Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth (original) (raw)
Abstract
We have developed a microfluidic device that allows the isolation and genome amplification of individual microbial cells, thereby enabling organism-level genomic analysis of complex microbial ecosystems without the need for culture. This device was used to perform a directed survey of the human subgingival crevice and to isolate bacteria having rod-like morphology. Several isolated microbes had a 16S rRNA sequence that placed them in candidate phylum TM7, which has no cultivated or sequenced members. Genome amplification from individual TM7 cells allowed us to sequence and assemble >1,000 genes, providing insight into the physiology of members of this phylum. This approach enables single-cell genetic analysis of any uncultivated minority member of a microbial community.
Keywords: environmental microbiology, metagenomics, microfluidics, single-cell analysis
The earth contains enormous microbial diversity. Microbes colonize a wide variety of environmental niches, creating complex ecosystems and communities. Despite the marvelous progress in microbiology over the past century, we have only scratched the surface of this microbial world. It has been estimated that <1% of bacterial species have been axenically cultured, and fewer than half of the recognized bacterial phyla include cultivated representatives (1). This can be viewed as biology's “dark matter” problem: just as astronomers can only indirectly infer the existence of a large amount of as-yet-undetected mass in the universe, microbiologists can only estimate microbial diversity by using techniques such as comparative 16S ribosomal RNA (rRNA) gene analysis (2), community DNA hybridization efficiency (3), and metagenomic gene inventories (4, 5). Although these techniques are useful, the cell, which is the ultimate unit of biological organization, is lost as a distinct informational entity.
Two general approaches have been used in addressing this problem. The first is to work on simple communities that contain only a few microbial species, in which case genome sequences can be reconstructed computationally after sequencing bulk DNA purified from the community (4). The second approach is to isolate individual cells by fluorescence-activated cell sorting, micromanipulation, or serial dilution, followed by genomic DNA amplification using techniques such as multiple-strand displacement amplification (MDA) (6, 7). The latter approach has been used successfully to perform genomic analysis of the cultivated and abundant marine bacterium Prochlorococcus MIT9312 (7). However, this approach remains difficult for two primary reasons: (i) the confidence needed to assert the presence of single cells in microliter volumes and (ii) the meticulous reagent cleaning and sample handling required to suppress background amplification in microliter-volume MDA (6). Those hurdles become even greater when complex environmental samples are used. The number of species present requires substantial reagent consumption and expensive postamplification screening, and the probability of contamination is much higher because of the presence of free DNA. Neither approach has been validated with a complex ecosystem.
We have designed and fabricated a microfluidic chip to address these limitations. This device provides the ability to perform parallel isolation of single bacteria by steering them to any one of eight individually addressable chambers, followed by lysis and amplification of their individual genomes in 60-nl volumes. By using nanoliter volumes, the specific template concentration is increased by three orders of magnitude, as suggested previously (8, 9). To demonstrate the potential of this approach in microbial ecology, we performed a selective survey of microbes found in the human subgingival crevice, followed by whole-genome amplification (WGA) and high-throughput sequencing. The 16S rRNA gene-based phylogeny of several of these microbes placed them within the candidate phylum TM7, for which no cultivated or sequenced members exist (10), thereby providing unique genetic information about oral representatives of the TM7 phylum.
Results and Discussion
The microfluidic strategy for microbe isolation and genome amplification (Fig. 1) was validated on Escherichia coli. More than two dozen amplifications on single E. coli cells were performed, with a success rate of >90%. Subsequent PCR analysis of 10 genomic loci distributed over the E. coli chromosome showed that the amplification achieved excellent coverage and was able to amplify sequences with equal efficacy independent of their location on the genome [see supporting information (SI) Fig. 4]. Control experiments with only culture fluid in the chamber showed no significant amplification.
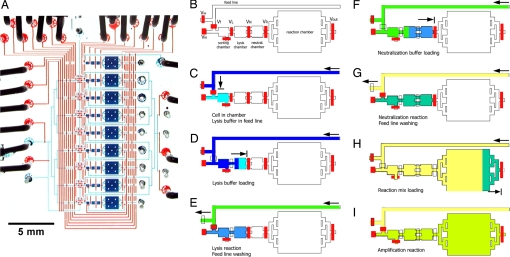
Fig. 1.
