. was supported by fellowships from the Japan Society for the Promotion of Science for Japanese Junior Scientists and the Japan Research and Development Corporation.
ABBREVIATIONS
BB
bacterial blight
NBS
nucleotide binding sites
LRR
leucine-rich repeat(s)
Xoo
Xanthomonas oryzae pv. oryzae
DAI
days after inoculation
IAI
immediately after inoculation
PEG
polyethylene glycol
RACE
rapid amplification of cDNA ends
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB002266).
References
- 1.Mew T W. Annu Rev Phytopathol. 1987;25:359–382. [Google Scholar]
- 2.Kinoshita T. Rice Genet Newsl. 1992;8:2–37. [Google Scholar]
- 3.Khush G S, Mackill D J, Sidhu G S. Bacterial Blight of Rice. Manila, Philippines: International Rice Research Institute; 1989. pp. 207–217. [Google Scholar]
- 4.Sakaguchi S. Bull Natl Inst Agr Sci Ser. 1967;D16:1–18. (in Japanese with English summary). [Google Scholar]
- 5.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 6.Jones D A, Thomas C M, Hammond-Kosack P J, Balint-Kurti K E, Jones J D G. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- 7.Dixon M, Jones D A, Keddile J S, Thomas C M, Harrison K, Jones J D G. Cell. 1996;84:451–459. doi: 10.1016/s0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence G J, Finnegan E J, Ayliffe M A, Ellis J G. Plant Cell. 1995;7:1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson P A, Lawrence G J, Morrish B C, Ayliffe M A, Finnegan E J, Ellis J G. Plant Cell. 1997;9:641–651. doi: 10.1105/tpc.9.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. Plant Cell. 1997;9:521–532. doi: 10.1105/tpc.9.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker J E, Coleman M J, Szabo V, Frost L N, Schmidt R, van der Biezen E A, Moores T, Dean C, Daniels M J, Jones J D G. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin G B, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 13.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Stascawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 14.Mindrinos M, Katagiri F, Yu G-L, Ausubel M. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 15.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangle J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 16.Song W Y, Wang G-L, Chen L-L, Kim H-S, Pi L-Y, Holsten T, Gardner J, Wang B, Zhai W-X, Zhu L-H, Fauquet C, Ronald P. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 17.Whitham S, Dinesh-Kumar S P, Choi D, Hehl R, Corr C, Baker B. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 18.Cai D, Kleine M, Kifle S, Harloff H-J, Sandal N N, Marcker K A, Klein-Lankhorst R M, Salentijn E M J, Lange W, Stiekema W J, Wyss U, Grundler F M W, Jung C. Science. 1997;275:832–834. doi: 10.1126/science.275.5301.832. [DOI] [PubMed] [Google Scholar]
- 19.Kauffman H E, Reddy A P K, Hsieh S P Y, Merca S D. Plant Disease Rep. 1973;57:537–541. [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Toki S, Takamatsu S, Nojiri C, Ooba S, Anzai H, Iwarta M, Christensen A H, Quail P H, Uchimiya H. Plant Physiol. 1992;100:1503–1507. doi: 10.1104/pp.100.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negrutiu I, Shillito R D, Potrykus I, Biasini G, Sala F. Plant Mol Biol. 1987;8:363–373. doi: 10.1007/BF00015814. [DOI] [PubMed] [Google Scholar]
- 23.Goto F, Toki S, Uchimiya H. Transgen Res. 1993;2:300–305. [Google Scholar]
- 24.Datta S K. In: Gene Transfer to Plants. Potrykus I, Spangenberg G, editors. Berlin: Springer; 1995. pp. 66–74. [Google Scholar]
- 25.Wu C, Shimamoto K. In: Gene Transfer to Plants. Potrykus I, Spangenberg G, editors. Berlin: Springer; 1995. pp. 93–98. [Google Scholar]
- 26.Murray M G, Thompson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshimura S, Umehara Y, Kurata N, Nagamura Y, Sasaki T, Minobe Y, Iwata N. Theor Appl Genet. 1996;93:117–122. doi: 10.1007/BF00225736. [DOI] [PubMed] [Google Scholar]
- 28.Churchill G A, Giovannoni J J, Tanksley S D. Proc Natl Acad Sci USA. 1993;90:16–20. doi: 10.1073/pnas.90.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuboki Y, Yoshimura S, Yano M. Rice Genome. 1995;4:3. [Google Scholar]
- 30.Boyes D C, McDowell J M, Dangl J L. Curr Biol. 1996;6:634–637. doi: 10.1016/s0960-9822(09)00435-7. [DOI] [PubMed] [Google Scholar]
- 31.Jones J D G. Nature (London) 1997;385:397–398. [Google Scholar]
- 32.Ellis J, Lawrence G, Ayliffe M, Anderson P, Collins N, Finnegan J, Frost D, Luck J, Pryor T. Annu Rev Phytopathol. 1997;35:271–291. doi: 10.1146/annurev.phyto.35.1.271. [DOI] [PubMed] [Google Scholar]