The Conserved Spc7 Protein Is Required for Spindle Integrity and Links Kinetochore Complexes in Fission Yeast (original) (raw)
Abstract
Spc7, a member of the conserved Spc105/KNL-1 family of kinetochore proteins, was identified as an interaction partner of the EB1 homologue Mal3. Spc7 associates with the central centromere region of the chromosome but does not affect transcriptional silencing. Here, we show that Spc7 is required for the integrity of the spindle as well as for targeting of MIND but not of Ndc80 complex components to the kinetochore. Spindle defects in spc7 mutants were severe ranging from the inability to form a bipolar spindle in early mitosis to broken spindles in midanaphase B. spc7 mutant phenotypes were partially rescued by extra α-tubulin or extra Mal2. Thus, Spc7 interacts genetically with the Mal2-containing Sim4 complex.
INTRODUCTION
The precise segregation of chromosomes is a complex process that requires the coordinated interaction between spindle and kinetochores. Kinetochores are macromolecular structures that assemble on centromeric DNA and fulfill multiple functions: they mediate the bipolar attachment of sister chromatids to spindle microtubules, maintain this attachment during dynamic microtubule behavior, and generate the spindle checkpoint signaling required for anaphase onset. These functions are essentially conserved although the composition and morphology of kinetochores can differ greatly among various organisms. In particular, the centromeric DNA requirements vary dramatically from the simple 125-base pair (bp) “point” centromeres of the budding yeast Saccharomyces cerevisiae with the three CDEI-III protein-binding motifs to the “regional” centromeres that are more complex, carry repetitive sequences, and can encompass millions of base pairs. Such centromeres exist in plants, metazoans, and fungi such as the fission yeast Schizosaccharomyces pombe (reviewed in Pidoux and Allshire, 2000, 2004; Cleveland et al., 2003). The centromere DNA of S. pombe is 40–100 kb in size and is composed of a central core region (cnt) that is flanked by inverted repeat elements (imr). These elements are surrounded by outer repeat elements (otr; reviewed in Clarke, 1998). The heterochromatic outer repeats are needed for sister centromere cohesion and might help in the orientation of the multiple kinetochore microtubule attachment sites (reviewed in Pidoux and Allshire, 2004), whereas the central region acts as a platform for the association of kinetochore complexes required for microtubule–kinetochore interaction (Saitoh et al., 1997; Pidoux et al., 2003; Hayashi et al., 2004; Kerres et al., 2004).
The simplest and best studied kinetochore, that of S. cerevisiae, contains more than 60 proteins, whereas mammalian kinetochores are predicted to contain ≥100 protein components (McAinsh et al., 2003; Fukagawa, 2004). These proteins, which exist in subcomplexes, can be classed as regulatory or structural components. The latter group of proteins are required as a connecting bridge between the centromeric DNA and the microtubules of the spindle. Interestingly, S. cerevisiae kinetochore subcomplexes that link centromere DNA-binding proteins to microtubule binding proteins have been conserved in evolution (Meraldi et al., 2006). Kinetochore components found in “point” and “regional” centromeres include the COMA complex member Mcm21p, the Ndc80, and MIND complexes and the Spc105p protein, implying the central importance of these components in kinetochore function (Ortiz et al., 1999; Wigge and Kilmartin, 2001; Euskirchen, 2002; De Wulf et al., 2003; Nekrasov et al., 2003; Westermann et al., 2003; Meraldi et al., 2006). In S. cerevisiae Ndc80, COMA and MIND complexes share a function in kinetochore capture by the side of spindle microtubules (Tanaka et al., 2005).
In S. pombe these conserved proteins are constitutive kinetochore components and exist in two biochemically separable complexes: the Mcm21p ortholog Mal2 is part of the 13 component Sim4 complex, whereas the four-component MIND subcomplex, the four-member Ndc80 complex, and the Spc105p ortholog Spc7 make up the Ndc80-MIND-Spc7 kinetochore complex (Obuse et al., 2004; Liu et al., 2005). The essential Mal2 protein associates with the central centromere region and is required for the transcriptional silencing and the specialized chromatin structure of this region (Jin et al., 2002). Mutations in mal2+ and other components of the Sim4 complex give rise to extreme missegregation of chromosomes (Saitoh et al., 1997; Jin et al., 2002; Pidoux et al., 2003; Hayashi et al., 2004; Kerres et al., 2006). Interestingly, the Sim4 complex plays a role in the incorporation of the kinetochore-specific histone H3 variant CENP-A and functions as a loading dock for the DASH complex (Takahashi et al., 2000; Pidoux et al., 2003; Liu et al., 2005; Sanchez-Perez et al., 2005). The nonessential fission yeast DASH complex is required for biorientation of sister chromatids (Liu et al., 2005; Sanchez-Perez et al., 2005). Members of the Ndc80-MIND-Spc7 complex are also associated with the central centromere and maintain the special chromatin architecture of this region but are not involved in CENP-A targeting (Goshima et al., 1999; Hayashi et al., 2004; Kerres et al., 2004; Liu et al., 2005). The Ndc80 complex in fission yeast and other organisms plays an important role in kinetochore-microtubule association and is needed for spindle checkpoint signaling (He et al., 2001; Janke et al., 2001; Nabetani et al., 2001; Wigge and Kilmartin, 2001; McCleland et al., 2003; Saitoh et al., 2005). Very recently, the Ndc80 complex and the Spc7 ortholog KNL-1 have been implicated in direct microtubule binding (Cheeseman et al., 2006; DeLuca et al., 2006). We had shown previously that Spc7 plays an important part at the microtubule–kinetochore interface as spc7+ was isolated as a suppressor of a mal3 mutant (Kerres et al., 2004). Mal3 is the fission yeast member of the EB1 microtubule-plus-end-tracking protein family, which regulates microtubule dynamics and mediates the interaction between different cellular complexes (reviewed in Gundersen and Bretscher, 2003; Mimori-Kiyosue and Tsukita, 2003; Vaughan, 2005). Mal3 is required for genome stability among others by preventing monopolar attachment of sister chromatids (Beinhauer et al., 1997; Asakawa et al., 2005). EB1 family members are targeted to kinetochores on polymerizing microtubules and play a role in kinetochore capture (Fodde et al., 2001; Kaplan et al., 2001; Tirnauer et al., 2002; Tanaka et al., 2005). Overexpression of the constitutive Spc7 kinetochore protein rescued all mitotic phenotypes of mal3 mutants and the Spc7 and Mal3 proteins interact physically. However, in contrast to the loss of the nonessential mal3+, loss of spc7+ results in inviability due to severe chromosome missegregation (Beinhauer et al., 1997; Kerres et al., 2004). This finding implies that the interaction with Mal3 is just one of the tasks of the Spc7 protein. We have thus extended our analysis of Spc7 function in mitosis.
MATERIALS AND METHODS
Strains and Media
The yeast strains used in this study are listed in Table 1. All new strains were obtained by crossing the appropriate strains followed by tetrad or random spore analysis and genotyping. At least three double mutants were tested per cross. Tetrad analysis of 16 tetrads of the cross _nuf2-1_× spc7-23 revealed that spores carrying both mutations were able to germinate and divide twice, indicating synthetic lethality. Double mutants between spc7-23 and a null allele of a component of the DASH complex, namely _duo1_Δ, were inviable. Tetrad analysis of 16 tetrads revealed that double mutants germinated and then died. Strains carrying the cold-sensitive nda3-KM311 allele were arrested by incubating them for 10 h at 20°C. Strains were grown in rich media (YE5S) or minimal media (EMM or MM) with supplements (Moreno et al., 1991). MM with 5 μg/ml thiamine repressed the nmt promoters. For high-level expression from nmt promoters cells were grown in thiamine-less media for 22–48 h at 25°C or 18–24 h at 30 or 32°C. Nitrogen starvation experiments (two independent experiments/strain) were carried out as described (Jin et al., 2002). Synchronous cultures were monitored microscopically and the mitotic index (35–40%) was determined. Resistance to G418 was tested on YE5S plates containing 100 mg/l G418; increased sensitivity to thiabendazole (TBZ) on YE5S plates containing 6–7 μg/ml TBZ. Transcriptional silencing assays were carried out as described (Jin et al., 2002; Pidoux et al., 2003).
