PU.1 can participate in an active enhancer complex without its transcriptional activation domain (original) (raw)
Abstract
The transcription factor PU.1 is necessary for the development of multiple hematopoietic lineages and contributes to the activity of the immunoglobulin κ 3′ enhancer. A variety of proteins bind to the 3′ enhancer (PU.1, PIP, ATF1, CREM, c-Fos, c-Jun, and E2A), but the mechanism of 3′-enhancer activity and the proteins necessary for its activity are presently unclear. We show here that PU.1 participates with other transcription factors in forming a higher-order complex with 3′-enhancer DNA sequences. Each protein is necessary for formation of this complex. Individually, transcription factors that bind to the 3′ enhancer do not appreciably stimulate transcription in a cell type in which the 3′ enhancer is normally silent (NIH 3T3). However, mixture of multiple transcription factors (PU.1, PIP, c-Fos, and c-Jun) can greatly activate the enhancer. PU.1 is necessary for maximal enhancer activity, but mutants of PU.1 that lack the transcriptional activation domain are nearly as efficient at stimulating enhancer activity as the wild-type PU.1 protein. PU.1 apparently can activate transcription by playing an architectural role in interactions with other transcription factors.
Keywords: immunoglobulin κ, transcription, protein interaction
PU.1 is an ets domain transcription factor (1) implicated in cell growth and differentiation (2–5). PU.1 can immortalize erythroblasts and overexpression of the PU.1 gene (Spi-1) by retroviral insertion appears to cause erythroleukemia (4, 6–9). Inactivation of the mouse PU.1 gene by targeted homologous recombination results in lethality at about 18 days of gestation apparently due to loss of B cells, T cells, and myeloid cells (5). Several domains within the PU.1 protein are associated with specific functions. The carboxyl-terminal region contains the 89 amino acid ets domain necessary for DNA binding (1). The amino-terminal 100 amino acids contain the PU.1 transcriptional activation domain (10–13), and a PEST domain (rich in proline, glutamic acid, serine, and threonine) lies between amino acids 118 and 160.
We previously demonstrated that PU.1 binds to the B cell-specific immunoglobulin kappa (Igκ) 3′ enhancer and can control transcriptional activity (14, 15). The 3′ enhancer contributes to transcriptional regulation, as well as to somatic recombination and somatic mutation of the κ locus (16–19). Therefore, understanding the mechanism of 3′-enhancer function is important. Binding of PU.1 to its target DNA sequence in the 3′ enhancer results in the cooperative recruitment of a second transcription factor, PIP (previously called NF-EM5), to an adjacent DNA site (14, 15). Recruitment of PIP to DNA requires both specific protein–protein interactions with PU.1 and specific protein–DNA interactions with Igκ enhancer sequences (14). PU.1 sequences 118 to 160 (the PEST domain) are required for PIP recruitment, and PU.1 serine residue 148 must be phosphorylated (15). PU.1 and PIP also bind in a similar fashion to the immunoglobulin λ2-4 enhancer (20).
In addition to PU.1 and PIP, the Igκ 3′-enhancer core (which contains most of the enhancer activity) binds to the transcription factors ATF1, CREM, and the E2A gene products (14, 21, 22). ATF1 and CREM bind to a region termed the κE3′–CRE (22). The κE3′–CRE sequence contains two AP-1 half binding sites and c-Fos and c-Jun can bind to this sequence and can activate a κE3′–CRE-dependent reporter plasmid (see below). Site-specific mutation of the κE3′–CRE, PU.1, PIP, or E2A binding sites results in greatly reduced enhancer activity (22), suggesting that these proteins cooperatively interact to stimulate enhancer activity. We show here that PU.1 participates with proteins that bind to the PIP, E2A, and κE3′–CRE binding sites in the formation of a higher-order protein–DNA complex. This complex is not formed with PU.1 mutants that cannot recruit PIP to bind to DNA. None of the enhancer binding proteins alone can significantly activate the 3′ enhancer. However, cotransfection of a mixture of enhancer binding proteins results in highly synergistic levels of transcription. PU.1 mutants that lack the PU.1 transactivation domain also stimulate enhancer activity in the presence of the other enhancer binding proteins. This indicates that PU.1 can stimulate enhancer activity by playing an architectural role in the assembly of a higher-order protein–DNA complex.
MATERIALS AND METHODS
Plasmid Constructs.