Single-cell genome amplification device. (A) Photograph of a single-cell isolation and genome amplification chip capable of processing eight samples in parallel. To visualize the architecture, the channels and chambers have been filled with blue food coloring, and the control lines to actuate the valves have been filled with red food coloring. (Scale bar, 5 mm.) (B) Schematic diagram of a single amplification unit. The feed line is used to bring reagents into the chambers when the VR valve is open and to the waste when the Vw valve is open. The Vin valve allows deposition of a single bacterium into the sorting chamber. The lysis (3.5 nl), neutralization (3.5 nl), and reaction chambers (50 nl) are used in sequence and are separated by individual valves VL, VN, and VR, respectively. Valve Vout allows recovery of the amplified genomic material from the chip into an individual microfuge tube. (C) After a cell is trapped in the chamber, the feed line is filled with lysis buffer. (D) The lysis buffer is used to push the cell into the lysis chamber. (E) While the lysis buffer is mixing with the cell solution by diffusion, the feed line is flushed. (F) Neutralization buffer is loaded into the feed line and used to push the cell lysate into the neutralization chamber. (G) While the neutralization reaction is mixing by diffusion, the feed line is flushed. (H) The WGA reagents are loaded into the feed line and used to push the neutralized cell lysate into the reaction chamber. (I) The amplification reaction proceeds in a closed system comprising sorting, lysis, neutralization, and reaction chambers.
We then demonstrated the ability to select, isolate, and amplify the genomes of single bacteria from the human oral microbiota. The number of species in the human mouth is estimated to be ≈700 (11, 12). Because of the challenges of removing intact biofilm samples, rather than performing a comprehensive survey of this complex community our purpose was instead to target an unexplored phylum and a relatively rare subset of the oral microbiota, TM7. By selecting microbial cells with a rod-like morphotype, we expected to enrich for the candidate phylum TM7 (13, 14). Little is known about the TM7 lineage. On the basis of comparative analyses of 16S rRNA genes, TM7 is one of a number of prominent “candidate” bacterial phyla lacking any cultivated representatives but comprising >50 phylotypes (1). rRNA gene sequences from the TM7 phylum have been found in a variety of habitats, ranging from deep sea hydrothermal vents to the healthy human mouth (10, 13, 14). In addition, sequence types within this phylum have been associated with chronic periodontitis in humans (13, 14). Fluorescence in situ hybridizations specific for TM7 showed that 0.7–1.9% of the subgingival microbiota belong to the TM7 phylum (13). A significant subset of this phylum has a peculiar morphology, characterized by long, thick filaments (up to 50 × 4 μm), making these cells good candidates for a morphotype-based selection (10, 13).
To identify the isolated rod-like cells, we performed PCR on the 16S rRNA gene from amplified genomic DNA, using primer sequences conserved across most species of the bacterial domain. Positive results for 16S rDNA PCR were obtained for 34 of 35 captured, single cells. After gel purification, 30 of these amplicons were directly sequenced; 28 gave unique sequences that were compared against the National Center for Biotechnology Information database by using BLAST (15). Fig. 2 shows a phylogenetic tree based on 16S rRNA gene sequences of most recognized bacterial phyla, with annotations for isolates from the present survey. The 28 sequences from this study are associated with five different bacterial phyla, with most sequences located in the phylum Fusobacteria and specifically related to the genus Leptotrichia.
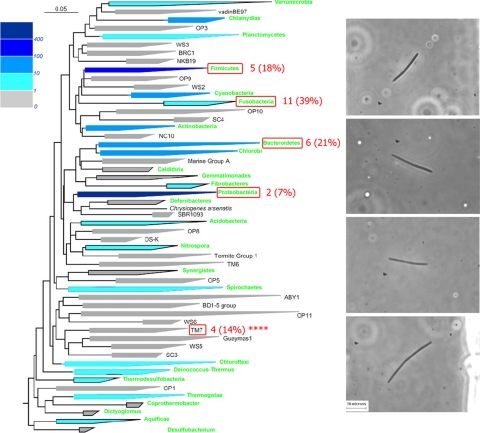
Fig. 2.
Results of rod-like morphotype survey. (Left) Phylogenetic tree showing bacterial phyla based on 16S rRNA gene analysis (adapted from ref. 1). Green text indicates that at least one member of the phylum has been cultivated; different shades of blue indicate the number of genome sequencing projects in a particular phylum that were completed or in progress as of May 2006. Red numbers and percentages indicate the results of our single-cell survey of the human subgingival crevice, in which filamentous bacteria with rod-like morphotypes were isolated and lysed and their genomes were amplified. [Reprinted with permission from ref. 1 (Copyright 2003, Annual Reviews, www.annualreviews.org).] (Right) Optical micrographs of the four TM7 cells isolated in this survey.