Table 1.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| UFY1267 | h_−_mis12-537 spc7-23/his3+ade6-M216 leu1-32 | This study |
| UFY1028 | h+spc7-23/his3+his3-D1 ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1029 | h+spc7-24/his3+his3-D1 ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1027 | h+spc7-30/his3+his3-D1 ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1163 | h_−_nuf2-GFP/ura4+spc7-23/his3+ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1249 | h_−_mis12-GFP/LEU+spc7-23/his3+leu1-32 | This study |
| UFY1307 | h+mis14-GFP spc7-23/his3+ura4_−_leu1-32 | This study |
| UFY1266 | h+spc24-GFP/Kan R spc7-23/his3+_ade6-M216 his_−_leu1-32 ura4_− | This study |
| UFY1069 | h+mal2-GFP/Kan R spc7-23/his3+_his3-D1 ade6-M216 leu1-32 ura4_− | This study |
| UFY1258 | h_−_sim4-GFP/Kan R spc7-23/his3+arg3-D4 ade6-M210 ura4-leu1-32 his3-D1 | This study |
| UFY1187 | h_−_dad1-GFP/Kan R spc7-23/his3+leu1-32 ura4-D18 | This study |
| UFY1260 | h_−_spc7-23/his3+mis15-68 ade6-M216 | This study |
| UFY1256 | h+spc7-23/his3+mis17-362 ura4-D18 | This study |
| UFY1264 | h+spc7-23/his3+sim4-193 his3-D1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1088 | h+spc7-23/his3+mal2-1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1196 | h+spc7-23/his3+mis6-302 leu1-32 ura4-D18 | This study |
| UFY1085 | h_−_fta2-291/his3+spc7-23/his3+leu1-32 ade6-M210 ura4-D18 his3-D1 | This study |
| UFY1175 | h+spc7-23/his3+_mad2_Δ::ura4+ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1177 | h_−_spc7-23/his3+_mph1_Δ::ura4+ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1062 | h_−_spc7-23/his3+cnt1(NcoI):arg3 his3-D1 ade6-M210 leu1-32 ura4_−_arg3-D4 | This study |
| UFY1081 | h+spc7-23/his3+otr2(HindIII):ura4+ura4-DS/E leu1-32 ade6-M216 arg3-D4 | This study |
| UFY1222 | h+spc7-23/his3+his7+::lacI-GFP lys1+::_LacOP leu1_−_ura4_− | This study |
| UFY1060 | h_−_spc7-30/his3+Kan R -nmt81-GFP-atb2+leu1-32 | This study |
| UFY1254 | h+mis6-3xHA/LEU+spc7-23/his3+ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1228 | h_−_spc7-HA/Kan R Kan R -nmt81-GFP-atb2+leu1-32 | This study |
| UFY1244 | h+spc7-23-GFP/Kan R /his3+his3-D1 ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1248 | h_−_spc7-GFP/Kan R nuf2-1/ura4+ura4_−_ade6-M210 his3-D1 | This study |
| UFY1269 | h+spc7-GFP/Kan R mis12-537 leu1-32 | This study |
| UFY1224 | h+spc7-23/his3+_mal3_Δ::his3+his3_−_ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1288 | h_−_spc7-23/his3+_alp14_Δ::kan R ura4_−_leu1-32 | This study |
| UFY1262 | h+spc7-23/his3+peg1-1 ura4-D18 leu1-32 | This study |
| UFY1307 | h+mis14-GFP/ura4+spc7-23/his3+ura4_−_leu1-32 | This study |
| UFY1342 | h_−_spc7-GFP/Kan R nda3-KM311 ade6-210 ura_−_leu1-32 | This study |
| UFY1340 | h+spc7-23-GFP/Kan R /his3+_nda3-KM311 leu1-32 ura4-D18 ade6_− | This study |
| UFY1030 | h_−_spc7-23/his3+Kan R -nmt81-GFP-atb2+ade6-M216 leu1-32 | This study |
| UFY1033 | h_−_spc7-24/his3+Kan R -nmt81-GFP-atb2+leu1-32 | This study |
| UFY617 | h_−_spc7-HA/Kan R ade6-M210 leu1-32 ura4-D6 Ch _16_[_ade6-M216_] | U. Fleig |
| UFY724 | h_−_spc7-GFP/Kan R mal2-1 ade6-M210 ura_−_Ch _16_[_ade6-M216_] | U. Fleig |
| UFY210 | h+spc7-GFP/Kan R ade6-M210 ura4-D6 Ch _16_[_ade6-M216_] | U. Fleig |
| UFY1048 | h_−_fta2-291/his3+his3_−_leu1-32 ura4-D18 ade6-M210 | U. Fleig |
| UFY135 | h+_mal3_Δ::his3+_ade6-M210 leu1-32 ura4-D18 his3_Δ | U. Fleig |
| h+_alp14_Δ::kan R _ura4_− | T. Toda | |
| IH1563 | h− peg1-1 leu1-32 ura4-D18 | I. Hagan |
| FY4540 | h_−_sim4-193 cnt1(NcoI):arg3 cnt3(NcoI):ade6 otr2(HindIII):ura4 tel1L:his3 ade6-M210 leu1-32 ura4-D18 arg3-D4 his3-D1 | R. Allshire |
| FY648 | h+_swi6_Δ::his1+otr1R(SphI)::ura4+ura4-DS/E leu1-32 ade6-M210 | R. Allshire |
| KG425 | _h_−_ade6-M210 leu1-32 his3_Δ ura4-D18 | K. Gould |
| ANF251-9A | h+nuf2-1/ura4+ura4-D18 | Y. Hiraoka |
| SS638 | _h_−_mad2_Δ::ura4+leu1-32 ura4-D18 ade6-M210 | S. Sazera |
| SS560 | _h_−_mph1_Δ::ura4+leu1-32 ura4-D18 ade6-M216 | S. Sazer |
| h_−_mis12-537 leu1-32 | M. Yanagida | |
| h_−_mis14-GFP/ura4+_leu1_−_ura4_− | M. Yanagida | |
| h_−_Kan R -nmt81-GFP-atb2+leu1-32 | T. Toda | |
| h+spc24-GFP/Kan R _ade6-M210 leu1-32 ura4_−_his_− | J. Kilmartin |
Generation of spc7ts Alleles and DNA Methods
A pBSK-based plasmid containing the last 2028 bp of the 4095-bp-long spc7+ open reading frame (ORF) followed 3′ by the his3+ gene was used as a template for a mutagenic PCR reaction. A 3896-bp long DNA fragment containing the last 1664 bps of the spc7+ ORF and the his3+ selection marker were transformed into strain KG425. His+ transformants that grew at 25°C but not at 36°C were identified and correct integration of the mutagenized DNA fragments was tested via PCR. A spc7+ containing plasmid was able to fully rescue the temperature sensitivity of these strains, which were backcrossed twice. DNA sequence analysis showed point mutations at the following positions in the ORF: spc7-24 [3340 (A to T); 3959 (T to G)], spc7-23 [3887 (T to G); 4079 (G to T)], and spc7-30 [3026 (G to A); 3914 (A to G); 4067 (G to T)]. We generated an endogenous spc7-23-gfp fusion via PCR-based gene targeting using the KanR cassette (Bahler et al., 1998). spc7-n-gfp and spc7-c-gfp fusions were constructed by homologous recombination in S. cerevisiae, cut out of vector pRS316 and cloned behind the nmt1+ promoter in plasmid pJR2-3XL (Moreno et al., 2000). spc7-n and spc7-c contain the first 2460 or the last 1632 bp of the spc7+ ORF.
Microscopy
Photomicrographs of fixed cells were obtained using a Zeiss Axiovert200 fluorescence microscope (Carl Zeiss, Jena, Germany) coupled to a CCD camera (Hamamatsu, Herrsching, Germany; Orca-ER) and Openlab imaging software (Improvision, Coventry, United Kingdom). Immunofluorescence microscopy was carried out as described previously (Hagan and Hyams, 1988; Bridge et al., 1998). For tubulin staining the monoclonal anti-tubulin antibody TAT1 was used as primary antibody followed by fluorescein isothiocyanate–conjugated goat anti-mouse antibodies (Sigma-Aldrich, St. Louis, MO). HA or GFP fusion proteins were observed by indirect immunofluorescence using mouse anti-HA antibody (Covance, Princeton, NJ) or rabbit anti-GFP antibodies (Invitrogen, Carlsbad, CA), respectively. Cy3-conjugated sheep anti-mouse antibodies or Cy3-conjugated sheep anti-rabbit antibodies (Sigma-Aldrich) were used as secondary antibodies. Before mounting, cells were stained with 4,6-diamidino-2-phenylindole (DAPI). Images of living cells expressing an integrated version of GFP.nmt81.atb2 were obtained using a DeltaVision RT Imaging System (Applied Precision, Issaquah, WA) with a Peltier-cooled CCD Coolsnap HQ Camera (Roper Scientific, Tucson, AZ). Optical sections were recorded every 0.3 μm in a volume totalling 6 μm every 10 s for 30 min. All images were analyzed using Imaris (Zurich, Switzerland; Bitplane) software. Transformed cells were grown in liquid EMM supplemented media without thiamine for 36–48 h at 25°C before analysis.
Immunoprecipitations
For immunoprecipitations or coimmunoprecipitations strains expressing Spc7-HA, Spc7-GFP, Spc7-23-GFP, Spc7-C-GFP, Mis12-GFP, Spc24-GFP, GFP-Atb2, or a combination of these tagged proteins were grown at 25 or 30°C in YE5S or MM overnight and then shifted to the restrictive temperature followed by protein extraction and immunoprecipitation as described previously (Kerres et al., 2004). Spc7 variants did not run at the expected size: for example, Spc7-GFP was detected at 110 instead of 181 kDa. Eluates were boiled, resolved on a SDS-8%-polyacrylamide gel, and blotted. Blots were probed with anti-HA antibody (monoclonal mouse; Roche Diagnostics, Alameda, CA) or anti-GFP antibody (polyclonal rabbit, Invitrogen) followed by the secondary antibody [peroxidase-conjugated AffiniPure goat anti-mouse IgG (H+L); Jackson ImmunoResearch Laboratories or peroxidase-conjugated donkey anti-rabbit IgG; GE Healthcare, respectively]. Immobilized antigens were detected using the ECL Advance Western blotting kit (GE Healthcare, Waukesha, WI).
RESULTS
Spc7 Is Not Required for Transcriptional Silencing of the Central Centromere Region
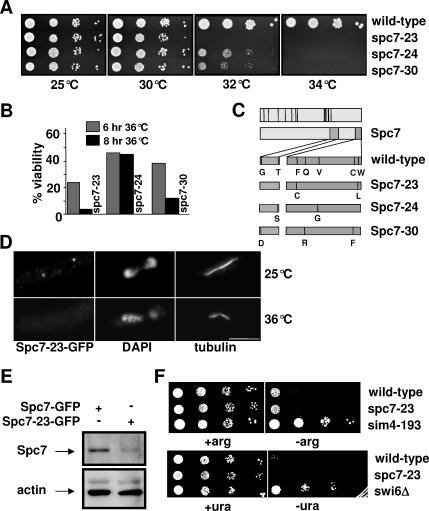
To better understand the function of Spc7 in mitosis, we generated temperature-sensitive (ts) spc7 alleles by mutating the 3′ part of the spc7+ ORF (Materials and Methods). The three spc7 mutant strains, named spc7-23, spc7-24, and spc7-30, that showed the tightest ts phenotype were analyzed in greater detail (Figure 1, A and B). DNA sequence analysis revealed that each of these strains carried several point mutations in the spc7+ ORF leading to the amino acid changes shown in Figure 1C. The mutations leading to ts spc7 alleles lie in two nonconserved regions at the very 3′end of the spc7+ ORF, implying that these regions are important for Spc7 protein function (Figure 1C; Desai et al., 2003; Nekrasov et al., 2003; Cheeseman et al., 2004; Meraldi et al., 2006). The endogenous spc7-23 ORF was tagged with gfp to determine the subcellular location of the mutant protein. The mutant Spc7-23 protein was detected at the kinetochore when cells were incubated at 25°C but not upon incubation at ≥32°C probably because of compromised Spc7-23 protein levels (Figure 1, D and E).