Plasmid CMVΔ33-100 contains the PU.1 coding sequences (deleted of residues 33–100) inserted into the _Eco_RI site of cytomegalovirus (CMV) expression plasmid pCB6+. Plasmid CMVΔ7-30 contains the PU.1 coding sequences (deleted of residues 7–30) inserted at the _Eco_RI–_Hin_dIII sites of pCB6+. Multimers (four copies) of either the PU.1 plus PIP sites or the κE3′–CRE sequence were inserted at the _Bam_HI–_Bgl_II sites upstream of the liver bone kidney (LBK) alkaline phosphatase promoter driving expression of the chloramphenicol acetyltransferase (CAT) gene to produce (PU.1 plus PIP)4LBKCAT and (κE3′–CRE)4LBKCAT, respectively (21). CoreLBKCAT contains enhancer sequences 391–523 (23) inserted into the _Bam_HI–_Bgl_II sites upstream of the promoter sequences of LBKCAT by blunt-end ligation. Plasmids CMV–PU.1, ΔPEST, and Δ33-100 have been described (14, 15). Plasmid CMV–PIP was supplied by H. Singh (University of Chicago). CMV–Fos and CMV–Jun were supplied by F. Rauscher (Wistar Institute, Philadelphia). SV40CREMτ was supplied by P. Sassone-Corsi (Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France), and CMVATF1 was supplied by M. Yoshida (University of Tokyo). CMV-E2A (E2-5) was supplied by T. Kadesch (University of Pennsylvania).
Electrophoretic Mobility-Shift Assays (EMSAs).
EMSA was performed with ≈0.1 ng of labeled DNA probe (4000 cpm) in a 20 μl reaction mixture containing 4 μg poly(dI-dC), 10 mM Tris·HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, and 8 μg nuclear extract. S194 nuclear extract was prepared by the method of Dignam et al. (24). For immunodepletions, 16 μg nuclear extract was incubated with 5 μg specific antibody for 2 hr at room temperature. Antibodies included anti-c-Fos, anti-c-Jun, anti-E2A (all from PharMingen); anti-CREM (Santa Cruz Biotechnology); and anti-ATF1 (Upstate Biotechnology, Lake Placid, NY). Immune complexes were removed by addition of protein A-agarose and centrifugation. Proteins made by in vitro transcription and translation were prepared from RNAs transcribed in vitro from cDNA plasmids (PU.1, Δ33-100, ΔPEST, and S148A; refs. 14 and 15) by either T7 or T3 RNA polymerases (Stratagene). Proteins were translated in vitro using nuclease-treated RNA-dependent rabbit reticulocyte lysates (Promega) at 30°C for 60 min. Oligonucleotide competitors (100 ng) were added to reactions before addition of the labeled probe. Oligonucleotides used in these studies are as follows: PU.1 plus PIP, CTTTGAGGAACTGAAAACAGAACCT; κE3′–CRE, AGCAACTGTCATAGCTACCGTCACA; E2A, ACATCTGTTGCTTTCGCTCCCATCC; oligo 3, TACCGTCACACTGCTTTGATCAAGA; and N.S., CATTGCACAATCTA. Samples were electrophoresed on 4% polyacrylamide gels in 6.7 mM Tris·HCl (pH 7.5), 3.3 mM NaAc, and 1 mM EDTA.
Transfections.
NIH 3T3 cells were grown in DMEM supplemented with 10% fetal calf serum. Transfections were performed by the calcium phosphate method of Graham and Van der Eb (25). Transfections contained 5 μg reporter plasmid, 3 μg of each effector plasmid, and 1 μg of the β-galactosidase expression plasmid pCH110 (26) to normalize for transfection efficiencies. Total DNA concentration was kept at 21 μg by inclusion of plasmid pCB6+. Transfections were harvested at 44 hr, and CAT assays were performed according to Gorman et al. (27). Data shown are averages of 3–7 independent transfections. For metabolic labeling of transfected proteins, cells were washed twice with media lacking cysteine and methionine and incubated for 10 min at 37°C 24 hr after transfection, and then they were incubated with [35S]methionine and [35S]cysteine (0.2 mCi/ml; 1 Ci = 37 GBq) for 2 hr. Cells were lysed in 20 mM Tris·HCl (pH 7.4), 0.5% SDS, 0.5% deoxycholate, 0.5% aprotinin, leupeptin, pepstatin, and phenylmethylsulfonyl fluoride, and cell lysates were then incubated with antibodies listed in the figure legend for immunoprecipitation.
