The Genomic Landscape of Short Insertion and Deletion Polymorphisms in the Chicken (Gallus gallus) Genome: A High Frequency of Deletions in Tandem Duplicates (original) (raw)
Abstract
It is increasingly recognized that insertions and deletions (indels) are an important source of genetic as well as phenotypic divergence and diversity. We analyzed length polymorphisms identified through partial (0.25×) shotgun sequencing of three breeds of domestic chicken made by the International Chicken Polymorphism Map Consortium. A data set of 140,484 short indel polymorphisms in unique DNA was identified after filtering for microsatellite structures. There was a significant excess of tandem duplicates at indel sites, with deletions of a duplicate motif outnumbering the generation of duplicates through insertion. Indel density was lower in microchromosomes than in macrochromosomes, in the Z chromosome than in autosomes, and in 100 bp of upstream sequence, 5′-UTR, and first introns than in intergenic DNA and in other introns. Indel density was highly correlated with single nucleotide polymorphism (SNP) density. The mean density of indels in pairwise sequence comparisons was 1.9 × 10−4 indel events/bp, ∼5% the density of SNPs segregating in the chicken genome. The great majority of indels involved a limited number of nucleotides (median 1 bp), with A-rich motifs being overrepresented at indel sites. The overrepresentation of deletions at tandem duplicates indicates that replication slippage in duplicate sequences is a common mechanism behind indel mutation. The correlation between indel and SNP density indicates common effects of mutation and/or selection on the occurrence of indels and point mutations.
ALTHOUGH insertion and deletion mutations (indels) contribute significantly to the genetic divergence between species (Britten 2002; Britten et al. 2003; Chimpanzee Sequencing and Analysis Consortium 2005), the rate, pattern, and evolutionary implications of indels generally have been less well characterized compared to that of nucleotide substitutions. This is partly because indels, at least when defined in a broad sense, represent a heterogenous class of mutations, including transposition and retrotransposition, duplication, length change in tandem repetitive DNA, as well as other types of genetic change. Recently, advance has been made in understanding the mutational properties of some of these types. For instance, the temporal activity and mutational mechanisms of retrotransposons, such as Alu elements, have been investigated in some detail (Price et al. 2004), and the same applies to tandem repetitive DNAs like microsatellites (Ellegren 2004) and minisatellites (Bois 2003). Moreover, whole-genome sequencing has illuminated the role of segmental duplications in genome evolution and organization (Samonte and Eichler 2002).
However, for indels that do not represent any of the specific categories listed above, knowledge is more limited. Gross chromosomal deletions can be analyzed by cytogenetic techniques, and insertions and deletions in coding sequence in some cases are uncovered by particular phenotypes, as is the case with human disease genes (Kondrashov and Rogozin 2004). However, neither approach is useful for large-scale and unbiased studies of mutational events involving a small number of nucleotides, the dominating type of insertion and deletion. Comparative genomics offers a means for genomewide analysis of the incidence and character of short indels (Makova et al. 2004; Ogurtsov et al. 2004; Taylor et al. 2004; Yang et al. 2004). Unfortunately, producing proper alignments of divergent sequences with a high density of indels, in particular in noncoding DNA, is notoriously difficult and is sensitive to parameters of the alignment model. As a consequence, alignments may be ambiguous with respect to the number and length of gaps corresponding to indel mutations (Holmes 2005). Preferably, comparative genomic studies of indels should therefore be based on sequence alignments of closely related species or, even better, from intraspecific detection of polymorphism, e.g., through resequencing (Mills et al. 2006).
Given the contribution of indels to genetic divergence, it is likely that they represent an important source of phenotypic divergence both within and between species (Chen et al. 2005a,b). Understanding the process of indel mutation is also important in other contexts. The relative incidence of insertions and deletions affects genome size and has been recognized as a key parameter governing genome size evolution (Gregory 2005). Moreover, analyses of the genomic occurrence of indels can reveal constraints in, e.g., regulatory regions associated with, at least in part, length dependence rather than sequence dependence (Ometto et al. 2005). Finally, there is a growing interest in using indels as unique event markers for phylogenetic reconstruction, thus avoiding the inherent problems of homoplasy and convergence in phylogenetic analysis based on nucleotide substitutions (Hamilton et al. 2003; Kawakita et al. 2003; Fain and Houde 2004; Müller 2006).
The International Chicken Polymorphism Map Consortium (2004) performed partial shotgun sequencing at 0.25× coverage of three different chicken strains. In combination with the chicken genome reference sequence obtained from a red jungle fowl (International Chicken Genome Sequencing Consortium 2004), this revealed a total of 2.8 million polymorphisms. This number is particularly significant if one considers that the size of avian genomes is only 30–40% of that of mammals; it corresponds to a mean density of about one polymorphism every 350 bp across chicken chromosomes. Approximately 10% of these polymorphisms represent length variants, which, in contrast to single nucleotide polymorphisms (SNPs) (nucleotide substitutions), were not closely examined by International Chicken Polymorphism Map Consortium (2004). Here, we reanalyze 140,000 short indels detected in unique sequence of the chicken genome and we use these data to address the character and rate of indels in the chicken genome and, by using outgroup sequences, the accumulation of indels over evolutionary timescales in birds.