Figure 1.
Spc7 is not involved in centromere silencing. Serial dilution patch tests (104 to 101 cells) of wild-type, spc7-23, spc7-24, and spc7-30 strains grown at the indicated temperatures for 3–4 d. (B) Viability of wild-type and spc7 mutant strains incubated at 36°C for 6 or 8 h. (C) Top diagram, black bars indicate conserved Spc7 residues; bottom diagram, amino acid changes found in the Spc7 mutant proteins in comparison to wild-type Spc7. (D) Kinetochore localization of Spc7-23. Cells expressing the mutant Spc7-23-GFP protein were incubated at 25°C (top lane) or at 36°C for 6 h (bottom lane), fixed, and stained with DAPI, anti-tubulin antibody, and anti-GFP-antibody. Bar, 5 μm. (E) Endogenous Spc7-GFP or Spc7-23-GFP protein isolated from cells grown asynchronously at 32°C for 6 h. Protein extracts prepared from these strains were used for immunoprecipitations using an anti-GFP antibody, followed by Western blotting using the same antibody. Protein extracts used had a similar protein concentration. Actin was used as a loading control. Spc7-23 protein is reduced relative to the wild-type protein by 70%. (F) Serial dilution patch tests (104 to 101 cells) of wild-type, spc7-23, and sim4-193 cells that have the promoter-crippled arg3+ gene inserted at cen1 (top panels) or ura4+ inserted at otr2 (wild-type, spc7-23, and _swi6_Δ cells). Cells were incubated on selective medium with (+arg) or without (−arg) arginine or with (+ura) and without (−ura) uracil at 25°C for 6 d.
We have shown previously that Spc7 is a constitutive kinetochore component that is associated with the central core region of the centromere (Kerres et al., 2004). Wild-type strains that carry a marker gene inserted at a centromeric region are auxotroph for this specific marker due to transcriptional silencing of the centromeric DNA (Pidoux and Allshire, 2000). Defective kinetochore components lead to alleviation of this transcriptional silencing (Allshire et al., 1995; Jin et al., 2002; Pidoux et al., 2003). To analyze if Spc7 was required for transcriptional repression, we tested if marker genes inserted at the otr2 (otr region of centromere 2) or cnt1 (cnt1 region of centromere 1) regions were expressed in the spc7-23 mutant strain (Partridge et al., 2000; Pidoux et al., 2003). Wild-type strains that contain the arg3+ gene inserted at the cnt1 region showed poor growth on arginine-minus medium, whereas kinetochore mutants such as sim4-193 allow growth on this medium (Figure 1F). Interestingly, the presence of spc7-23 did not alleviate transcriptional silencing of the central cnt region at 25°C (Figure 1F). Raising the incubation temperature to 30°C gave the same result (data not shown). Furthermore, transcriptional repression of the otr regions was unaffected in the spc7-23 mutant strain (Figure 1F). Consistent with this finding, centromere association of the histone H3 variant Cnp1 (CENP-A) was unaffected in spc7 mutant cells (data not shown). Because Spc7 is part of the Ndc80-MIND-Spc7 complex, we tested if other members of this complex were required for transcriptional silencing of the central cnt1 region. We found that in mutant nuf2-1 (Ndc80 component) and mis12-537 (MIND component) cells transcriptional silencing at cnt1 still occurred (data not shown). Thus the constitutive Ndc80-MIND-Spc7 kinetochore complex is not required for transcriptional silencing of the central centromere region.
spc7 Mutants Show Severe Defects in Chromosome Segregation and Spindle Attachment
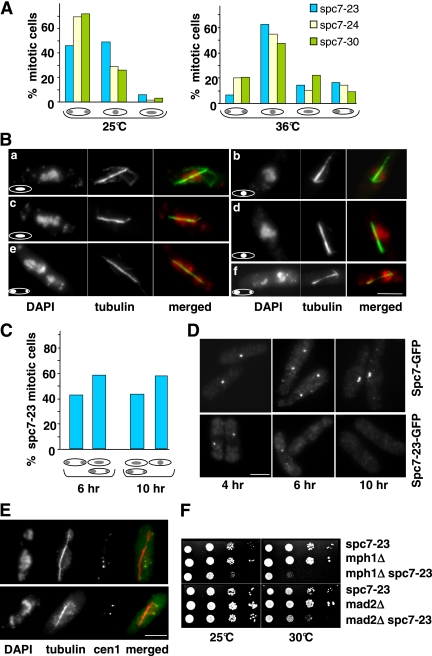
The mutations in the spc7+ ORF lead to aberrant chromosome segregation. At the permissive temperature, the majority of spc7-24 and spc7-30 mitotic cells appeared to segregate chromosomes equally, whereas expression of the spc7-23 allele at this temperature led to an increased number of abnormal mitosis (Figure 2A, left panel). Incubation of these strains at the nonpermissive temperature gave rise to over 70% of mitotic cells with severe chromosome segregation defects (Figure 2A, right panel). All three mutant spc7 strains showed the following abnormal chromosome resolution phenotypes: 1) no separation of highly condensed chromatin on an elongating spindle (Figure 2B, b and d), 2) condensed chromatin that was smeared along the spindle (Figure 2B, a and c), and 3) unequally or partially separated chromatin (Figure 2B, e and f). Spindle structure was often aberrant (see later). Next, we synchronized wild-type spc7+-gfp and spc7-23-gfp cells in the G1 phase of the cell cycle by nitrogen starvation at 25°C (Jin et al., 2002). Release into rich medium at the nonpermissive temperature was followed by microscopic analysis of chromatin and spindles at various time points (0–11 h, Materials and Methods). Entry into mitosis was similar for wild-type and spc7-23 cells and the majority of mitotic cells was present at 6 and 10 h (first and second mitosis after release, respectively; Jin et al., 2002). Although no mitotic defects were observed in wild-type cells, chromosome segregation was severely affected in spc7-23 cells. In the first mitosis, 58% of anaphase cells showed unequally segregated chromatin or condensed chromatin smeared along an elongating spindle (Figure 2C). The phenotype of the cells in the second mitosis was even more severe: we found no anaphase cells that showed equal chromatin segregation (Figure 2C). Instead, cells with an elongating spindle showed no separation of highly condensed chromatin or smeared or unequally segregated chromatin. The differences in mitotic phenotypes seen for the first and second mitosis can be explained by the amount of Spc7-23 present at the kinetochore. Synchronized cells undergoing a first mitosis showed a very reduced or no Spc7-23-GFP signal (57 and 43%, respectively; Figure 2D, middle panel), whereas no Spc7-23-GFP signal could be detected in cells in the second mitosis (Figure 2D, right panel).
Figure 2.
Temperature-sensitive spc7 mutants have severe mitotic defects. (A) Chromatin distribution in mitotic spc7 mutant cells with an elongating spindle incubated at the permissive (25°C) or restrictive temperature (6 h at 36°C). N/strain = 100. (B) Photomicrographs of spc7-23 cells incubated at 36°C. Fixed cells were stained with DAPI and anti-tubulin antibody. Shown are the three main phenotypes observed: smeared chromatin (a and c), nonseparated chromatin (b and d), and unequally/partially segregated chromatin (e and f) on an elongating spindle. Bar, 5 μm. (C) Diagrammatic representation of spc7-23 anaphase phenotypes at 6 and 10 h after the release from G1 arrest and incubation at 36°C. N/time point = 300. (D) Photomicrographs of cells expressing endogenous Spc7-GFP or Spc7-23-GFP. Synchronized G1 cells were released into the cell cycle and incubated at 36°C. Cells were fixed and stained with anti-GFP antibody. Cells shown at the 4-h time point are also representative of earlier time points. (E) Photomicrographs of cen1.gfp spc7-23 cells incubated at 36°C for 6 h. Fixed cells were stained with anti-GFP antibody, DAPI, and anti-tubulin antibody. The merged images show cen1.GFP plus spindle staining. Bar, 5 μm. Thirteen of 28 cells analyzed showed this phenotype. (F) spc7-23 interacts genetically with components of the spindle checkpoint pathway. Serial dilution patch tests of spc7-23, _mph1_Δ, _mad2_Δ, and the respective spc7-23 double mutants grown at the indicated temperatures for 3 d.
The very high frequency of spc7 mitotic cells with nonseparated chromatin or condensed chromatin smeared along the elongating spindle can be caused by compromised kinetochore–microtubule interactions. We therefore assayed if centromeres were associated with the mitotic spindle by determining colocalization of centromere 1 marked with GFP (cen1-gfp) and the spindle in the spc7-23 strain (Nabeshima et al., 1998). In cells with an elongating spindle but smeared or nonseparated chromatin, 46% of the cen1-GFP signals were not spindle associated, implying that the microtubule–kinetochore interactions were severely affected in these cells (Figure 2E, data not shown; Kerres et al., 2004). Furthermore in 50% of anaphase cells with unequally segregated chromatin, the cen1-GFP sister centromeres cosegregated, indicating that Spc7 is also required for bipolar chromosome orientation. In these cells, we measured the distance between the two green fluorescent protein (GFP) signals and found that in 8 of 15 cells the distance between the signals was significantly greater than 0.6 μm. Our data thus imply that cosegregation of sister chromatids is not simply due to a nondisjunction event (Nabeshima et al., 1998).