We identified four members of the phylum TM7 from the amplified cells, of which three were closely related to a known oral TM7 clone (>99.6%; GenBank accession no. AY144355) (14) and a fourth clone related to a more distant lineage in the phylum (97.3%; AY134895) (16). To verify that the genome of a unique sequence type was amplified, the 16S rRNA amplicon of one TM7 sample (TM7a) was cloned, and 24 clones were sequenced; 23 of these had >99.5% sequence identity to the directly sequenced PCR product. To provide insight into the biology of the TM7 phylum and to investigate the ability to recover whole genome sequences from single uncultivated cells, we used the amplified genomic DNA from this sample for pyrosequencing and genome assembly. The resulting genome sequence data set was loaded into the Integrated Microbial Genomes with Microbiomes (IMG/M) database (17) to facilitate comparative analysis.
The assembly of TM7a genomic sequence resulted in the generation of 3,245 genes and gene fragments distributed across 1,825 scaffolds, totaling 2.86 megabases (Mb). Genome size estimates based on approaches such as the Lander–Waterman equation (18) or the characterization of known, conserved, single-copy genes (19) rely on random sampling of the genome. Single-cell amplification introduces a bias in read sampling such that we could not reliably estimate TM7 genome size. The assembly was fairly fragmented, with only 60% of the genes on multigene scaffolds. This suggested that there is multiple representation of some genes in the assembly and that the actual number of sampled genes in TM7a is somewhat smaller. If one applies a more conservative filter and only includes genes from large contigs (defined as those having three or more genes), then one is left with 1,474 genes on 288 scaffolds. This is probably a better estimate of the number of unique sampled genes in TM7a. Approximately 43% of genes were assigned a predicted function based on homology to published sequences, and 44% of the genes were mapped to clusters of orthologous groups (Table 1). We tested the validity of the assembly by choosing five regions of the genome with an average size of 1 kb. We designed PCR primers and successfully amplified all five regions from aliquots of the amplified TM7a genomic DNA (see SI Fig. 5).
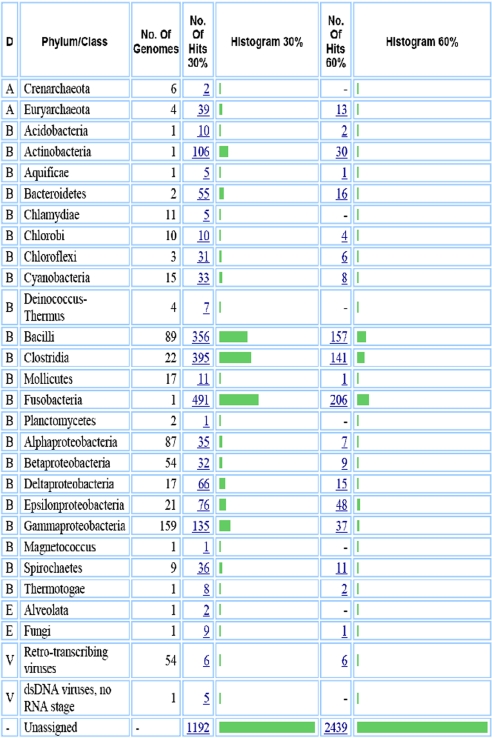
Table 1.
Statistics characterizing TM7a assembly and annotation, derived from IMG/M
| Item | No. | % of total |
|---|---|---|
| DNA bases | ||
| Total | 2,864,887 | 100.0 |
| Coding | 1,160,954 | 40.5 |
| G + C | 981,862 | 34.3 |
| DNA scaffolds | 1,825 | 100.0 |
| Genes | ||
| Total | 3,245 | 100.0 |
| Protein coding | 3,160 | 97.4 |
| With function prediction | 1,389 | 42.8 |
| Without function prediction | 1,771 | 54.6 |
| Assigned to enzymes | 530 | 16.3 |
| Connected to KEGG pathways | 400 | 12.3 |
| Not connected to KEGG pathways | 2,760 | 85.1 |
| In clusters of orthologous groups | 1,422 | 43.8 |
| In protein families | 1,221 | 37.6 |
Sequence similarity-based mapping showed that most of the TM7a genes are not closely related to genes from representatives of any known phyla. For example, 80% of the predicted TM7 proteins have <60% sequence identity to proteins from other sequenced organisms (Table 2). With this approach, a full third (33%) of the TM7 genes have <30% protein sequence identity to genes from any known phylum. This result is consistent with other cases of genome sequencing in previously uncharacterized phyla. For example, _Rhodopirellula baltica_ was the first sequenced representative of the _Planctomycetes_ phylum, and 89% of its proteins have <60% identity to proteins from other known organisms; 20% have no matches with >30% identity. In contrast, a survey of 13 bacterial species in phyla with multiple sequenced representatives showed that, on average, only 15% of the proteins have <60% identity to proteins in other organisms, and 3% are unassigned at the 30% cutoff (see SI Table 3).