RESULTS
PU.1, PIP, E2A, and κE3′–CRE Binding Proteins Can Form a Higher-Order Protein–DNA Complex.
The κE3′–CRE and E2A regions flank the PU.1 and PIP binding sites on their 5′ and 3′ sides, respectively (Fig. 1). As mentioned above, mutation of any one protein binding site in the 3′ enhancer greatly reduces enhancer activity (Fig. 1), suggesting that these proteins functionally interact as a complex (22). To study functional interactions between the proteins that bind to the 3′ enhancer, we performed EMSAs with the entire 132-bp core as probe. A complex EMSA pattern was obtained using S194 plasmacytoma cell nuclear extract (Fig. 2A, lane 1). Because PU.1 can cooperatively bind to DNA with PIP (14, 15), we sought to determine whether addition of exogenous PU.1 to the S194 cell nuclear extract would assist in the formation of protein–DNA interactions. PU.1 bound to the core probe and yielded the expected EMSA complex (lane 2). Addition of PU.1 to the S194 extract resulted in increased intensity of the PU.1 and PU.1 plus PIP complexes (14) as well as the appearance of a new very slowly migrating complex (complex A, compare lanes 1 and 3). Rabbit reticulocyte lysate alone had no effect on the EMSA complexes (lane 4). This new complex contains proteins that bind to the κE3′–CRE, PU.1 plus PIP, and E2A binding sites because inclusion of unlabeled competitor oligonucleotides containing these binding sites abolished complex A (Fig. 2B, lanes 2, 4, and 5). An oligonucleotide that partially overlaps the κE3′–CRE but extends further 3′ of this sequence (oligo 3) abolished some higher mobility complexes, but not complex A (lane 3). A nonspecific oligonucleotide had no effect on any complexes (lane 6). PU.1 appears to be the limiting component of complex A because addition of other proteins that bind to the enhancer core did not result in appearance of this complex (data not shown).
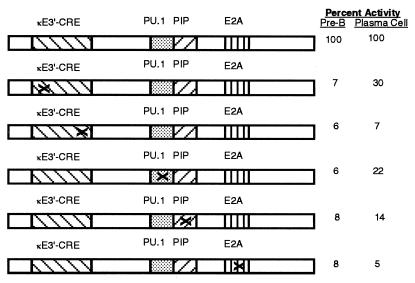
Figure 1.
Mutation of any protein binding site in the Igκ 3′-enhancer core region reduces enhancer activity. Shown is the 3′-enhancer core region spanning nucleotides 391–523 (23). The relative positions of the κE3′–CRE, PU.1, PIP, and E2A binding sites are indicated. The consequences of mutation of each protein binding site on enhancer activity in pre-B and plasmacytoma cells is shown on the right. These data are summarized from Pongubala and Atchison (22) and represent mutants LSB, LSD, LSF, LSH, and LSJ, respectively.
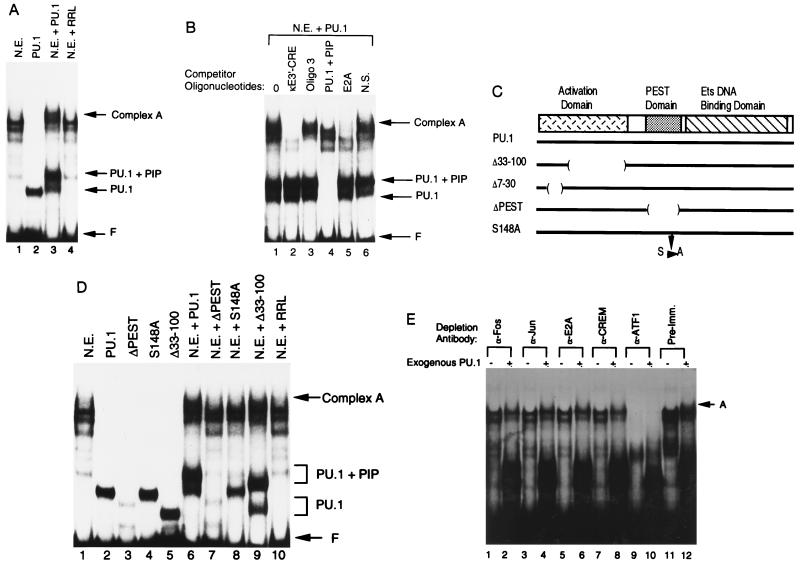
Figure 2.