The nonessential 10-subunit DASH complex coordinates bipolar chromosome attachment in S. pombe (Liu et al., 2005; Sanchez-Perez et al., 2005). We therefore attempted to construct double-mutant strains of spc7-23 with a null allele of duo1+, which encodes a component of the DASH complex (Materials and Methods). Such double mutants were inviable indicating that spc7 mutants require the presence of a functional DASH complex for survival at the permissive temperature.
Compromised microtubule–kinetochore interactions should lead to the activation of the spindle assembly checkpoint that detects unattached, monotelic, or syntelically attached kinetochores (reviewed in Musacchio and Hardwick, 2002; Cleveland et al., 2003). Indeed, double mutants of spc7-23 with null alleles of mph1+ and mad2+, which code for conserved spindle checkpoint components (He et al., 1997, 1998) showed reduced growth in comparison to the single-mutant strains (Figure 2F).
Spc7 Is Required for Kinetochore Targeting of the MIND Complex
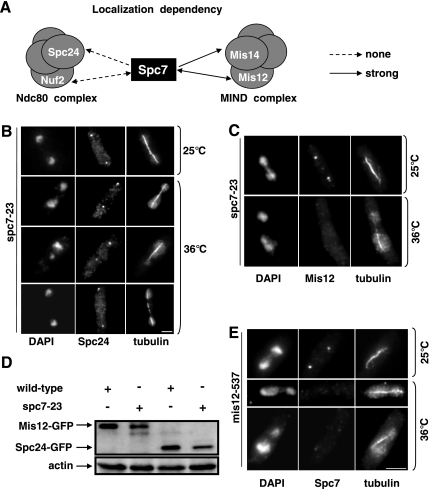
Spc7 is part of the Ndc80-MIND-Spc7 complex (Obuse et al., 2004; Liu et al., 2005). To analyze the role of Spc7 within this complex, we determined the subcellular localization of the Ndc80 complex components Spc24 and Nuf2 and the MIND complex components Mis14 and Mis12 in the spc7-23 ts strain (Goshima et al., 1999; Nabetani et al., 2001; Wigge and Kilmartin, 2001; Obuse et al., 2004). Immunofluorescence analysis of the _gfp_-tagged fusion protein Spc24 revealed that this protein was localized correctly in the spc7-23 ts mutant incubated at the nonpermissive temperature (Figure 3, A and B). Spc24 kinetochore targeting was also unaffected in synchronous spc7-23 populations incubated at the restrictive temperature (data not shown). In addition, kinetochore association of the Spc7-GFP fusion protein was unaffected in the nuf2-1 ts strain and vice versa (Figure 3A), indicating that components of the Ndc80 complex and Spc7 localize to the kinetochore independent of each other. We then analyzed kinetochore localization of Mis12-GFP and Mis14-GFP fusion proteins in the spc7-23 mutant. Although Mis12 kinetochore localization was unaffected in a spc7-23 strain grown at the permissive temperature, kinetochore localization of this protein was severely reduced or absent in the majority of fixed spc7-23 cells incubated at the nonpermissive temperature (Figure 3, A and C). However the Mis12 protein was still present in _spc7_ts cells (Figure 3D). Mis14 kinetochore localization was also affected in a spc7-23 strain (Figure 3A). Furthermore the Spc7-GFP fusion protein was severely reduced (40% cells; Figure 3E, bottom panel) or absent (Figure 3E, middle panel) in the mis12-537 mutant incubated at the restrictive temperature, implying that the kinetochore localization of Spc7 and components of the MIND complex are dependent on each other.
Figure 3.
Interaction between spc7+ and other components of the Ndc80-MIND-Spc7 kinetochore complex. (A) Diagrammatic representation of the kinetochore localization of Spc7-GFP in nuf2-1 and mis12-537 ts mutants and that of the Spc24-GFP, Nuf2-GFP, Mis12-GFP, and Mis14-GFP fusion proteins in the spc7-23 ts mutant. (B and C) Photomicrographs of spc7-23 cells expressing Spc24-GFP or Mis12-GFP. The strains were incubated at 25°C or for 6 h at 36°C, fixed, and stained with DAPI, anti-tubulin antibody, and anti-GFP antibody. Bar, 5 μm. (D) Mis12-GFP and Spc24-GFP in spc7-23 cells. Protein extracts prepared from wild-type or spc7-23 strains that expressed Mis12-GFP or Spc24-GFP endogenously were used for immunoprecipitations using an anti-GFP-antibody, followed by Western blotting using the same antibody. Protein extracts used had a similar protein concentration. Actin was used as a loading control. The strains were incubated at 36°C for 6 h before protein extraction. (E) Photomicrographs of mis12-537 expressing endogenous Spc7-GFP. The strain was treated as described in B.
Next, we analyzed the growth phenotypes of spc7-23 and mis12-537 and nuf2-1 double mutants. spc7-23 mis12-537 double mutants were viable but showed slightly reduced growth compared with the single mutant strains at temperatures below 28°C. They were inviable at 28°C (Supplementary Figure 1B). We were unable to construct a spc7-23 nuf2-1 double mutant by tetrad analysis (Material and Methods, Supplementary Figure 1A). Microscopic analysis of spc7-23 nuf2-1 spores showed that such spores germinated and divided twice. We then tested if overexpression of spc7+ could rescue the ts phenotype of the Ndc80 component nuf2-1 or the MIND component mis12-537. Extra spc7+ resulted in reduced growth of the nuf2-1 strain at 30°C and could not suppress the nongrowth phenotype of this strain at higher temperatures (Supplementary Figure 1C). However, overexpression of spc7+ partially rescued the ts phenotype of the mis_12-537_ strain and extra mis12+ rescued the nongrowth phenotype of the spc7-23 strain at 32°C (Supplementary Figure 1D; Obuse et al., 2004). Thus, Spc7 and MIND show a tight functional interaction, whereas kinetochore targeting of Spc7 and components of the Ndc80 complex do not depend on each other.
Spc7 Interacts Genetically with the Sim4 Complex Component Mal2
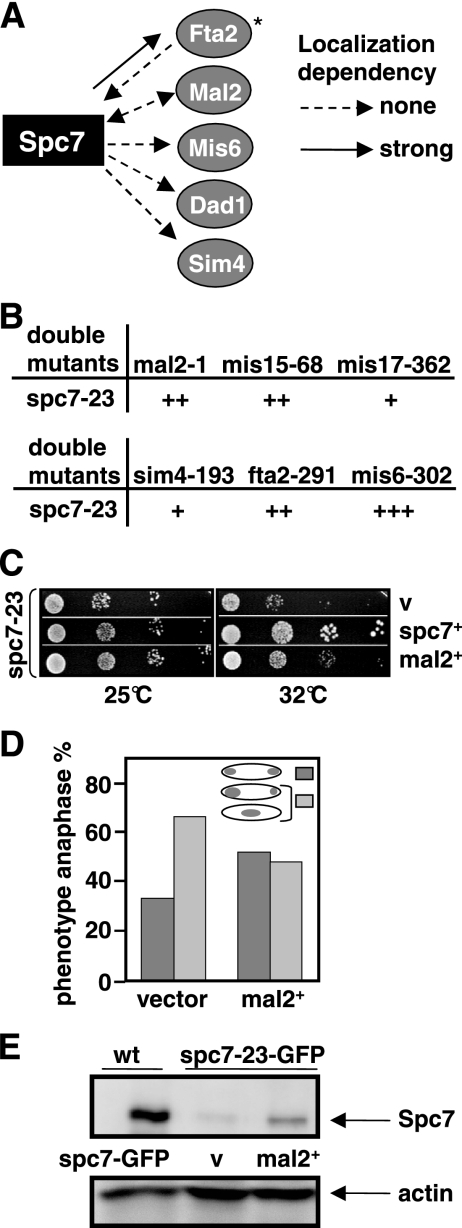
The Sim4 kinetochore complex is a 13-component protein complex that exists independently of the Ndc80-MIND-Spc7 complex (Liu et al., 2005). However, we have shown previously that the kinetochore localization of the Sim4 complex component Fta2 was reduced in a spc7 mutant strain (Kerres et al., 2006). This dependency seems to be specific for Fta2 as other Sim4 complex components such as Mal2-GFP, Mis6-HA, Sim4-GFP, and Dad1-GFP were localized correctly in a spc7-23 mutant background at the restrictive temperature (Figure 4A). Next, we analyzed the growth phenotype of double mutants of spc7-23 with components of the Sim4 complex. All double mutants were viable at 25°C but showed different degrees of growth inhibition at higher temperatures (Figure 4B). spc7-23 mis6-302 double mutants showed the most severe growth reduction, whereas a slight synthetic effect was observed for spc7-23 sim4-193 and spc7-23 mis17-362 double-mutant strains.
Figure 4.
spc7 interacts with components of the Sim4 complex. (A) Diagrammatic representation of the kinetochore localization of Fta2-GFP, Mal2-GFP, Mis6-GFP, Dad1-GFP, and Sim4-GFP in the _spc7-23_ts mutant and Spc7-GFP in a mal2-1 and fta2-291 mutants incubated at the nonpermissive temperature. *Data taken from Kerres et al. (2006). (B) Representation of the genetic interactions between spc7-23 and mutant components of the Sim4 complex. + to +++; weak to strong genetic interaction. (C) Serial dilution patch test of spc7-23 transformants grown on selective medium at the indicated temperatures for 3 (32°C) or 4 (25°C) days. Vector control (v) indicates plasmid without insert, mal2+ and spc7+ denote the presence of wild-type mal2+ or spc7+ expressed from the wild-type promoter or from the thiamine-repressible nmt41+ promoter in the absence of thiamine, respectively. (D) Diagrammatic representation of anaphases observed in spc7-23 cells transformed with a vector control or a plasmid overexpressing mal2+. Cells were incubated for 6 h at 32°C before fixation. N/strain = 100. (E) Immunoprecipitations of GFP-tagged Spc7 proteins. Protein extracts prepared from a wild-type (wt) strain expressing Spc7-GFP from a plasmid and a spc7-23-gfp strain transformed with a vector control (v) or a plasmid expressing mal2+ were analyzed by Western blot analysis using an anti-GFP antibody. Strains were grown for 6 h at 32°C. Actin was used as a loading control.