MATERIALS AND METHODS
The analyses were performed using a pipeline set up as a number of perl scripts, and all data were stored either as text files or in a MySQL database. All statistical tests were done using the R statistics environment (R Development Core Team 2006).
Sequence and polymorphism data:
Information on polymorphisms in chicken, both indels and SNPs, originally identified by International Chicken Polymorphism Map Consortium (2004), were downloaded through the table browser interface at the UCSC Genome Browser (http://genome.ucsc.edu). Version 1.0 of the chicken genome was downloaded from the Washington University School of Medicine Genome Sequencing Center (http://genome.wustl.edu/). Fully sequenced bacterial artificial chromosomes (BAC) clones of turkey, generated by the National Institutes of Health Intramural Sequencing Center (http://www.nisc.nih.gov/), were downloaded from GenBank. The Ensembl chicken gene build of December 2005 was downloaded from the Ensembl website in January 2006 (http://www.ensembl.org/).
SNP data filtering:
A total of 459,618 length variants were observed in the genomewide polymorphism screening of three domestic chicken breeds (Broiler, Layer, and Silkie) made by the International Chicken Polymorphism Map Consortium (2004). It has been acknowledged that many 1-bp indels in homonucleotide runs of this data set are erroneous due to problems with the base caller (International Chicken Polymorphism Map Consortium 2004). These incorrectly called bases thus appear as single nucleotide gaps in shotgun-sequencing reads when aligned to the reference chicken sequence, which was obtained by different sequencing technology and with much higher sequence coverage and accuracy (International Chicken Genome Sequencing Consortium 2004). Such possibly erroneous indels are flagged in the data tables provided by the International Chicken Polymorphism Map Consortium (2004) based on sequence context and quality scores. These possibly erroneous indels were not considered for further analysis, giving an initial data set of 272,820 length polymorphisms. This filtering procedure would imply that the actual occurrence of single-base-pair gaps is somewhat underestimated. For indel rate estimates, we assumed that there should be as many true single-base-pair gaps in the genomes of birds used for shotgun sequencing as in the chicken genome reference sequence. This number was therefore added to the number of indels left after filtering when estimating indel rates.
As the focus of the study was on insertions and deletions in nonrepetitive, unique sequence, two filtering methods for microsatellite sequences were applied. First, the longer allele of all indels, including 200 bp of flanking sequence on each side, were scanned using Tandem Repeats Finder (Benson 1999). This algorithm applies a method of fuzzy matching that will also pick up cases of degenerate microsatellites. Second, an in-house written script was used to detect if the indel was part of a short perfect tandem repeat of three or more units. For example, instances of “unique sequence[AGT][AGT][AGT]unique sequence” in the longer allele and “unique sequence[AGT][AGT][---]unique sequence” in the shorter allele would be excluded, while the observation of “unique sequence[AGT][AGT]unique sequence” and “unique sequence [AGT][---]unique sequence” would be included.
Data analysis:
Indel density was determined as both number of indel events per base pair and number of indel bases per base pair. These figures were averaged over the three strains that were screened for polymorphisms. As the screened strains were sequenced using a sparse shotgun approach, all density figures for indels and SNPs were based on the actual number of bases covered by shotgun reads in each strain. To calculate the expected numbers of 2- to 5-bp indel words, the genomic background frequencies of words were determined. The frequency of duplet tandem repeats was obtained according to the same principles.
The Ensembl tables over known and predicted genes in the chicken genome contain many alternatively spliced genes with multiple transcripts. To account for this (and the fact that genes can reside within short distances of other genes) in the analysis of indel density in relation to genes, the Ensembl table of transcripts was collapsed to a canonical table, where sequences were assigned to be coding sequence, untranslated region (UTR), first intron, other intron, or up- or downstream flanking at increasing distances to a gene, with priority following the mentioned order. If a sequence, for instance, was both coding sequence and first intron in two transcripts, it was categorized as coding sequence.
Chicken–turkey comparison:
Placement of turkey BAC clone sequences on the chicken genome was determined by blastn searches (Altschul et al. 1997) and alignments were done using MAVID (Bray and Pachter 2004). These alignments were scanned for indels, where indels classified as microsatellites using the same methods as above were excluded. The chicken–turkey alignments were also used to determine the ancestral state of indels segregating in chicken. This was done by realigning 400 bp surrounding the indel to orthologous turkey sequence using mcalign (Keightley and Johnson 2004), parameterized with chicken polymorphism data.
RESULTS
Overall density of indel events:
The International Chicken Polymorphism Map Consortium (2004) identified a total of 272,830 length variants in the chicken genome, and these data formed the basis for this study. This set of polymorphisms consists of length variation in unique DNA as well as in tandem repetitive DNA; the latter includes numerous microsatellite (simple repeat) loci. To be able to focus on the former, we subsequently filtered the data from microsatellite structures. The filtering was performed down to a level of excluding all cases where the particular sequence motif absent in the shorter allele was tandemly iterated three or more times in the longer allele. The resulting final data set used for all further analysis contained 140,484 indels.