Formation of a higher-order complex over the enhancer core. (A) EMSA was performed with the 3′-enhancer core sequence (residues 391–523) as a probe. Assays were performed with either S194 plasmacytoma nuclear extract (N.E.), PU.1 prepared by in vitro transcription and translation (PU.1), nuclear extract plus PU.1 (N.E. plus PU.1), or nuclear extract incubated with unprogrammed rabbit reticulocyte lysate (N.E. plus RRL). The positions of free probe (F), PU.1–DNA complex (PU.1), PU.1 plus PIP–DNA complex (PU.1 plus PIP), and complex A are indicated by the arrows at the right. (B) Complex A contains multiple enhancer binding proteins. EMSA was performed with the enhancer core probe and S194 nuclear extract plus exogenous PU.1. The various unlabeled competitor oligonucleotides added to the reactions are listed above each lane. The positions of various protein–DNA complexes are indicated by the arrows at the right. (C) Diagram of the PU.1 protein. The PU.1 transcriptional activation, PEST, and Ets DNA binding domains are indicated. Various PU.1 mutants used in this study are diagrammed below. (D) Complex A contains PIP. EMSA was performed with the enhancer core probe in the presence of various recombinant proteins alone or with S194 nuclear extract. Proteins included in each reaction are indicated above the lanes. (E) Identification of other proteins in complex A. EMSA was performed with the enhancer core probe and immunodepleted S194 nuclear extract. Antibodies used for immunodepletion are indicated above each lane. The presence or absence of exogenous PU.1 is indicated by a + or −, respectively.
Competition with a PU.1 plus PIP oligonucleotide cannot prove whether PIP is present in complex A because PIP cannot bind to DNA in the absence of PU.1 (14, 20). To determine whether PIP is a necessary component of complex A, we performed EMSA with mutants of PU.1 (Fig. 2C) that either retain or lose their ability to recruit PIP to bind to DNA. PU.1 mutant Δ33-100 can recruit PIP to bind to DNA whereas mutants ΔPEST and S148A cannot (14, 15). Each of these proteins can individually bind to the enhancer core (Fig. 2D, lanes 2–5). Addition of the wild-type PU.1 protein or the Δ33-100 PU.1 mutant to S194 nuclear extract resulted in appearance of complex A (Fig. 2D, lanes 6 and 9), whereas mutants ΔPEST and S148A did not (Fig. 2D, lanes 7 and 8). Therefore, PIP is very likely to be a component of complex A.
To further characterize the proteins in complex A, we used antibodies specific for c-Fos, c-Jun, E2A, CREM, and ATF1 to deplete S194 nuclear extracts. Depleted extracts were analyzed by EMSA with the enhancer core in the absence or presence of exogenous PU.1 (Fig. 2E). When compared with depletion by preimmune sera, all specific antibodies reduced the appearance of complex A (Fig. 2E). Although conclusive proof will require additional studies, these studies suggest that c-Fos, c-Jun, E2A, CREM, and ATF1 may participate in the formation of complex A. In summary, our results indicate that a higher-order complex (complex A) can form on the 3′-enhancer probe, and this complex contains proteins that bind to the κE3′–CRE, PU.1, PIP, and E2A binding sites. Loss of any one of these protein components results in loss of complex A.
Enhancer Activation Requires Synergy Between Multiple Transcription Factors.