Interestingly, in a first screen for multicopy suppressors of the spc7 ts phenotype we identified the mal2+ ORF. When present on a plasmid mal2+ expressed from its wild-type promoter can partially rescue the nongrowth phenotype of spc7 mutant strains (Figure 4C) by reducing the number of aberrant mitosis (Figure 4D) and increasing the amount of Spc7-23 protein in the cell (Figure 4E). The converse, i.e., the rescue of the mal2-1 ts mutant phenotype by extra spc7+ was not observed (data not shown). Overexpression of other components of the Sim4 complex, such as the Mal2-interaction partner Fta2 or the Sim4 protein cannot rescue the ts phenotype of the spc7-23 strain at the nonpermissive temperature (data not shown). Thus, the specific rescue of spc7-23 mutants by overexpression of mal2+ implies that Mal2 and Spc7 share some function and point to an interaction between the Sim4 and Ndc80-MIND-Spc7 complexes.
spc7 Mutants Exhibits Defects in Spindle Formation and Function
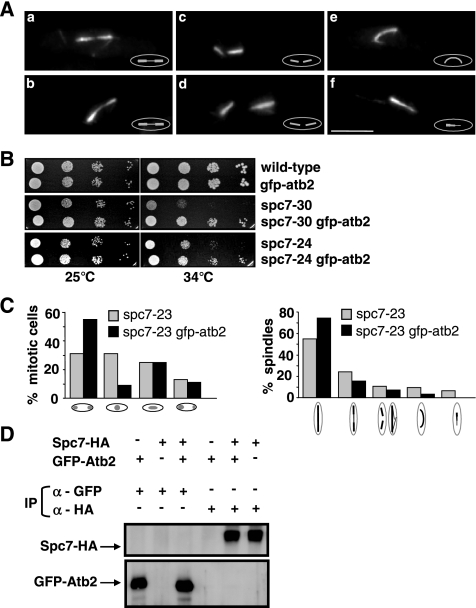
Immunofluorescence analysis of fixed _spc7_ts mitotic cells revealed that all three _spc7_ts mutants gave rise to abnormal spindle structures. For example, in the spc7-23 mutant strain grown at the restrictive temperature 47% of all spindles analyzed were aberrant. Immunofluorescence staining of spc7-23 spindles showed the following phenotypes: 1) elongating spindles with very thinly staining midzones (Figure 5A, a and b), 2) disintegration of the spindle evidenced by spindle fraying and/or two separated half-spindles present in one cell (Figure 5A, c and d), 3) elongating anaphase spindles that were bent (Figure 5Ae), and 4) unequally stained spindles (Figure 5Af). The latter phenotype comprised approximately equal proportions of monopolar and bipolar spindles as determined by the subcellular localization of the spindle pole body component Cut12 (Bridge et al., 1998; data not shown). The other spc7 ts mutants such as spc7-30 showed similar spindle abnormalities (Supplementary Figure 2).
Figure 5.
Spc7 is required for the integrity of the spindle. (A) Photomicrographs of spindle defects observed in spc7-23 cells incubated at 36°C for 6 h: thin spindle midzones (a and b), two half-spindles per cell (c and d), bent spindle (e), and unequally stained spindle (f). Fixed cells were stained with anti-tubulin antibody. (B) Serial dilution patch tests (104 to 101 cells) of wild-type, spc7-30, and spc7-24 cells carrying the integrated nmt81-GFP-atb2. Strains were grown at the indicated temperatures for 5 d under derepressed conditions. (C) Chromosome segregation and spindle phenotypes observed in spc7-23 and spc7-23 nmt81-GFP-atb2 (spc7-23 gfp-atb2) strains grown at 34°C. Right diagram (from left to right), wild-type anaphase spindle, anaphase spindles with thinly staining midzone, disintegrating/broken anaphase spindles, bent spindles and unequally stained spindles. N/strain = 100. (D) Immunoprecipitations of Spc7-HA and GFP-Atb2 proteins. Protein extracts prepared from strains expressing Spc7-HA, GFP-Atb2, or both were used for immunoprecipitations (IP) with an anti-GFP or anti-HA antibody, followed by Western blot analysis using anti-GFP and anti-HA antibodies.
Interestingly, the synchronous spc7-23 culture experiments showed that only 6% of cells undergoing the first mitosis had spindle defects, whereas 52% of cells in the second mitosis had abnormal spindles. Thus spindle abnormalities arise when Spc7-23 can no longer be detected at the kinetochore. We next wanted to analyze these phenotypes in live cells with fluorescence microscopy and thus generated spc7 ts mutant strains that harbored an integrated version of the α-tubulin atb2+ ORF tagged with gfp and driven by the nmt81 promoter (gfp.nmt81.atb2; Garcia et al., 2001). However, presence of the extra α-tubulin partially rescued the nongrowth phenotypes of spc7-24 and spc7-30 mutant strains (Figure 5B) and reduced the number of aberrant mitosis and abnormal spindle structures in all spc7 mutants (Figure 5C, Supplementary Figure 2). We thus tested if these two proteins could interact physically by performing coimmunoprecipitations in exponentially growing strains expressing Spc7-HA and GFP-Atb2 but failed to find an interaction (Figure 5D). As aberrant spindle phenotypes in the spc7 ts GFP.nmt81.atb2 mutants were infrequent, we analyzed spindle structure in wild-type GFP.nmt81.atb2 cells that overexpressed the dominant negative spc7-c variant (Kerres et al., 2004). Overproduction of this C-terminal part of the spc7+ ORF affects chromosome segregation and leads to similar types of spindle defects as those observed for the spc7 ts mutants (see below; Kerres et al., 2004). Spc7-C is able to associate with the kinetochore (see Figure 8A) and does not appear to interact with wild-type Spc7 as the two proteins cannot be coimmunoprecipitated (Figure 6A). The kinetochore appears to be assembled in _spc7-c_–overexpressing cells, as the Spc24, Spc7, Mis14, and Mal2 kinetochore proteins are correctly localized (Figure 6B; Kerres et al., 2004). Examination of 30 spindles in wild-type GFP.nmt81.atb2 cells that overexpressed spc7-c revealed that 53% of these spindles were abnormal. Defects were found at all stages of spindle formation. In three cells formation of a bipolar spindle was defective as microtubules emanated from a single focus or microtubules coming from separated spindle pole bodies were unable to form a stable bipolar spindle (Figure 7C). 7/30 spindles showed a prolonged delay at the metaphase/anaphase A to anaphase B transition probably due to an activated spindle control checkpoint (Figure 7B). This phenotype was observed for only 2 of 14 wild-type GFP.nmt81.atb2 cells transformed with the vector control. Three cells were unable to switch to the phase III spindle stage, i.e., spindle elongation in anaphase B in the time frame measured (Figure 7, D and H; Nabeshima et al., 1998). Whether spindle phase III was abolished in these cells or just severely delayed is yet unclear. Intriguingly, two spindles were assembled and started to elongate with apparently wild-type dynamics and then collapsed in midanaphase B. One of these spindles showed rejoining of the two spindle halves and further spindle elongation (Figure 7E).
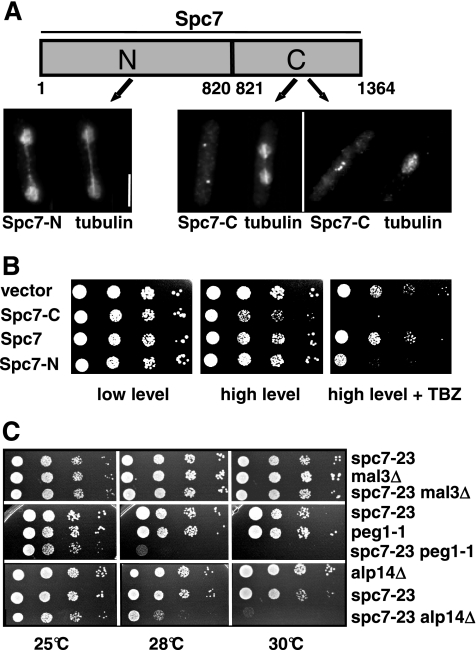
Figure 8.
Interaction of spc7 with components of the mitotic spindle. (A) Subcellular localization of the Spc7 variants fused to GFP and expressed on a plasmid under the control of the nmt1+ promoter. Cells were grown under promoter-derepressing conditions for 24 h at 30°C, fixed, and stained with anti-tubulin and anti-GFP antibodies. Bar, 5 μm. (B) Overexpression of Spc7-N or Spc7-C in a wild-type strain leads to TBZ hypersensitivity. Panels show serial dilution patch tests of wild-type cells expressing low (left panels) and high (middle and right panels) amounts of the indicated Spc7 variants. Cells shown in the right panels were grown on medium containing TBZ. (C) Serial dilution patch tests (104 to 101 cells) of spc7-23, _mal3_Δ, peg1-1, _alp14_Δ, and the respective double mutants grown on YE5S at the indicated temperatures for 3–6 d.
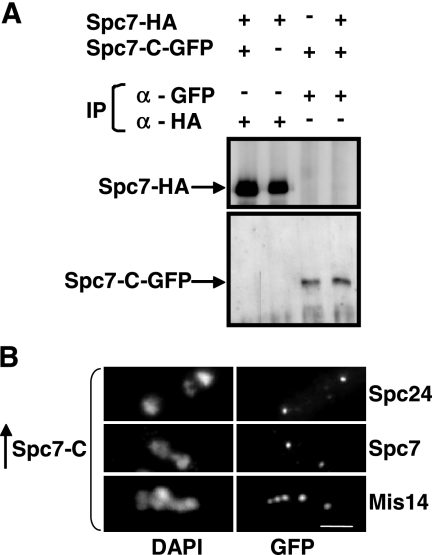
Figure 6.