Table 2.
BLAST-based mapping of the genes in the TM7a assembly by using IMG/M shows that the majority of TM7a genes are unlike those of any previously sequenced organism
Although the majority of genes in the TM7a assembly are only distantly related to genes found in other organisms, a minority have a relatively high sequence similarity (>60% identity) to genes found in members of the classes Bacilli, Clostridia, or Fusobacteria. The presence of these genes may be the result of extensive lateral transfer between species in the mouth, as has been postulated for other oral bacteria (20), or may be due to the presence of contaminating DNA in our samples, perhaps from free DNA that entered the microfluidic amplification reactor with the TM7 cell, either in solution or bound to the cell membrane. If the presence of these genes was due to contaminant DNA, one would expect them to cluster together by organism in the assembly. The data show that in many cases the opposite is true; genes with putative relationships to disparate organisms assemble onto the same contig. The TM7a assembly does contain at least some exogenous DNA. Examination of the raw sequencing reads shows that >40 reads assembled into the TM7a 16S rRNA gene sequence, whereas 4 reads assembled onto a separate small contig with the 16S rRNA gene sequence belonging to Leptotrichia species. Extrapolating from the ratio between these raw reads, we estimate that the proportion of Leptrichia contamination is <10%. Because it is difficult to assign a more precise estimate, one avenue of analysis is to interpret the TM7a assembly as a metagenome that is highly enriched for a TM7 bacterium.
We also sequenced a second TM7 cell, TM7b, with an identical 16S rRNA gene sequence to TM7a, that had been isolated on a separate day on a separate chip. Ten megabases of sequence data were obtained, which was not enough to provide a complete assembly but does represent a broad sampling of the genome. These sequence data were analyzed with BLAST (15) to independently confirm the TM7a genome assembly and to facilitate identification of bona fide TM7 genes. The results are shown in Fig. 3. The vast majority of TM7b sequence reads could be mapped to contigs in TM7a with high statistical significance. As a control experiment, we also aligned the TM7b reads to Fusobacterium nucleatum (the only sequenced organism in the phylum Fusobacteria, to which Leptotrichia belongs) and Chloroflexus aurantiacus (the sequenced organism with the closest 16S rRNA gene sequence to TM7 in Fig. 2). Neither of the latter demonstrated substantial sequence identity to the TM7b sequence assembly. Sequencing multiple representatives of an unexplored phylum is, therefore, a useful approach for identifying bona fide target phylum genes in metagenomic samples containing exogenous DNA, which may be an unavoidable limitation associated with amplification of single cells removed from multispecies samples.
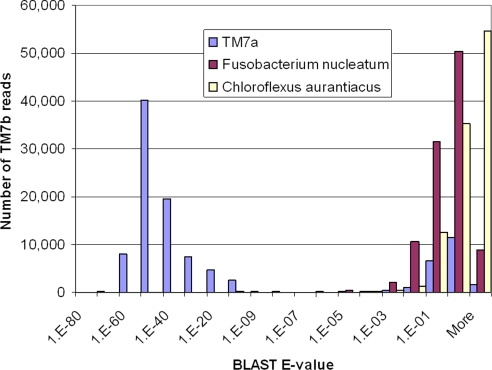
Fig. 3.
TM7b has much higher sequence similarity to the TM7a assembly than to the F. nucleatum or C. aurantiacus genomes. Mapping was performed by using BLAST (21); ≈100,000 individual sequence reads with average length 104 bp were mapped onto each genome. The histogram shows E-values returned by BLAST, which indicate the statistical significance with which the read can be mapped onto the genome.