Individually, PU.1 can stimulate a reporter plasmid containing a multimerized (four copies) PU.1 binding site (Fig. 3A, lane 2). As expected, PIP alone cannot stimulate transcription but can synergize with PU.1 to stimulate this reporter plasmid (Fig. 3A, lanes 3 and 4). Similarly, the κE3′–CRE-binding proteins (ATF1, CREMτ, c-Fos plus c-Jun) can stimulate expression of a reporter plasmid containing a multimerized κE3′–CRE binding site (Fig. 3A, lanes 5–8). However, as mentioned above, our previous work indicated that in the context of the 3′ enhancer, all of these protein binding sites must be intact for maximal enhancer activity (ref. 22; Fig. 1). One might therefore predict that none of the enhancer binding proteins alone would greatly stimulate the 3′ enhancer. However, in the presence of multiple enhancer binding proteins the enhancer might be greatly activated. A 3′ enhancer-dependent reporter plasmid was transfected into NIH 3T3 cells either alone or in the presence of various plasmids expressing enhancer-binding proteins. The 3′ enhancer is B cell-specific and, as expected, the enhancer core was inactive in NIH 3T3 cells (Fig. 3B, lane 1). Addition of PU.1, PU.1 plus PIP, c-Fos plus c-Jun, or E2A did not stimulate the enhancer (lanes 2–4 and 7). CREMτ and ATF1 each weakly stimulated enhancer activity (lanes 5 and 6). However, no synergism in activity was observed upon mixture of PU.1 and PIP with ATF1 or CREMτ in the presence or absence of E2A (lanes 8, 9, 12, and 13). Similarly, no synergism was observed when mixing PU.1, PIP, c-Jun, and CREMτ (lane 11). However, a very dramatic induction in enhancer activity (20-fold) was observed upon mixture of PU.1, PIP, c-Fos, and c-Jun (lane 10). Inclusion of E2A did not increase this induction (lane 14). The enhancer induction by PU.1, PIP, c-Fos, and c-Jun was particularly impressive because neither PU.1 plus PIP nor c-Fos plus c-Jun showed any activity (lanes 3 and 4). No enhancer activity was observed in the absence of κE3′–CRE binding proteins (i.e., transfections containing PU.1, PIP, and E2A; lane 15) or in the absence of PU.1 and PIP proteins (transfections containing c-Fos, c-Jun, and E2A; lane 16). None of the factors, either alone or mixed together, activated the parent vector lacking the 3′ enhancer core sequences (data not shown).
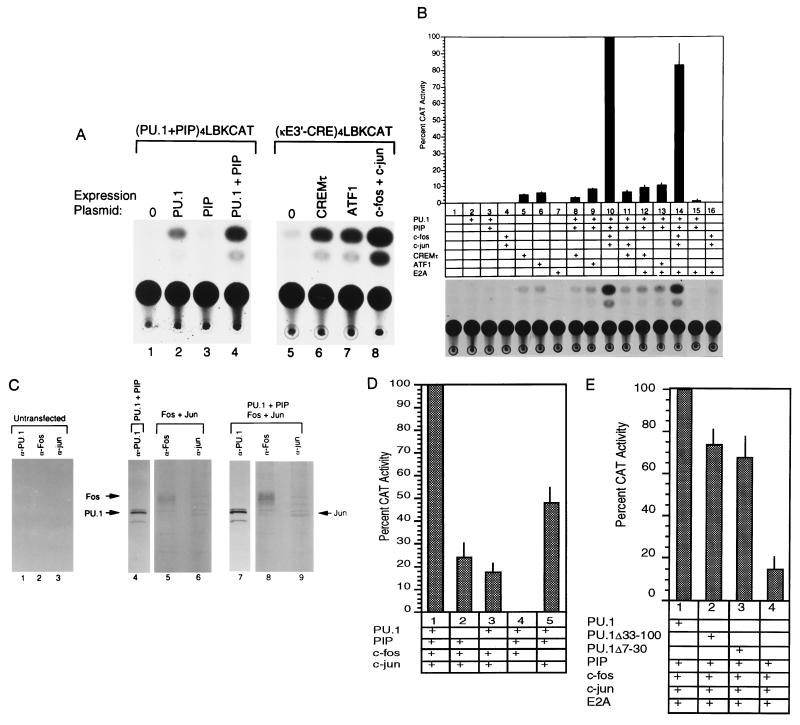
Figure 3.