Overexpression of spc7-c does not affect localization of members of the Ndc80-MIND-Spc7 complex. (A) Immunoprecipitations of Spc7-HA and Spc7-C-GFP proteins. Protein extracts prepared from a wild-type and a Spc7-HA strain expressing plasmid-borne Spc7-C-GFP under the control of the nmt41 promoter were used for immunoprecipitations using anti-GFP or anti-HA antibodies. The immunoprecipitates were halved and analyzed by Western blot analysis using anti-GFP and anti-HA antibodies. (B) Photomicrographs of Spc24-GFP, Spc7-GFP, and Mis14-GFP strains overexpressing spc7-c from the nmt1+ promoter for 24 h at 30°C. Fixed cells were stained with DAPI and anti-GFP antibody. Bar, 5 μm.
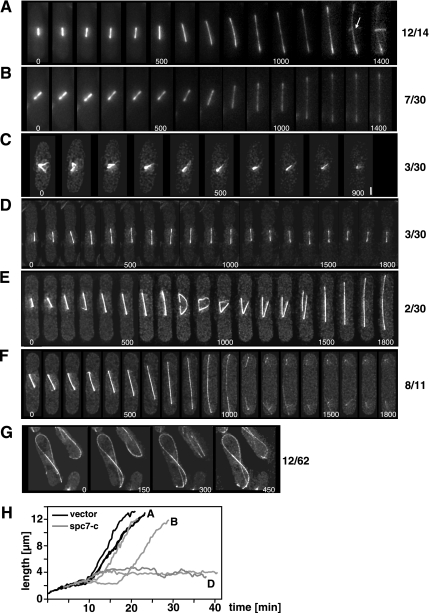
Figure 7.
Mitosis in living _spc7-c_–expressing cells. Time-lapse images of mitosis in wild-type nmt81-GFP-atb2 cells transformed with a vector control (A) or overexpressing spc7-c (B–G) from the nmt1+ promoter for 36–48 h at 25°C. Time interval between images was 100 s (A–F) or 150 s (G). Numbers beside photomicrographs indicate the number of spindles with this phenotype. (A) Normal spindle elongation. The appearance of the PAA is indicated by an arrow. (B) Delay in spindle elongation possibly caused by an active spindle checkpoint. (C) Inability to form a bipolar spindle. The separated spindle pole bodies (second panel) collapse into a single fluorescent signal. Bar, 1.5 μm. (D) Failure of bipolar spindle elongation. In the time frame measured the spindle shows cycles of spindle elongation (up to 4.7 μm) followed by shrinkage. (E) Elongating spindle that collapses in midanaphase B, followed by fusion of the elongating spindle fragments and further elongation of the bipolar spindle. (F) Normal elongation of the spindle and delay in PAA appearance. The cell shown did not have a PAA in the time measured as shown for the wild-type mitosis in A. (G) Aberrant interphase microtubule cytoskeleton in _spc7-c_–overexpressing cells. Cells showed fewer microtubule bundles, which often curved around the cell. (H) Quantitation of spindle length for nmt81-GFP-atb2 cells transformed with a vector control (black graphs, 3 spindles) or overexpressing spc7-c (gray graphs, 4 spindles). The graphs marked with A, B, and D represent the spindles shown in A, B, and D, respectively.
After anaphase the postanaphase array (PAA), which is nucleated by the microtubule-organizing center [equatorial (e)MTOC], forms at the cell equator (reviewed in Hagan and Petersen, 2000). In wild-type GFP.nmt81.atb2 cells transformed with a vector control this structure was observed in 12/12 cells analyzed (Figure 7A). However in 8/11 GFP.nmt81.atb2 cells overproducing the Spc7 variant the PAA was not observed in the time frame measured, although the spindle had elongated fully followed by spindle breakdown (Figure 7F). Intriguingly, extra spc7-c also influenced the interphase microtubule cytoskeleton. In wild-type cells, interphase microtubules grow out from the nucleus, continue growth until they reach the cell tip, and then depolymerize (Drummond and Cross, 2000). In 20% of _spc7-c_–overexpressing cells or spc7 ts mutants the interphase microtubules continued to grow when they reached the cell tip and thus curled around the cell tip (Figure 7G, data not shown).
Thus, the Spc7 kinetochore protein is required for the integrity of the mitotic spindle and also seems to influence the interphase microtubule cytoskeleton. We therefore determined if Spc7 was able to associate with the microtubule cytoskeleton. Wild-type Spc7 is associated with the kinetochore and we were unable to detect colocalization with the mitotic spindle even in an overexpression situation (Kerres et al., 2004; data not shown). The same holds true for the Spc7-C variant. Overexpression of a Spc7-C-GFP fusion protein from a plasmid gave rise to a kinetochore-specific fluorescence signal (Figure 8A). We then looked at the intracellular localization of a Spc7-N variant (Figure 8A). In contrast to Spc7-C, overexpression of the N-terminal part of the Spc7 protein in a wild-type strain does not lead to obvious growth defects but results in an increased sensitivity to the microtubule poison thiabendazole (Figure 8B). Spc7-N-GFP–expressing cells showed GFP fluorescence present in the entire nucleus possibly because the C-terminal nuclear export sequence is no longer present (Matsuyama et al., 2006). In addition to staining of the nucleus, Spc7-N showed colocalization with the mitotic spindle during mitosis (Figure 8A). Thus Spc7 appears to have the potential to colocalize with the mitotic spindle. This localization is independent of the presence of the microtubule plus-end–associated protein Mal3 (data not shown). We next determined the subcellular localization of Spc7-GFP, Spc24-GFP, and Mis12-GFP in cells overexpressing Spc7-N. We found that these kinetochore proteins localized in a wild-type manner, indicating that Spc7-N did not cause mislocalization of other kinetochore proteins (data not shown).
Genetic Interaction between spc7+ and Genes Coding for Microtubule Plus-End–associated Proteins
spc7+ was isolated originally as a suppressor of a mal3 mutant. This suppression appears to be specific for mal3 as extra spc7+ cannot rescue the mutant phenotypes of dis1-288, _dis1_Δ, _alp14_Δ, or peg1-1 mutant strains (data not shown; Kerres et al., 2004). Dis1 and Alp14/Mtc1 are members of the TOG/XMAP215 family, whereas Peg1 is the fission yeast member of the CLASP family (Ohkura et al., 1988; Nabeshima et al., 1995; Garcia et al., 2001; Nakaseko et al., 2001; Grallert et al., 2006). Double-mutant strains between spc7 and _alp14_Δ, peg1-1 and _mal3_Δ showed that the absence of mal3+ in a spc7-23 mutant does not lead to an increased phenotype of that strain, possibly indicating that the two proteins act in the same pathway (Figure 8C). _spc7-23 alp14_Δ and spc7-23 peg1-1 double mutants showed no enhanced phenotype at 25°C but were unable to grow at 30°C (Figure 8C). Tetrad analysis at incubation temperatures of 25, 28, or 30°C revealed that dis1-288 spc7-23 and _dis1_Δ spc7-23 double mutants could not be generated. Altogether 52 tetrads with four germinating spores were analyzed without recovering dis1 spc7-23 double mutants that were able to form a colony. These findings show that spc7 mutants require the presence of the dis1+ wild-type gene product for survival at the permissive temperature.
DISCUSSION
During mitosis, the correct interaction of the kinetochore with spindle microtubules is essential for the precise segregation of the duplicated sister chromatids. We had proposed previously that the Spc7 protein plays a direct role at the microtubule–kinetochore interface as Spc7 was identified as a suppressor of the EB1 family member Mal3 and interacted with this protein genetically as well as physically (Kerres et al., 2004). Very recently the Spc7 ortholog from C. elegans, the KNL-1 protein, has been shown to have a specific microtubule-binding activity, suggesting that this protein family is part of the core microtubule-binding site of the kinetochore (Cheeseman et al., 2006).
In this work, we have extended our analysis of Spc7 function and have shown 1) Spc7 is required for formation/function of the spindle in vivo, 2) Spc7 is required for kinetochore association of the MIND complex, and 3) Spc7 provides a genetic link between the two S. pombe kinetochore complexes Sim4 and Ndc80-MIND-Spc7 as it is required for kinetochore targeting of the Sim4 complex component Fta2 and extra expression of the Sim4 complex component Mal2 rescues the spc7 mutant phenotypes (Kerres et al., 2006).
Extra Spc7 can partially rescue the temperature sensitivity of the mis12 and mis14 mutant strains (Supplementary Figure 1; Obuse et al., 2004). Furthermore, overexpression of mis12+ can partially suppress the nongrowth phenotype of a _spc7_ts mutant. In addition kinetochore targeting of Spc7 and MIND components was dependent on each other, whereas members of the Ndc80 complex associated with the kinetochore independent of functional Spc7. Overall, these experiments suggest that within the Ndc80-MIND-Spc7 complex Spc7 and the MIND complex show a tight functional interaction. The findings that MIND but not Ndc80 requires Spc7 for kinetochore targeting are in contrast to those observed for KNL-1, the C. elegans homologue of Spc7. In that organism the Ndc80 complex components Ndc80 and Spc25KBP-3 require KNL-1 for kinetochore association, whereas kinetochore targeting of the MIND complex component Mis12 is only slightly affected in KNL-1–depleted cells (Desai et al., 2003; Cheeseman et al., 2004). Kinetochore targeting of other Spc7 homologues remains to be determined: however, independent kinetochore association of MIND and Ndc80 complexes has been shown for a number of organisms (De Wulf et al., 2003; Emanuele et al., 2005; Saitoh et al., 2005).