Indel density can be given either as the number of indel events per base pair (IDE/bp) or the number of base pairs inserted or deleted (ID/bp) per base-pair sequence covered by the polymorphism screening. In our data, the mean genomewide, pairwise density of short indels in unique sequence was 1.9 × 10−4 IDE/bp or 6.7 × 10−4 ID/bp. The International Chicken Polymorphism Map Consortium (2004) reported the genomewide nucleotide diversity (π), the pairwise sequence heterozygosity with gaps excluded, to be 4–5 × 10−3 in comparisons within and between breeds as well as in comparisons between breeds and the red jungle fowl. These data indicate that segregating short indels in unique sequence of the chicken genome are on average ∼5% as common as SNPs. By extrapolation, and given a genome size of 1 Gb, it may be expected that two random copies of the chicken genome differ at ∼5 million sites, 670,000 of which would be represented by short indels in unique sequence. To this should be added differences due to longer indels and duplications and to length variation in tandem repetitive DNA.
Character of mutation:
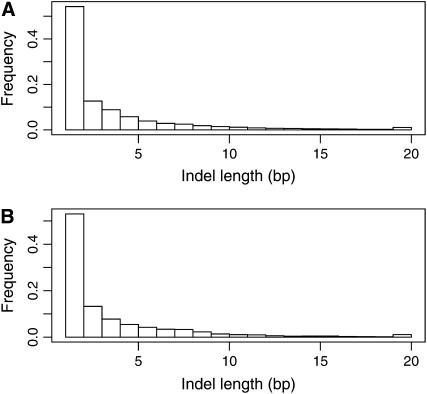
Shotgun sequencing limits the size of detectable indels to below the typical length of sequence reads. Moreover, the algorithm used to align shotgun sequence reads to a reference sequence introduces a further limit, well below the length of individual reads, a limit that will vary depending on sequence context and location of the indel within the read. The longest indel identified in our data set was 69 bp. With this caveat in mind, Figure 1A shows the observed distribution of indel lengths in the chicken genome. Clearly, single-base-pair insertions and deletions represent the predominant class. The mean length of indels was 3.6 bp with a median of 1 bp.
Figure 1.—
Size distribution of indels. (A) The size distribution of indels segregating in the chicken genome and (B) the size distribution of those observed in the chicken–turkey comparison.
To analyze the sequence motifs of short indels, the frequencies of 2- to 5-bp indel words were compared with their background genomic frequencies. There were significant deviations from random expectations for all size classes investigated, generally within the range of a twofold excess or deficit. Among 2-bp words, AT and AG were overrepresented while AA, CC, GC, and GA were underrepresented (Table 1); the low frequency of AA and CC is likely due to the filtering of homonucleotide arrays. Among 3 bp (Table 2) and longer (supplemental Table S1 at http://www.genetics.org/supplemental/) words, A-rich motifs showed clear evidence for overrepresentation at indel sites. For instance, 6 of 8 overrepresented 3-bp words consisted of two A's while none of 14 underrepresented words had two A's.
TABLE 1.
Occurrence of the 10 different canonical 2-bp indel motifs and indel flanking sequence
| Word frequencies | Identical flank | |||||
|---|---|---|---|---|---|---|
| Sequence | Observed | Expected | P | Observed | Expected | P |
| AA | 1969 | 2402 | 0 | 0 | 0 | NA |
| AC | 3424 | 3316 | 0.189 | 1218 | 452 | 0 |
| AG | 5555 | 4596 | 0 | 2948 | 736 | 0 |
| AT | 3630 | 2261 | 0 | 925 | 435 | 0 |
| CA | 4923 | 4875 | 0.49 | 2084 | 662 | 0 |
| CC | 1007 | 1756 | 0 | 0 | 0 | NA |
| CG | 315 | 344 | 0.23 | 22 | 14 | 0.047 |
| GA | 3184 | 3756 | 0 | 949 | 445 | 0 |
| GC | 753 | 1564 | 0 | 83 | 18 | 0 |
| TA | 2012 | 1902 | 0.045 | 387 | 218 | 0 |
TABLE 2.