Metabolic analysis of TM7 was performed by pooling sequence data from TM7a and TM7b, along with data from a third TM7 cell (TM7c). TM7c assembled into 474 kb and 632 genes but was not used as an independent reference because a sample-handling error during sequencing caused commingling with genomic DNA from TM7a. We binned the metagenome on the basis of similarities between the three TM7 samples and phylogenetic markers by selecting contigs that have phylogenetically unique marker genes. On the basis of the presence of recognizable signature genes, the oral TM7 cells are predicted to be capable of a range of common metabolic processes, such as glycolysis (3-phosphoglycerate kinase, phosphoglycerate mutase triosephosphate isomerase, and pyruvate kinase), the tricarboxylic acid cycle (succinyl-CoA synthetase), nucleotide biosynthesis (dihydroorotate dehydrogenase, uridylate kinase, guanylate kinase, aerobic-type ribonucleoside diphosphate reductase, and thymidylate synthase), and some amino acid biosynthesis and salvage pathways (cysteine synthase and glycine hydroxymethyltransferase). We identified several genes coding for glycosyl hydrolase family enzymes distantly related to α-amylases and oligo-1,6-glucosidases, suggesting that oral TM7 cells may be capable of using oligosaccharides as growth substrates. Arginine is another potential growth substrate because of the presence of genes from the arginine deiminase pathway (arginine deiminase, ornithine carbamoyltransferase, and carbamate kinase). We also identified genes for ABC transporters that are likely responsible for oligopeptide uptake, suggesting that TM7 cells may be capable of using other amino acids as well.
It is an open question whether these bacteria have attributes associated with virulence and might be capable of contributing to oral disease. We noted the presence of genes for type IV pilus biosynthesis, including one similar to that which encodes the Vibrio vulnificus type IV pilin (21). Although type IV pili may facilitate the adherence of bacteria to epithelial cells, and contribute to biofilm formation, in Gram-positive cells, type IV pili have been shown to be responsible for an unusual communal form of gliding motility (22). TM7 cells from a sludge bioreactor appeared to have typical Gram-positive cell envelopes by electron microscopy (10). Therefore, if the TM7 are Gram-positive, their type IV pili may be involved in gliding motility.
We also investigated genes that might participate in cell envelope biosynthesis and found a gene predicted to encode a novel sortase, distantly related to those of Firmicutes and Actinobacteria, and a gene predicted to encode a UDP-_N_-acetylmuramyl tripeptide synthetase related to those of the bifidobacteria, suggesting a specific relationship of the TM7 cells to the Gram-positive lineages (see SI Fig. 6). Interestingly, in bifidobacteria the latter enzyme is predicted to add an atypical amino acid (ornithine or lysine instead of the more common diaminopimelate) to the growing peptidoglycan chain producing an A4α/β type peptidoglycan. This peptidoglycan type has been implicated in chronic granulomatous inflammation (23) and may serve as a virulence factor for oral TM7.
In conclusion, we have isolated single bacterial cells from a complex human microbial community and sequenced their DNA to provide genetic insights into the TM7 phylum. The cell selection process described here used morphology as the basis for selection of the targeted bacteria. It would also have been possible to achieve the same results from an unbiased survey of the environmental sample; this simply would have required processing of a larger number of cells. Given that the cells were isolated from a complex bacterial biofilm (24) with no manipulation other than pipetting and dilution, many environmental microbial ecosystems should be amenable to this technique. We predict that, as genomes from the microbial dark matter are sampled by using techniques such as single-cell amplification, a much richer tapestry of microbial evolution will emerge.
Materials and Methods
Microfluidic Chip Fabrication.
Microfluidic chips (Fig. 1) were fabricated as described previously (25), using the “push up” geometry with the following adjustments. The flow molds contained two layers: one for feeding lines and valves (SPR220; 7 μm high) and one for the reaction chambers (SU8 2025; 25 μm high). The control molds contained two layers: one layer for hydration channels under the reaction chamber (SU8 2015; 10 μm high) and one for the control lines (SU8 2025; 25 μm high).
Sample Collection and Isolation.