Transcriptional activation by enhancer binding proteins. (A) PU.1, PU.1 plus PIP, CREMτ, ATF1, and c-Fos plus c-Jun can activate reporter plasmids containing multimers of their binding sites. Transfections were performed in NIH 3T3 cells with reporter plasmids (PU.1 plus PIP)4LBKCAT (lanes 1–4) or (κE3′–CRE)4LBKCAT (lanes 5–8). Expression plasmids included in each assay are indicated above the lanes. Fold activation levels are as follows: PU.1, 3.7 ± 1.6; PIP, 0; PU.1 plus PIP, 18.4 ± 8.1; CREMτ, 6.6 ± 3.4; ATF1, 8.1 ± 2.6; c-Fos plus c-Jun, 40.5 ± 20. (B) Maximal transcriptional synergy requires PU.1, PIP, c-Fos, and c-Jun. The CoreLBKCAT reporter plasmid was transfected into NIH 3T3 cells either alone (lane 1), or in the presence of various expression plasmids (lanes 2–16). (Bottom) CAT assays. (Top) Histogram of the data averaged from 3 to 5 independent experiments. Thin lines represent standard deviations. (Middle) Expression plasmids included in each transfection. (C) Expression of transfected proteins. NIH 3T3 cells were either untreated (lanes 1–3), or transfected with PU.1 and PIP (lane 4), c-Fos and c-Jun (lanes 5 and 6), or PU.1, PIP, c-Fos, and c-Jun (lanes 7–9). After transfection, cells were metabolically labeled with [35S]methionine and [35S]cysteine and then lysed. Lysates were immunoprecipitated with α-PU.1, α-c-Fos, or α-c-Jun antibodies then subjected to SDS/PAGE. The identity of the antibody used is shown above each lane, and the positions of PU.1, c-Fos, and c-Jun proteins are shown by arrows. (D) Omission of PU.1, PIP, c-Fos, or c-Jun expression plasmid reduces enhancer activity. (Lower) NIH 3T3 cells transfected with the various expression plasmids. (Upper) Histograms of enhancer activity for each transfection. Activity with PU.1, PIP, c-Fos, and c-Jun was defined as 100%. Thin lines represent standard deviations. (E) The PU.1 transactivation domain is not necessary for stimulation of enhancer activity. NIH 3T3 cells were transfected with PIP, E2A, c-Fos, and c-Jun in the presence of either wild-type PU.1 or PU.1 transactivation domain mutants (Δ33-100 and Δ7-30). The histogram shows relative enhancer activities with the activity observed with PU.1, PIP, c-Fos, c-Jun, and E2A defined as 100%. Thin lines represent standard deviations.
To be certain that the strong enhancer activation observed with PU.1, PIP, c-Fos, and c-Jun (Fig. 3B, lane 10) was not due to differential expression of transfected proteins, we assayed protein levels in transfected cells (Fig. 3C). Cells transfected with either PU.1 and PIP (Fig. 3C, lane 4), c-Fos and c-Jun (lanes 5 and 6), or PU.1, PIP, c-Fos, and c-Jun (lanes 7–9) were metabolically labeled and protein levels were assayed by immunoprecipitation with antibodies specific for PU.1, c-Fos, or c-Jun (antisera for PIP is not currently available). These studies showed very similar levels of each transcription factor in each transfection (Fig. 3C). Therefore, altered protein levels cannot account for the very high enhancer activity observed with PU.1, PIP, c-Fos, and c-Jun.
To more systematically discern the role of PU.1, PIP, c-Fos, and c-Jun in activating the enhancer core, transfections were performed in which each factor was individually omitted. Removal of any one factor reduced enhancer activity (Fig. 3D). Removal of c-Jun totally abolished activation (lane 4). Removal of PIP (lane 3) lowered activity to 17% of maximal activity. Removal of PU.1 (lane 2) resulted in 24% activity. This was particularly surprising because in the absence of PU.1, PIP has not previously been observed to bind to DNA (14, 20). However, the weak but detectable activation by PIP in the presence of c-Fos and c-Jun suggests that PIP may interact with other factors in addition to PU.1. Deletion of c-Fos had the least dramatic affect on enhancer induction (48%; Fig. 3D, lane 5).
PU.1 Can Stimulate 3′-Enhancer Activity in the Absence of its Transcriptional Activation Domain.
The fact that enhancer activation could not be achieved by any of the individual proteins and that maximal activation required PU.1, PIP, c-Jun, and c-Fos, suggested that these factors cooperate as a complex. One possibility is that an important function of PU.1 is to assist in the formation of this complex. If this is true, PU.1 mutants that lack the transcriptional activation domain may still stimulate enhancer activity. Transfections were performed (Fig. 3E) with PIP, c-Fos, c-Jun, and E2A in the presence of either wild-type PU.1, or PU.1 mutants (Δ33-100, Δ7-30; Fig. 2C) lacking the transcriptional activation domain (13). Interestingly, both transactivation domain mutants were nearly as efficient as the wild-type PU.1 protein at stimulating enhancer activity (Fig. 3E, lanes 1–3). Addition of either of the two transactivation domain deletion mutants resulted in a 5-fold transcriptional activation compared with the activity observed with PIP, c-Jun, c-Fos, and E2A alone (Fig. 3E). Therefore, PU.1 mutants lacking the transcriptional activation domain can still stimulate transcription apparently by assisting in the formation of a higher order protein complex over the Igκ 3′ enhancer.