Occurrence of 27 different canonical 3-bp indel motifs and indel flanking sequence
| Word frequencies | Identical flank | |||||
|---|---|---|---|---|---|---|
| Sequence | Observed | Expected | P | Observed | Expected | P |
| AAC | 1139 | 729 | 0 | 617 | 44 | 0 |
| AAG | 1735 | 851 | 0 | 1024 | 90 | 0 |
| AAT | 791 | 774 | 0.54 | 290 | 35 | 0 |
| ACA | 1022 | 729 | 0 | 547 | 40 | 0 |
| ACC | 264 | 417 | 6.7 × 10−13 | 61 | 10 | 0 |
| ACG | 220 | 407 | 0 | 52 | 12 | 0 |
| AGA | 1810 | 851 | 0 | 1083 | 94 | 0 |
| AGC | 217 | 407 | 0 | 62 | 12 | 0 |
| ATA | 821 | 774 | 0.23 | 340 | 36 | 0 |
| CAA | 900 | 749 | 2.5 × 10−7 | 345 | 33 | 0 |
| CAC | 438 | 632 | 1.5 × 10−13 | 108 | 16 | 0 |
| CAG | 289 | 515 | 0 | 74 | 16 | 0 |
| CCA | 447 | 632 | 1.7 × 10−12 | 121 | 17 | 0 |
| CCG | 66 | 102 | 0.0016 | 4 | 2 | 0.20 |
| CGA | 276 | 516 | 0 | 64 | 16 | 0 |
| CGC | 64 | 102 | 0.00090 | 2 | 2 | 0.85 |
| CTA | 871 | 1197 | 0 | 276 | 28 | 0 |
| GAA | 1015 | 823 | 2.25 × 10−10 | 479 | 57 | 0 |
| GAC | 284 | 553 | 0 | 66 | 12 | 0 |
| GAG | 727 | 604 | 3.6 × 10−6 | 274 | 34 | 0 |
| GCA | 320 | 552 | 0 | 70 | 11 | 0 |
| GCC | 198 | 397 | 0 | 14 | 2 | 0 |
| GGA | 1431 | 1249 | 2.0 × 10−6 | 478 | 66 | 0 |
| GTA | 900 | 1202 | 0 | 316 | 29 | 0 |
| TAA | 707 | 756 | 0.23 | 257 | 30 | 0 |
| TAC | 428 | 634 | 2.9 × 10−15 | 155 | 14 | 0 |
| TAG | 404 | 630 | 0 | 144 | 13 | 0 |
To further characterize the sequence context of indels, the immediate flanking sequence of all length variants were examined. Specifically, we asked whether flanking sequences were identical to the motif being inserted or deleted. The observed number of cases of such identities was then compared to the expected number based on the genomic averages of word frequencies and a random genomic distribution of words. There was a vast excess of identical motifs immediately preceding or following the words of indels; that is, sequences being inserted or deleted were likely to be part of tandem duplicates. The relative excess increased with the length of the indel motif, with up to a 3-fold excess for dinucleotides (Table 1), up to a 10-fold excess for trinucleotides (Table 2), and more than a 10-fold excess for tetra- and pentanucleotides (supplemental Table S1 at http://www.genetics.org/supplemental/).
Distribution of indels across the chicken genome:
There was significant heterogeneity in indel density among chromosomes (ANOVA, P < 10−16) with a trend for lower densities in smaller chromosomes (Table 3); the median density in the large macrochromosomes (chromosomes 1–5) was 20% higher (1.89 × 10−4 IDE/bp) than in the minute microchromosomes (11–32; 1.50 × 10−4 IDE/bp, P = 0.030, Mann–Whitney U-test). The Z chromosome showed significantly fewer indels than autosomes, with a density of 1.44 × 10−4 IDE/bp, 30% lower than that of macrochromosomes (P = 0.024, Wilcoxon test). This would be compatible with the lower effective population size of the Z chromosome compared to autosomes; under random mating, the effective population size of Z is three-fourths that of autosomes.
TABLE 3.
Mean indel frequency (×10−4) across chromosomes in the chicken genome
| Chromosome | Total frequency | Frequency in nongenic DNA |
|---|---|---|
| 1 | 1.89 | 1.96 |
| 2 | 1.89 | 1.92 |
| 3 | 1.67 | 1.96 |
| 4 | 1.89 | 2.07 |
| 5 | 1.89 | 2.04 |
| 6 | 2.00 | 2.19 |
| 7 | 2.00 | 2.09 |
| 8 | 1.78 | 1.89 |
| 9 | 2.11 | 2.19 |
| 10 | 1.78 | 1.92 |
| 11 | 1.44 | 1.73 |
| 12 | 2.00 | 2.07 |
| 13 | 1.67 | 1.90 |
| 14 | 1.67 | 1.90 |
| 15 | 1.33 | 1.69 |
| 17 | 1.44 | 1.70 |
| 18 | 1.78 | 2.09 |
| 19 | 1.44 | 1.67 |
| 20 | 1.56 | 1.77 |
| 21 | 1.56 | 1.69 |
| 22 | 1.11 | 1.04 |
| 23 | 1.33 | 1.63 |
| 24 | 1.56 | 1.77 |
| 26 | 1.33 | 1.76 |
| 27 | 1.67 | 1.99 |
| 28 | 1.67 | 1.83 |
| 32 | 1.44 | 1.76 |
| Z | 1.44 | 1.44 |
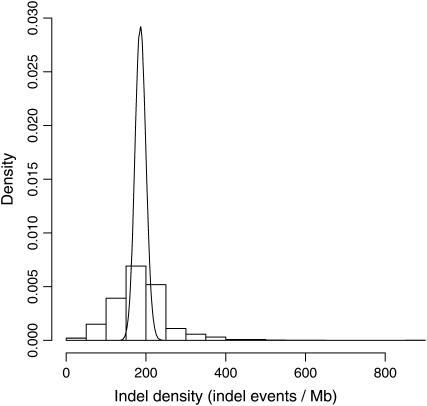
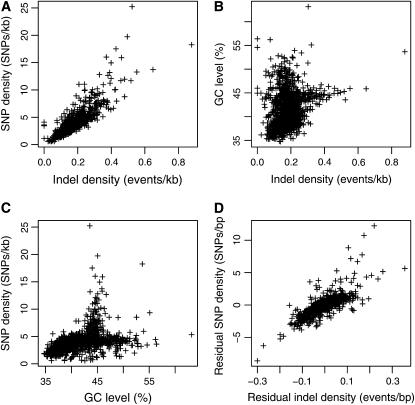