Samples were collected from periodontal pockets by scraping subgingival tooth surfaces of a healthy individual (male, 40 years of age) after 5 days without tooth brushing. These biofilm specimens were dispersed, suspended, and washed twice in 1× PBS buffer and then resuspended in 1× PBS 0.2% Tween 20 before loading onto the chip. The chip was placed on an optical microscope, and the sample was pumped through a sorting channel. When a single rod-shaped cell or a filament with the appropriate morphology (13) was visually detected in front of each processing unit, an isolation valve was closed and the cell was examined with a higher magnification. If the cell satisfied the selection criteria, the sorting valve was opened and the cell was pumped into the sorting chamber. Otherwise, the isolation valve was reopened and another cell was selected. This operation was repeated for seven processing units of the chip; the eighth unit was used for a negative control, having only suspension fluid inside. The chip also contains an independent processor with a separate, nonaddressable input that was filled with a mixture of lysed cells as a positive control. Every template chamber was then carefully checked for the number of bacterial cells, and a high-magnification image was recorded for every cell (Fig. 2). Of 42 processing units (six chips) used, 35 contained only one visible cell or filament.
Cell Lysis and WGA.
Lysis, neutralization, and WGA were performed with the REPLI-g kit (Qiagen, Valencia, CA), using the recommended protocol except for on-chip WGA, for which the reaction mix was supplemented by 0.2% Tween 20 and one additional volume of polymerase. Once all of the chambers were loaded with cells, a 1-h-long lysozyme treatment was applied, using 1× PBS with 0.2% Tween 20 and 100 units/μl lysozyme (Epicentre, Madison, WI). This procedure was performed by taking advantage of the gas-permeability of polydimethylsiloxane to dead-end fill the feeding lines with the lysis buffer (Fig. 1C) and by opening the feeding valve to push the contents of the sorting chamber into the lysis chamber (Fig. 1D). Lysis and DNA denaturation reagents were allowed to incubate for 30 min. During this time, the feeding lines were washed first with air and then with the neutralization buffer (Fig. 1E). After completion of the lysis, the feeding valve was reopened, and neutralization buffer was pushed into the unit by dead-end filling of the neutralization chamber (Fig. 1F). After 15–20 min, the feeding line was washed again, this time with the WGA reaction mix (Fig. 1G). The feeding valve was reopened, and the reaction mix was used to dead-end fill the reaction chamber. With each WGA reaction isolated by closed valves, the chip was placed on a hotplate set at 32°C. The on-chip amplification took place for 10 to 16 h, after which samples were retrieved from the chip. The amount of amplified DNA after this step was estimated to be ≈50 ng. A second, off-chip amplification was performed with the REPLI-g kit to obtain micrograms of DNA, the amount required for sequencing.
16S rRNA Gene Amplification, Cloning, and Sequencing.
Gene PCR of 16S rRNA was performed on amplified genomic DNAs by using broad-range bacterial primers 8FM (5′-AGAGTTTGATCMTGGCTCAG-3′; adapted from ref. 26) and 1391R (5′-GACGGGCGGTGTGTRCA-3′; adapted from ref. 11). These primers amplify approximately >90% of the full-length bacterial 16S rRNA coding sequence. PCR mixtures were composed of 1× PCR buffer II (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2, 0.05% Triton X-100, 20 mM tetramethylammonium chloride, 0.1 mM of each deoxyribonucleoside triphosphate, 0.4 μM of each primer, 2.5 units of AmpliTaq DNA polymerase (Applied Biosystems), and 1 μl of amplified DNA in a final volume of 50 μl. PCRs included 5 min at 95°C and 35 cycles of 30 sec at 94°C, 30 sec at 55°C, and 90 sec at 72°C, followed by 8 min at 72°C. PCRs were sequenced (Geneway, Hayward, CA) directly after purification from agarose gel by using the QIAquick gel extraction kit (Qiagen) or after cloning by using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA).
Genome Sequencing and Assembly.
Pyrosequencing (454; Life Sciences, Branford, CT) was performed on randomly amplified genomic material from three TM7 cells: TM7a, TM7b, and TM7c. Each sequencing run yielded 10–39 Mb of raw data composed of ≈100-bp reads. The reads were assembled by using the 454 Newbler assembler and Forge whole-genome shotgun assembler (D. Platt, unpublished data). An initial assembly treating the coverage as a classic Poisson distribution indicated that the coverage of these genomes was quite uneven and that some regions were not joined because of either excess or very low coverage. The data were reassembled with Forge, using “metagenomic assumptions.” In this configuration, the assembler relaxes the Poisson depth assumption, which allows for much deeper coverage and exploration of low-coverage, less-certain overlaps between reads. All single-read, more highly error-prone contigs were excluded from the assembly. Genes were predicted on contigs greater than or equal in length to an average Sanger read (750 bp) by using fgenesb, as described previously (27), and then loaded into the IMG/M system (17) to facilitate comparative analysis.