DISCUSSION
Our results show that nucleation of multiple transcription factors on the 3′ enhancer core is necessary for enhancer activity. PU.1, PIP, c-Fos, and c-Jun appear to be particularly important for activity. These factors can activate the 3′ enhancer in fibroblasts, a cell in which the enhancer is normally silent. Fibroblasts contain E2A products, c-Jun and c-Fos, but lack PU.1 and PIP. Our previous EMSA studies with the κE3′–CRE sequence revealed identical complexes with NIH 3T3 and B cell nuclear extracts (oligo 2 in ref. 21), suggesting that the absence of PU.1 and PIP may be responsible for the lack of enhancer activity in fibroblasts. However, transfection of PU.1 and PIP alone was not sufficient to activate the enhancer indicating that other proteins are necessary.
It is interesting that transfections performed with various κE3′–CRE binding proteins yielded differing levels of enhancer activity. CREMτ and ATF1 yielded low levels of activity whereas c-Fos and c-Jun caused significantly higher activity. The CREM gene can express numerous isoforms of CREM by differential RNA processing, promoter usage, or translational start site (28–31). Some of these isoforms are activators, while others are repressors. One repressor isoform, CREMα, can repress 3′-enhancer activity (22). Therefore, one mechanism for controlling enhancer activity could involve the differential expression of proteins that bind to the κE3′–CRE.
The role of c-Fos in 3′-enhancer activity is somewhat unclear. While highest enhancer activity was observed in the presence of c-Fos (i.e., transfections containing PU.1, PIP, c-Fos, and c-Jun), absence of c-Fos lowered enhancer activity by only 50% (Fig. 3D). In addition, we were previously unable to detect c-Fos in EMSA complexes with the κE3′–CRE probe and S194 nuclear extracts (22). Due to the complex nature of the AP-1 and ATF transcription factor families, further studies will be required to determine the specific factors responsible for enhancer activity in B cells. However, the work presented here provides a framework for studying the assembly of enhancer binding proteins on the Igκ 3′ enhancer. The assortment of factors that can potentially control 3′ enhancer activity are summarized in Fig. 4.
Figure 4.
Summary of the influence of various enhancer binding factors on transcriptional activity of the 3′ enhancer.
It is curious that E2A did not significantly contribute to the enhancer activity observed here because the E2A binding site is important for enhancer activity in plasmacytoma cells (22). Several explanations are possible. First, E2A binds to DNA as a homodimer in B cells whereas it binds mainly as a heterodimer in other cell types (32). Differences in E2A DNA binding patterns in B cells compared with fibroblasts may alter its ability to induce 3′ enhancer activity in 3T3 cells. Second, endogenous E2A proteins in 3T3 cells may be high enough such that additional E2A is unnecessary for maximal activity. Finally, a different E box-binding protein may contribute to enhancer activity in B cells.
Transfections with PU.1 transactivation domain deletion mutants and our EMSA studies suggest that PU.1 plays an important role in the formation of a higher-order protein–DNA complex. Efficient enhancer activation depends upon the presence of PU.1 but not the PU.1 transactivation domain. Therefore, interaction of PU.1 with other proteins appears to be more important than the specific contribution of the PU.1 transactivation domain. This mode of regulation by nucleation of enhancer binding proteins may be a common mechanism. The T cell receptor α enhancer binds to factors LEF-1, Ets-1, PEBP2α, and ATF/CREB. Cooperative interactions between Ets-1 and PEBP2α are stabilized by the binding of ATF/CREB (33). Mutation of any of these protein binding sites greatly reduces enhancer activity (33). Similarly, activation of the interferon β gene requires a multiprotein complex consisting of IRF-1, NF-κB, ATF2/c-Jun, and HMGI (34).
It is interesting that the 3′ enhancer PU.1 binding site is also very important for somatic recombination of Igκ genes. Point mutation of the PU.1 site in the 3′ enhancer disrupts the tissue-specificity and developmental timing of Igκ gene rearrangement (14). Therefore, a protein complex over the 3′ enhancer containing PU.1 may be important for both enhancer activity and for the somatic recombination of Igκ genes. Based on our results here, we would predict that mutation of any protein binding site within the enhancer core region might have the same effect on somatic recombination.
Acknowledgments
We thank F. Rauscher, H. Singh, P. Sassone-Corsi, M. Yoshida, and T. Kadesch for expression plasmids. We thank R. Ricciardi, N. G. Avadhani, J. Pehrson, J. Perkel, and S. Maitra for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM42415 to M.L.A. J.M.R.P. is a Special Fellow of the Leukemia Society.