The density of indels in 1-Mb windows varied significantly across the chicken genome, a variation well beyond what is expected assuming a random distribution of indel events (Figure 2). Moreover, indel density in 1-Mb windows was strongly correlated with the density of SNPs (_r_2 = 0.69, P < 2.2 × 10−16; Figure 3A), indicating common effects of mutation and/or selection. Both SNP (_r_2 = 0.15, P < 2.2 × 10−16; Figure 3B) and indel density (_r_2 = 0.046, P = 2.3 × 10−13; Figure 3C) were correlated with local GC content. To test whether the correlation between indel and SNP density can be explained by GC, we fitted linear models of indel and SNP density against GC, respectively. The residuals from these models correlate at the same level as uncorrected indel and SNP densities (_r_2 = 0.69, P < 2.2 × 10−16; Figure 3D), indicating that GC content cannot explain the correlation alone.
Figure 2.—
Distribution of indel density in 1-Mb windows. Indel density was determined in 1-Mb windows across the chicken genome and normalized by shotgun read coverage. The smooth curve indicates the expected Poisson distribution.
Figure 3.—
Correlates of indel and SNP density in 1-Mb windows. (A) A significant correlation between indel density and SNP density. Both indel density (B) and SNP density (C) are correlated with GC level. However, correcting indel density and SNP density for GC does not remove the correlation (D), indicating that the same evolutionary forces act on indels and SNPs.
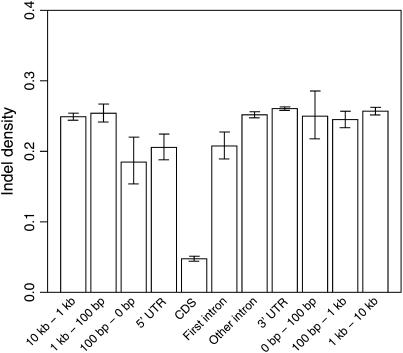
Variation in indel density should at least partly depend on functional constraint, with a lower density expected for some functional categories of the genome. Using the protein-coding gene set of the chicken genome defined by Ensembl, we investigated the frequency of indels in exons, UTRs, introns, and flanking noncoding regions at various distances from genes (Figure 4). Indels in protein-coding sequence occur at ∼10% of the background rate in noncoding DNA. Moreover, the density of indels in the first 100-bp upstream sequence, in the 5′-UTR, and in first introns was significantly lower than in the >100-bp upstream sequence, in introns other than the first intron, in the 3′-UTR, and in downstream sequences.
Figure 4.—
Indel density in different regions associated with protein-coding genes. Regions ar defined according to Ensembl annotations. Whiskers indicate the 95% confidence interval.
The difference in the occurrence of indels among functional categories could imply that differences in the abundance of functional categories among regions or chromosomes contribute to the overall heterogeneity in indel density. In particular, the observation that gene density is higher and intron size is shorter in microchromosomes than in macrochromosomes (International Chicken Genome Sequencing Consortium 2004) could potentially explain why we found fewer indels on microchromosomes. However, the contrasting density of indels in macrochromosomes and microchromosomes is still present when the 100-bp upstream sequence, 5′-UTR, exons, and first introns are excluded (Table 3; medians of 1.96 × 10−4 and 1.76 × 10−4 IDE/bp, P = 0.021).
Indels in the chicken–turkey comparison:
To enable a comparison of polymorphic and fixed indels, we aligned 5.7 Mb of genomic sequence from fully sequenced turkey (Meleagris gallopavo) BAC clones to orthologous parts of the chicken genome. Both chicken and turkey belong to the order Galliformes and show a neutral autosomal sequence divergence of ∼10% (Axelsson et al. 2005). We found 11,011 interspecific indels in the alignments after filtering for repetitive arrays using the same criteria as in the polymorphism data set. Their size distribution followed that of chicken indel polymorphisms, with an average length of 3.7 bp (Figure 1B). The incidence of indels in the chicken–turkey comparison of genomic sequence is 0.0019 IDE/bp, ∼2% of the nucleotide substitution divergence. Chicken and turkey are estimated to have diverged ∼40 million years ago (van Tuinen and Dyke 2004; Pereira and Baker 2006). This yields a molecular clock rate of 2.4 × 10−5 short indel events/million years. Note that this does not include length variants in repetitive structures.