We also investigated if Spc7 played a role in kinetochore targeting of Sim4 complex components. We found that in spc7 mutant cells kinetochore localization of the Sim4 complex component Fta2 is significantly reduced, whereas other tested components of this complex do not appear to be affected (Kerres et al., 2006, Figure 4A). In particular, kinetochore targeting of Mal2, a very close interaction partner of Fta2, appears unaffected in spc7 mutants. As kinetochore localization of Fta2 and Mal2 are dependent on each other, our data imply that the severely reduced amount of Fta2 at the kinetochore in spc7 mutants is sufficient for proper localization of Mal2 in these cells. Interestingly, extra mal2+ was able to suppress the nongrowth phenotype of the spc7-23 mutant at 32°C (Figures 4, C and D) implying that these two proteins share some function(s). Such interactions between Spc7 and Mal2 family members appear to be conserved. Affinity purification of proteins interacting with human CENP-O/Mcm21R (Mal2 ortholog) or the S. cerevisiae Mal2 homologue Mcm21p identified the Spc7 orthologues AF15q14 and Spc105p, respectively (De Wulf et al., 2003; Okada et al., 2006).
At present the mechanism by which extra mal2+ can suppress the spc7-23 mutant is unknown. In particular it is unclear how mal2+ overexpression can rescue the spc7-23 spindle defects, as Mal2 does not appear to be required for normal spindle structure (Jin et al., 2002). However, the mal2-1 mutant strain is hypersensitive to microtubule poisons and human cells depleted for the Mal2 ortholog Mcm21R/CENP-O show defects in spindle assembly (Jin et al., 2002; McAinsh et al., 2006).
In fission yeast three distinct spindle phases have been defined and spc7-23 mutants show defects in all phases (Nabeshima et al., 1998). Phase I involves formation of the bipolar spindle in prophase to prometaphase. In phase II, which encompasses metaphase chromosome alignment to the end of anaphase A, the spindle has a constant length, whereas spindle elongation occurs in the third phase (anaphase B) by sliding apart of antiparallel microtubules in the spindle midzone. Entry into phase III is accompanied by a change in microtubule dynamics leading to more stable microtubules and spindle elongation (Ding et al., 1993; Nabeshima et al., 1995, 1998; Mallavarapu et al., 1999; Sagolla et al., 2003; Khodjakov et al., 2004; Tolic-Norrelykke et al., 2004). Once the nuclei have been separated toward the cell ends, the spindle breaks down and the PAA appears in the cell middle (reviewed in Hagan, 1998; Figure 7A). We observed monopolar spindles or small aberrant bipolar spindles that collapsed into a single focal point indicating defects in spindle phase I. Such staining patterns have been observed in a wide variety of mutants, among them mutants with defects in mitotic motor proteins, spindle pole body components, or mitotic regulators such the Ran GTPase and the Aurora-related kinase Ark1 (Hagan and Yanagida, 1990, 1992, 1995; Bridge et al., 1998; West et al., 1998; Fleig et al., 2000; Petersen et al., 2001; Leverson et al., 2002). We were unable to assess defects in spindle phase II directly by microscopy. However the finding that cells overexpressing spc7-c showed a prolonged delay at the transition to phase III suggests that this spindle stage is also affected in spc7 mutants. Spindle stage III involves the rapid elongation of the spindle from 2 to 3 μm to 10–14 μm. Elongation in cells expressing spc7-c was discontinous as envisaged by the cycles of spindle elongation and spindle shortening (Figure 7, D and H), suggesting that the switch in microtubule dynamics that occurs at the onset of spindle phase III was defective in these cells (Mallavarapu et al., 1999). Similar phenotypes have been observed in S. cerevisiae cells expressing mutant version of the Cdc14p phosphatase or the Ndc10p kinetochore protein (Bouck and Bloom, 2005; Higuchi and Uhlmann, 2005). Cdc14p is required for changing microtubule dynamics at the onset of anaphase and targets the Ndc10p protein, which is needed for spindle stability to the plus ends of interpolar microtubules at the spindle midzone during anaphase (Goh and Kilmartin, 1993; Bouck and Bloom, 2005; Higuchi and Uhlmann, 2005).
spc7 mutant cells that could execute spindle phase III had a high proportion of anaphase B spindles with abnormal spindle midzones. The spindle midzone, which consists of overlapping antiparallel microtubules (Ding et al., 1993), stained very thinly in living and fixed spc7 mutants (Figure 5A). Reduced tubulin staining of the spindle midzone has also been observed for a number of S. cerevisiae kinetochore mutants, among them components of the Ndc80 complex (Wigge and Kilmartin, 2001; Le Masson et al., 2002; McCleland et al., 2003). Consistent with a abnormal spindle midzone, we observed fixed spc7 mutant cells with two spindle halves and elongating midanaphase B spindles that abruptly collapsed in the middle region in living cells (Figures 5A and 7E). In one case the two spindle halves were able to rejoin and continue spindle elongation. The latter phenotype has also been observed when the middle of medium-length spindles is cut by laser microsurgery or in mutants required for central spindle formation (Mitchison and Salmon, 2001; Khodjakov et al., 2004; Tolic-Norrelykke et al., 2004; Loiodice et al., 2005; Yamashita et al., 2005). Our data thus demonstrate that Spc7 is required for the integrity of the spindle midzone possibly by influencing the dynamics of the microtubule-plus ends. However we can at present not exclude that Spc7 regulates spindle function by some other means such as influencing microtubule bundling as similar phase III phenotypes have been observed in mutants with an ase1+ null allele (Loiodice et al., 2005; Yamashita et al., 2005). Fission yeast Ase1, which localizes to the spindle midzone in anaphase B, belongs to the conserved Prc1/MAP65 family of microtubule bundling proteins, that is required for central spindle formation and cytokinesis (Schuyler et al., 2003; Verni et al., 2004; Loiodice et al., 2005; Yamashita et al., 2005).
Taken together our results indicate that Spc7 plays a profound role in the formation and function of the spindle. How does Spc7 exert its influence on spindle integrity? Our immunofluorescence analysis of an endogenously expressed wild-type Spc7 fusion protein shows an exclusive kinetochore localization. However, it is possible that Spc7 also associates with the mitotic spindle but we fail to detect it either because the signal is below the threshold sensitivity of our imaging system or due to a highly transient association of the protein with the spindle. In this respect, the colocalization of the Spc7 variant, Spc7-N, with the mitotic spindle might argue that Spc7 has the potential to associate with spindle microtubules and that this association is regulated by the C-terminal part of the Spc7 protein. Interestingly, a component of the Ndc80 kinetochore complex in budding yeast, namely Ndc80p, was shown to be associated with spindle microtubules using immunoelectron microscopy (Muller-Reichert et al., 1998, 2003). An alternative, but not mutually exclusive possibility is that Spc7 could exert its influence on spindle microtubules by regulating proteins that localize to kinetochore and spindle. In S. cerevisiae the Cdc14p phosphatase is required for spindle localization of a number of proteins that affect spindle function among them the chromosomal passenger proteins aurora kinase Ipl1p and INCENP Sli15p as well as the kinetochore proteins Slk19p and Ndc10p (Pereira and Schiebel, 2003; Bouck and Bloom, 2005). We therefore looked at the localization of the S. pombe Cdc14p homologue Flp1/Clp1 (Cueille et al., 2001; Trautmann et al., 2001) in spc7 mutants and found a wild-type-like localization pattern (data not shown). Furthermore Spc7-N showed colocalization with the mitotic spindle in a _flp1_Δ strain (data not shown). We also found that spindle association of the chromosomal passenger protein Ark1 and the DASH component Dam1 is still possible in the spc7-23 mutant at the nonpermissive temperature (data not shown; (Petersen et al., 2001; Leverson et al., 2002; Liu et al., 2005; Sanchez-Perez et al., 2005). However in 40% of _spc7_ts cells with a late anaphase spindle and separated chromatin, Ark1 staining was not confined to the spindle midzone. Instead Ark1 was distributed as a broad signal over most of the spindle (data not shown). Whether this abnormal Ark1 localization is the cause or a consequence of the aberrant spindle midzone in _spc7_ts cells is at present unclear.
After anaphase, the PAA, nucleated by the eMTOC microtubule organizing structure, is seen by the time the spindle reaches its maximum length (Hagan and Petersen, 2000; Figure 7A). In the majority of spc7 mutant cells that went through anaphase B this structure was not observed, although breakdown of the spindle occurred (Figure 7F). Thus Spc7 function is needed for PAA formation. Failure to form a PAA has also been observed in mutants that affect γ-tubulin–complex function (Sawin et al., 2004; Venkatram et al., 2004; Samejima et al., 2005). Interestingly, components of the γ-tubulin complex, such as Alp4 and Alp6, are also required for a proper interphase microtubule cytoskeleton (Vardy and Toda, 2000). alp4 mutants have longer interphase microtubules that curve around the cell end: a phenotype very similar to what we have observed for spc7 mutants (Figure 7G). Intriguingly, a highly overexpressed Spc7-YFP fusion protein localizes to the kinetochore and as a single dot to the periphery at the site of septum formation in the middle of the cell (Matsuyama et al., 2006), thus raising the possibility that Spc7 and the eMTOC co-localize.
Supplementary Material
[Supplemental Material]
ACKNOWLEDGMENTS
We thank Agnes Grallert, Steve Bagley, and Iain Hagan (Paterson Institute for Cancer Research, Manchester) for their generous help with the photomicrographs shown in Figure 7. We thank Robin Allshire (Wellcome Trust Centre for Cell Biology, Edinburgh, United Kingdom), Iain Hagan (Paterson Institute for Cancer Research), Kathy Gould (Vanderbilt University, Nashville, TN), Keith Gull (University of Oxford, Oxford, United Kingdom), Yashushi Hiraoka (Kansai Advanced Research Center, Kobe, Japan), John Kilmartin (Medical Research Council Laboratory, Cambridge, United Kingdom), Jonathan Millar (University of Warwick, Coventry, United Kingdom), Viesturs Simanis (Swiss Institute for Experimental Cancer Research, Epaninges, Switzerland), Takashi Toda (Cancer Research, London, United Kingdom), Mitsuhiro Yanagida (Kyoto University, Kyoto, Japan) and the Yeast Genetic Resource Center (Osaka, Japan) for reagents; Shelley Sazer and Johannes Hegemann for reading of the manuscript; Johannes Hegemann for support; and Eva Walla for excellent technical assistance.