Footnotes
Abbreviations: Igκ, immunoglobulin κ; CMV, cytomegalovirus; LBK, liver bone kidney; CAT, chloramphenicol acetyltransferase; EMSA, electrophoretic mobility-shift assay.
References
- 1.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 2.Ben-David Y, Bernstein A. Cell. 1991;66:831–834. doi: 10.1016/0092-8674(91)90428-2. [DOI] [PubMed] [Google Scholar]
- 3.Schuetze S, Paul R, Gliniak B C, Kabat D. Mol Cell Biol. 1992;12:2967–2975. doi: 10.1128/mcb.12.7.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuetze S, Stenberg P E, Kabat D. Mol Cell Biol. 1993;13:5670–5678. doi: 10.1128/mcb.13.9.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott E W, Simon M C, Anastasi J, Singh H. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 6.Paul R, Schuetze S, Kozak S L, Kabat D. J Virol. 1989;63:4958–4961. doi: 10.1128/jvi.63.11.4958-4961.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul R, Schuetze S, Kozak S L, Kozak C A, Kabat D. J Virol. 1991;65:464–467. doi: 10.1128/jvi.65.1.464-467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreau-Gachelin F, Tavitian A, Tambourin P. Nature (London) 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 9.Moreau-Gachelin F, Ray D, Mattei M-G, Tambourin P, Tavitian A. Oncogene. 1989;4:1449–1456. [PubMed] [Google Scholar]
- 10.Hagemeier C, Bannister A J, Cook A, Kouzarides T. Proc Natl Acad Sci USA. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kominato Y, Galson D L, Waterman W R, Webb A C, Auron P E. Mol Cell Biol. 1995;15:58–68. [PMC free article] [PubMed] [Google Scholar]
- 12.Shin M K, Koshland M E. Genes Dev. 1993;7:2006–2015. doi: 10.1101/gad.7.10.2006. [DOI] [PubMed] [Google Scholar]
- 13.Klemsz M J, Maki R A. Mol Cell Biol. 1996;16:390–397. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pongubala J M R, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pongubala J M R, Van Beveren C, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 16.Hiramatsu R, Akagi K, Matsuoka M, Sakumi K, Nakamura H, Kingsbury L, David C, Hardy R R, Yamamura K-i, Sakano H. Cell. 1995;83:1113–1123. doi: 10.1016/0092-8674(95)90138-8. [DOI] [PubMed] [Google Scholar]
- 17.Sharpe M J, Milstein C, Jarvis J M, Neuberger M S. EMBO J. 1991;10:2139–2145. doi: 10.1002/j.1460-2075.1991.tb07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betz A G, Milstein C, Gonzalel-Fernandez A, Pannell R, Larson T, Neuberger M S. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 19.Maizels N. Cell. 1995;83:9–12. doi: 10.1016/0092-8674(95)90227-9. [DOI] [PubMed] [Google Scholar]
- 20.Eisenbeis C F, Singh H, Storb U. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 21.Pongubala J M R, Atchison M L. Mol Cell Biol. 1991;11:1040–1047. doi: 10.1128/mcb.11.2.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pongubala J M R, Atchison M L. J Biol Chem. 1995;270:10304–10313. doi: 10.1074/jbc.270.17.10304. [DOI] [PubMed] [Google Scholar]
- 23.Meyer K B, Neuberger M S. EMBO J. 1989;8:1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dignam D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham F L, Van der Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 26.Hall C V, Jacob P E, Ringold G M, Lee F. J Mol Appl Genet. 1983;2:101–110. [PubMed] [Google Scholar]
- 27.Gorman C M, Moffat L F, Howard B H. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foulkes N S, Borrelli E, Sassone-Corsi P. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 29.Foulkes N S, Mellstrom B, Benusiglio E, Sassone-Corsi P. Nature (London) 1993;355:80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- 30.Foulkes N S, Sassone-Corsi P. Cell. 1992;68:411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- 31.Delmas V, Laoide B M, Masquilier D, deGroot R P, Foulkes N S, Sassone-Corsi P. Proc Natl Acad Sci USA. 1992;89:4226–4230. doi: 10.1073/pnas.89.10.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen C-P, Kadesch T. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 34.Thanos D, Maniatis T. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]