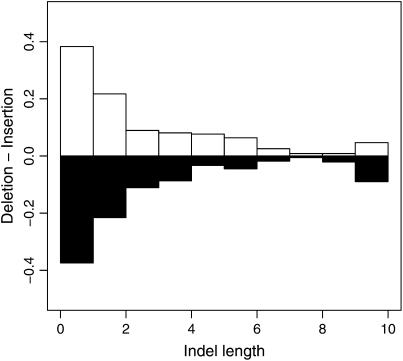
We used the available genomic sequence data from turkey to determine the ancestral state of indels segregating in chicken, thereby being able to distinguish between insertions and deletions. There were 334 deletions and 235 insertions, that is, a deletion bias of 1.42 (P = 3.3 × 10−5, χ2 test). Taking the length of mutation events into account (1246 bp deleted vs. 773 bp inserted), the deletion:insertion ratio was 1.61. Interestingly, the deletion bias was limited to macrochromosomes (278 deletions and 170 insertions; ratio 1.64, P = 3 × 10−7), while there was an equal number of deletions and insertions (56 of each) in microchromosomes. Insertions and deletions showed similar length distributions (Figure 5).
Figure 5.—
Size distributions of rooted insertions and deletions. Open bars represent insertions, while solid bars represent deletions.
For deletions, the incidence of flanking sequences in the ancestral allele identical to the deleted motif exceeded by far expectations based on the genomic distribution of duplicate words (dinucleotides: 35 observed, 8 expected, P < 10−99; trinucleotides: 16 observed, 2 expected, P < 10−99). In contrast, the incidence of duplicate words arising from insertion events did not show as pronounced deviation from expectations (dinucleotides: 7 observed, 4 expected, P = 0.036). The same trend is manifested in the different deletion:insertion ratio for cases where the longer allele was not part of a tandem duplicate (231 deletions, 188 insertions, ratio 1.23, P = 0.036) and cases where it was (99 deletions, 42 insertions, ratio 2.36, P = 1.6 × 10−6).
DISCUSSION
This study describes the genomewide occurrence of short insertions and deletions in chicken, the only bird yet to have been subject to whole-genome sequencing (International Chicken Genome Sequencing Consortium 2004). Focusing on mutations arising in unique sequence, frequencies of one indel polymorphism every 5 kb were observed, ∼5% of the SNP rate. Comparisons of the incidence of indels found in different studies are difficult because the observed frequencies depend on how length variation in tandem repetitive DNA is dealt with. The fact that the observed indel:SNP ratio in chicken is in the lower end of the range of ratios reported for other organisms is likely to be due to our stringent filtering of tandem repeats. Without tandem repeat filtering, the observed density is one indel every 2800 bp, 8% the SNP rate, a relationship more similar to that seen in, e.g., humans. Filtering and detection methods are also likely to affect the length distribution of indels. However, overall, the observed distribution in chicken is similar to that seen in other organisms (Petrov et al. 2000; Zhang and Gerstein 2003; Bhangale et al. 2005; Chimpanzee Sequencing and Analysis Consortium 2005; Mills et al. 2006).
While assessing the usefulness of individual markers was beyond the scope of this study, we acknowledge that the polymorphisms included in this data set have not been subject to PCR-based validation in population samples of chicken. However, the International Chicken Polymorphism Map Consortium (2004) validated a set of SNPs by resequencing and confirmed 94.5% in noncoding sequence. Moreover, the proportion of indels that we found (5% the SNP rate) is very similar to that seen in an extensive resequencing study of 50 unrelated chicken (Sundström and Ellegren 2004). In any case, the data should therefore be treated with due caution at the level of individual markers. Yet, as our aim was to analyze the broad-scale pattern of indel polymorphism in a nonhuman genome and since the genomewide, shotgun-based polymorphism screening in chicken presented by the International Chicken Polymorphism Map Consortium (2004) represents one of the most comprehensive diversity surveys of a large eukaryotic genome, the present data and results may have some general significance. We also note that, although chicken has been subject to strong artificial selection during domestication, the coalescence time of the great majority of polymorphisms seen in contemporary chicken populations dates back long before the initiation of domestication some 6000 years ago (International Chicken Polymorphism Map Consortium 2004).
Mechanism of indel mutation:
Together, several observations from our data highlight a potentially important mechanism behind indel mutation. First, duplicate sequences were consistently overrepresented at polymorphic indel sites of chicken. Moreover, inferred deletion mutations in the chicken–turkey comparison occurred four to eight times more often in duplicated sequence than expected, while a more modest bias was observed for insertion mutations generating tandem duplicates. Furthermore, the deletion:insertion ratio in the chicken–turkey comparison was twice as high for duplicated sequence as for nonduplicated sequence. This suggests a propensity for indel sites to represent deletion mutations in tandemly duplicated sequence. We refer to such mutation events as “2 → 1” (e.g., “unique sequence[CAG][CAG]unique sequence” → “unique sequence[CAG][---]unique sequence”), as opposed to the “1 → 2” type of insertion mutation generating tandem duplicates (e.g., “unique sequenceCTAGunique sequence” → “unique sequence[CTAG][CTAG]unique sequence”).