Abbreviations used:
ts
temperature sensitive
PAA
postanaphase array.
Footnotes
REFERENCES
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Asakawa K., Toya M., Sato M., Kanai M., Kume K., Goshima T., Garcia M. A., Hirata D., Toda T. Mal3, the fission yeast EB1 homologue, cooperates with Bub1 spindle checkpoint to prevent monopolar attachment. EMBO Rep. 2005;6:1194–1200. doi: 10.1038/sj.embor.7400540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [In Process Citation] [DOI] [PubMed] [Google Scholar]
- Beinhauer J. D., Hagan I. M., Hegemann J. H., Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D. C., Bloom K. S. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge A. J., Morphew M., Bartlett R., Hagan I. M. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., 3rd, Oegema K., Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Clarke L. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Cueille N., Salimova E., Esteban V., Blanco M., Moreno S., Bueno A., Simanis V. Flp1, a fission yeast orthologue of the s.cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 2001;114:2649–2664. doi: 10.1242/jcs.114.14.2649. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Musacchio A., Salmon E. D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:962–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Muller-Reichert T., Shevchenko A., Hyman A., Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., McDonald K. L., McIntosh J. R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D. R., Cross R. A. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 2000;10:766–775. doi: 10.1016/s0960-9822(00)00570-4. [DOI] [PubMed] [Google Scholar]
- Emanuele M. J., McCleland M. L., Satinover D. L., Stukenberg P. T. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell. 2005;16:4882–4892. doi: 10.1091/mbc.E05-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G. M. Nnf1p, Dsn1p, Mtw1p, and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae Eukaryot. Cell. 2002;1:229–240. doi: 10.1128/EC.1.2.229-240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig U., Salus S. S., Karig I., Sazer S. The fission yeast Ran GTPase is required for microtubule integrity. J. Cell Biol. 2000;151:1101–1112. doi: 10.1083/jcb.151.5.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde R., et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- Fukagawa T. Assembly of kinetochores in vertebrate cells. Exp. Cell Res. 2004;296:21–27. doi: 10.1016/j.yexcr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Garcia M. A., Vardy L., Koonrugsa N., Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P. Y., Kilmartin J. V. NDC10, a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Saitoh S., Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A., Beuter C., Craven R., Bagley S., Wilks D., Fleig U., Hagan I. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 2006;20:2421–2436. doi: 10.1101/gad.381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G. G., Bretscher A. Cell biology. Microtubule asymmetry. Science. 2003;300:2040–2041. doi: 10.1126/science.1084938. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. M. The fission yeast microtubule cytoskeleton. J. Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Hagan I. M., Hyams J. S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan I. M., Petersen J. The microtubule organizing centers of Schizosaccharomyces pombe. Curr. Top Dev. Biol. 2000;49:133–159. doi: 10.1016/s0070-2153(99)49007-6. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- He X., Jones M. H., Winey M., Sazer S. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J. Cell Sci. 1998;111:1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- He X., Patterson T. E., Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Rines D. R., Espelin C. W., Sorger P. K. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz J., Lechner J., Shevchenko A., Shevchenko A., Magiera M. M., Schramm C., Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q. W., Pidoux A. L., Decker C., Allshire R. C., Fleig U. The mal2p protein is an essential component of the fission yeast centromere. Mol. Cell. Biol. 2002;22:7168–7183. doi: 10.1128/MCB.22.20.7168-7183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan K. B., Burds A. A., Swedlow J. R., Bekir S. S., Sorger P. K., Nathke I. S. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat. Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- Kerres A., Vietmeier-Decker C., Ortiz J., Karig I., Beuter C., Hegemann J., Lechner J., Fleig U. The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol. Biol. Cell. 2004;15:5255–5267. doi: 10.1091/mbc.E04-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A., Jakopec V., Beuter C., Karig I., Pöhlmann J., Pidoux A., Allshire R., Fleig U. Fta2, an essential fission yeast kinetochore component, interacts closely with the conserved Mal2 protein. Mol. Biol. Cell. 2006;17:4167–4178. doi: 10.1091/mbc.E06-04-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., La Terra S., Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr. Biol. 2004;14:1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Le Masson I., Saveanu C., Chevalier A., Namane A., Gobin R., Fromont-Racine M., Jacquier A., Mann C. Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol. Microbiol. 2002;43:1431–1443. doi: 10.1046/j.1365-2958.2002.02844.x. [DOI] [PubMed] [Google Scholar]
- Leverson J. D., Huang H. K., Forsburg S. L., Hunter T. The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell. 2002;13:1132–1143. doi: 10.1091/mbc.01-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J. R., 3rd, He X. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I., Staub J., Setty T. G., Nguyen N. P., Paoletti A., Tran P. T. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A., Sawin K., Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol. 1999;9:1423–1426. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- McAinsh A. D., Tytell J. D., Sorger P. K. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- McAinsh A. D., Meraldi P., Draviam V. M., Toso A., Sorger P. K. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 2006;25:4033–4033. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M. L., Gardner R. D., Kallio M. J., Daum J. R., Gorbsky G. J., Burke D. J., Stukenberg P. T. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A. D., Rheinbay E., Sorger P. K. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Tsukita S. “Search-and-capture” of microtubules through plus-end-binding proteins (+TIPs) J. Biochem. (Tokyo) 2003;134:321–326. doi: 10.1093/jb/mvg148. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. Mitosis: a history of division. Nat. Cell Biol. 2001;3:17–21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- Moreno M. B., Duran A., Ribas J. C. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast. 2000;16:861–872. doi: 10.1002/1097-0061(20000630)16:9<861::AID-YEA577>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Muller-Reichert T., Chretien D., Severin F., Hyman A. A. Structural changes at microtubule ends accompanying GTP hydrolysis: information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha, beta)methylenediphosphonate. Proc. Natl. Acad. Sci. USA. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T., Sassoon I., O'Toole E., Romao M., Ashford A. J., Hyman A. A., Antony C. Analysis of the distribution of the kinetochore protein Ndc10p in Saccharomyces cerevisiae using 3-D modeling of mitotic spindles. Chromosoma. 2003;111:417–428. doi: 10.1007/s00412-002-0220-6. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K. G. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Kurooka H., Takeuchi M., Kinoshita K., Nakaseko Y., Yanagida M. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A. F., Murray A., Chikashige Y., Yamashita Y. M., Hiraoka Y., Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A., Koujin T., Tsutsumi C., Haraguchi T., Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y., Goshima G., Morishita J., Yanagida M. M phase-specific kinetochore proteins in fission yeast. Microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Nekrasov V. S., Smith M. A., Peak-Chew S., Kilmartin J. V. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004;6:1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- Ohkura H., Adachi Y., Kinoshita N., Niwa O., Toda T., Yanagida M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 1988;7:1465–1473. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:427–429. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J. F., Borgstrom B., Allshire R. C. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Petersen J., Paris J., Willer M., Philippe M., Hagan I. M. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 2001;114:4371–4384. doi: 10.1242/jcs.114.24.4371. [DOI] [PubMed] [Google Scholar]
- Pidoux A. L., Allshire R. C. Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol. 2000;12:308–319. doi: 10.1016/s0955-0674(00)00094-6. [DOI] [PubMed] [Google Scholar]
- Pidoux A. L., Allshire R. C. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- Pidoux A. L., Richardson W., Allshire R. C. Sim4, a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 2003;161:295–307. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagolla M. J., Uzawa S., Cande W. Z. Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 2003;116:4891–4903. doi: 10.1242/jcs.00796. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Ishii K., Kobayashi Y., Takahashi K. Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol. Biol. Cell. 2005;16:3666–3677. doi: 10.1091/mbc.E05-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Takahashi K., Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Samejima I., Lourenco P. C., Snaith H. A., Sawin K. E. Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol. Biol. Cell. 2005;16:3040–3051. doi: 10.1091/mbc.E04-11-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S. J., Crawley K., Karig I., Buck V., Meadows J. C., Franco-Sanchez A., Fleig U., Toda T., Millar J. B. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K. E., Lourenco P. C., Snaith H. A. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Schuyler S. C., Liu J. Y., Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E. S., Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C., Tanaka T. U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tirnauer J. S., Canman J. C., Salmon E. D., Mitchison T. J. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic-Norrelykke I. M., Sacconi L., Thon G., Pavone F. S. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr. Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Trautmann S., Wolfe B. A., Jorgensen P., Tyers M., Gould K. L., McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 2001;11:931–940. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Vardy L., Toda T. The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 2000;19:6098–6111. doi: 10.1093/emboj/19.22.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K. T. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J. Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram S., Tasto J. J., Feoktistova A., Jennings J. L., Link A. J., Gould K. L. Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell. 2004;15:2287–2301. doi: 10.1091/mbc.E03-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verni F., Somma M. P., Gunsalus K. C., Bonaccorsi S., Belloni G., Goldberg M. L., Gatti M. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr. Biol. 2004;14:1569–1575. doi: 10.1016/j.cub.2004.08.054. [DOI] [PubMed] [Google Scholar]
- West R. R., Vaisberg E. V., Ding R., Nurse P., McIntosh J. R. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell. 1998;9:2839–2855. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I. M., Anderson S., Yates J. R., 3rd, Drubin D. G., Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P. A., Kilmartin J. V. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere componente and has a function in chromosome segregation. J. Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Sato M., Fujita A., Yamamoto M., Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol. Biol. Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]