Identification of Putative TM7 Genes.
Putative TM7 genes were identified by comparing contigs and reads from the three TM7 data sets. Contigs >750 bp from cell TM7a or TM7b with a match (blastn, _e_-value 10_e_10, low-complexity filter off) to one or more reads from a different cell (TM7b or TM7a, respectively) were assigned to the TM7 metagenome if the match was >90% identity, had an alignment length >90% of the read length, and was at least 50 bp. This reciprocal comparison was also conducted on TM7b and TM7c. The rationale behind this binning is that TM7 cells with >99% 16S rRNA identity would be the only source of orthologs (between data sets) with >90% sequence identities, because contaminating exogenous DNA would presumably be randomly “sampled” from the oral microbiota. This strict identity threshold likely means numerous, more divergent TM7 orthologs would have been excluded. A total of 386 contigs with a combined length of 963 kbp were identified as putatively originating from TM7 genomes. These contigs encoded 850 ORFs, of which 481 could be assigned a putative function.
Supplementary Material
Supporting Information
Acknowledgments
This work was supported by National Institutes of Health (NIH) Director's Pioneer Awards (to S.R.Q. and D.A.R.) and by NIH Grant 1R01 HG002644-01A1 (to S.R.Q. and Y.M.). T.L. was supported by the Stanford Dean's Postdoctoral Fellowship/Aaron Fund. This work was performed in part under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program; and by the University of California, Lawrence Livermore National Laboratory under Contract W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract DE-AC02-05CH11231, and Los Alamos National Laboratory under Contract W-7405-ENG-36.
Abbreviation
WGA
whole genome amplification.
Footnotes
The authors declare no conflict of interest.
References
- 1.Rappe MS, Giovannoni SJ. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt TM, Relman DA. Methods Enzymol. 1994;235:205–222. doi: 10.1016/0076-6879(94)35142-2. [DOI] [PubMed] [Google Scholar]
- 3.Palmer C, Bik EM, Eisen MB, Eckburg PB, Sana TR, Wolber PK, Relman DA, Brown PO. Nucleic Acids Res. 2006;34:e5. doi: 10.1093/nar/gnj007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 6.Raghunathan A, Ferguson HR, Jr, Bornarth CJ, Song W, Driscoll M, Lasken RS. Appl Environ Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 8.McBride L, Lucero M, Unger M, Nassef HR, Facer G. 2005. US Patent Appl 20050019792A1.
- 9.Hutchison CA, III, Smith HO, Pfannkoch C, Venter JC. Proc Natl Acad Sci USA. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Appl Environ Microbiol. 2001;67:411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouverney CC, Armitage GC, Relman DA. Appl Environ Microbiol. 2003;69:6294–6298. doi: 10.1128/AEM.69.10.6294-6298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Appl Environ Microbiol. 2003;69:1687–1694. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paster BJ, Russell MK, Alpagot T, Lee AM, Boches SK, Galvin JL, Dewhirst FE. Ann Periodontol. 2002;7:8–16. doi: 10.1902/annals.2002.7.1.8. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz VM, Ivanova N, Palaniappan K, Szeto E, Korzeniewski F, Lykidis A, Anderson I, Mavromatis K, Kunin V, Garcia Martin H, et al. Bioinformatics. 2006;22:e359–e367. doi: 10.1093/bioinformatics/btl217. [DOI] [PubMed] [Google Scholar]
- 18.Lander ES, Waterman MS. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 19.Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mira A, Pushker R, Legault BA, Moreira D, Rodriguez-Valera F. BMC Evol Biol. 2004;4:50. doi: 10.1186/1471-2148-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paranjpye RN, Strom MS. Infect Immun. 2005;73:1411–1422. doi: 10.1128/IAI.73.3.1411-1422.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker RA, Melville SB. Mol Microbiol. 2006;62:680–694. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- 23.Simelyte E, Rimpilainen M, Zhang X, Toivanen P. Ann Rheum Dis. 2003;62:976–982. doi: 10.1136/ard.62.10.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 26.Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia Martin H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, Yeates C, He S, Salamov AA, Szeto E, et al. Nat Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information