The abundance of 2 → 1 mutations at indel sites is compatible with a scenario in which replication slippage frequently gives rise to deletions at tandem duplicates. With two immediate neighbors of the same sequence motif there is the possibility for out-of-frame reassociation of the two strands as the polymerase traverses the duplicate region during replication. Depending on how far the nascent strand has been synthesized, slippage can give rise to either 2 → 1 deletions or 2 → 3 insertions. Our preliminary analyses indicate that these events occur with roughly equal likelihood in the chicken genome (data not shown). In contrast, 1 → 2 insertion mutations are not easily conceived with standard models of replication slippage (Levinson and Gutman 1987). We propose that this asymmetry can, at least in part, explain the deletion bias seen in this study as well as in other studies of insertions and deletions across a wide range of organisms. The observation that A-rich motifs dominate among chicken indels is consistent with the lower thermal stability of AT-rich regions and the associated higher risk for strand dissociation—the first step in slippage—during replication. Petrov (2002b) suggested that a thermodynamic asymmetry can explain a deletion bias for mutations involving longer segments; long insertions require disassociation of an appreciable stretch of already replicated DNA, whereas deletions do not. While seemingly possible, it seems less evident that this could explain the deletion bias seen for very short indels.
If replication slippage is an important mechanism behind the generation of indel polymorphism, mutation and evolution of short indels as well as of microsatellite repeats might be viewed merely as two sides of the same coin. Moreover, in this perspective, the distinction between “indels” and “microsatellites/simple repeats” becomes somewhat arbitrary (cf. Weber et al. 2002). Comparative sequencing has shown that, over evolutionary timescales, long microsatellites can evolve from initially very short repetitive structures, including tandem duplicates (Messier et al. 1996; Primmer and Ellegren 1998). The stochastic process of nucleotide substitution will continuously generate simple repeat structures, the incidence of which is highest for the shortest motifs, which form the raw material for subsequent length polymorphism due to replication slippage (Zhu et al. 2000). Starting from a situation where a particular sequence motif is repeated two times, the subsequent evolution can be seen as a birth-and-death process for a microsatellite locus. A deletion caused by replication slippage, or a nucleotide substitution, will obliterate the repetitive nature of the sequence, blocking length expansion. However, a slippage-induced insertion will generate a three-repeat locus prone to further length expansion, reinforced by the tendency for microsatellite repeat insertions to be more common than deletions and the increase in mutation rate with repeat length (Ellegren 2004).
Correlation between rates of indels and SNPs:
Local genomic context may affect rates of evolution and levels of intraspecific polymorphism in various ways, often resulting in observations of correlation between several genomic parameters, including indel density (e.g., Hardison et al. 2003). There is significant variation in the rate of point mutation at the subchromosomal level, with evidence from rodents that the within-chromosomal variability in mutation rate exceeds that of the between-chromosomal variability with an order of magnitude (Gaffney and Keightley 2005). The causes of this variation remain elusive. For instance, although it has been suggested that recombination is mutagenic and that the local rate of recombination thereby affects mutability (Lercher and Hurst 2002; Hardison et al. 2003; Hellmann et al. 2003), recent studies indicate that the correlation between recombination and mutation is at most weak (Gaffney and Keightley 2005; Huang et al. 2005). In the avian genome, as well as in other organisms (e.g., Mouse Genome Sequencing Consortium 2002), local GC content is positively correlated with sequence divergence and diversity, possibly due to a combined effect of CpG mutability and biased gene conversion introducing fixation biases in GC-rich regions (Webster et al. 2006). However, GC is also correlated with the rate of recombination, illustrating the complex relationship between genomic parameters. Our observation of a strong correlation between the local densities of SNPs and indels in the chicken genome adds to this complexity and has implications for an issue arising from partly contradictory findings in studies of mammalian genomes, as described below.
Hardison et al. (2003) found the rate of nucleotide substitution in the human–mouse comparison to covary with the incidence of nonrepetitive, nonaligning sequence, interpreted as deletions. Mills et al. (2006) found hot-spot regions for indel variability in the human genome to often, but not always, overlap with regions of high SNP density; this data set contained indels in unique sequence as well as in microsatellite repeats. Similar observations have been made on smaller scales (e.g., Longman-Jacobsen et al. 2003). Moreover, the incidence of transposon insertion also covaries with nucleotide substitution divergence in mammalian genomewide alignments (Yang et al. 2004). In contrast, Cooper et al. (2004) found no clear correlation between the rates of short indels and point substitution in the mouse–rat comparison and concluded that contextual factors influence nucleotide substitutions and short indels differentially. Our observation based on polymorphism data from the chicken genome does not support this conclusion. Explanations for this difference are presently confined to speculation. One possibility could be lineage-specific differences in the character of local evolutionary forces that affect rates of nucleotide substitution and indels differently. A methodological aspect is that the identification of both nucleotide substitutions and, in particular, indels is more straightforward in data sets based on intraspecific shotgun sequencing, or resequencing, than in genomewide alignments of distantly related species.
GC content has been shown to correlate with the rate of nucleotide substitution (reviewed in Ellegren et al. 2003). The observation that both SNP and indel density in chickens are positively correlated with GC could indicate that the rates of both types of polymorphism are influenced by a similar mechanism, manifested in GC content. However, after factoring out the effect of GC by taking the residuals of a regression of SNP and indel density on GC and then computing the correlation between residuals, we still found the rates of SNPs and indels to covary. This, of course, can be interpreted as that the rates are indeed affected by a similar mechanism related, for instance, to replication, repair, or recombination, although this mechanism is unrelated to base composition. However, SNP and indel density could also covary due to the effects of selection on overall levels of genetic variability across the genome.
Indel density in genic sequences:
With the exception of events involving multiples of 3 bp (see Podlaha and Zhang 2003), indel mutations disrupt the reading frame of coding sequence and in most cases are likely to be deleterious. In line with studies of other organisms (Bhangale et al. 2005), we found a markedly lower frequency of indels in coding sequence, ∼10% of that in nongenic DNA. The actual ratio in fact may be even lower since the frequency estimates are based on unvalidated polymorphisms; any error rate in polymorphism identification will have the most pronounced effect on frequency estimates of rare categories of sequence variants (cf. International Chicken Polymorphism Map Consortium 2004). We also found a lower density of indels in the 100-bp upstream sequence, in 5′-UTR, and in first introns, but not in sequences farther upstream, in other introns, or in 3′-UTR. These observations mirror the constraint generally indicated by the rate of sequence polymorphism (Zhao et al. 2003; Tsunoda et al. 2004) and divergence in untranslated and promotor regions (Xie et al. 2005), as well as in first introns (Chamary and Hurst 2004). Indels can disrupt important motifs in regulatory regions and also alter the spacing between regulatory binding sites (Ludwig et al. 1998; Xie et al. 2005). Xie et al (2005) found a peak in the density of conserved transcription-factor-binding sites within 100 bp upstream of the transcription start; this concurs with our finding of low indel density in the same region.
Evolution of genome size:
Theories on the evolution of genome size are many but may be broadly characterized as reflecting either neutral or selective processes (reviewed in, e.g., Petrov 2002a; Gregory 2004). Several investigators have found a correlation between metabolic rate and genome size, as in the selectionist view interpreted as evidence for adaptive evolution of genome size (Vinogradov 1997; Kozlowski et al. 2003; Vinogradov and Anatskaya 2006). It has been argued that the small genome size of birds evolved as an adaptation to the energetic demands of flight (Hughes and Hughes 1995; Hughes and Piontkivska 2005), an idea that is controversial (Waltari and Edwards 2002). An alternative neutralist view is that genome size is determined by a mutational equilibrium where the insertion rate of transposable elements is balanced by a deletion bias of short indels (Petrov 2002b).
The deletion bias (1.4 times the insertion rate) indicated by rooted chicken indel polymorphisms is in the lower end of the range reported in studies of other species (0.8–5) (Ophir and Graur 1997; Comeron and Kreitman 2000; Petrov et al. 2000; Vinogradov 2002; Neafsey and Palumbi 2003; Zhang and Gerstein 2003; Cooper et al. 2004). To some extent, this can be due to the criteria for identification of indels as the insertion:deletion ratio may be affected by whether or not indels in repetitive structures are included (Ometto et al. 2005). Johnson (2004) studied the incidence of indels in a single intron of the β-fibrinogen gene across a suite of species of pigeons and doves. He observed a more pronounced deletion bias (six times the insertion rate; n = 50 mutation events); however, the sequences were not filtered for simple repeat structures and, upon closer inspection of the data, it is clear that many of those events represent microsatellite length variation rather than indels in unique sequence. If a limited deletion bias proved to be a general phenomenon across divergent avian lineages, short deletions may not be the primary determinant of the small genome size in birds through DNA loss. Given that indel mutations are relatively rare and usually involve a limited number of nucleotides, it is unlikely that a modifier of the rate and/or proportion of deletion mutation is selected because of its effect on genome size (Petrov 2002b). However, this in itself does not distinguish between adaptive scenarios and the mutation equilibrium hypothesis. The repeat content of the chicken genome is much lower than that of mammals (∼10% vs. 45% in humans) with an almost complete lack of SINE elements and with the most common LINE element, CR1, present mainly as short truncated copies (International Chicken Genome Sequencing Consortium 2004). This either could be taken as support for the neutral equilibrium model or be seen as avian genome size being governed by selection on reduced activity of transposable elements for adaptive reasons.
Conclusions:
Here we report the analysis of an extensive data set of short indel mutations in chicken (International Chicken Polymorphism Map Consortium 2004). After filtering our data for tandem repeats of three or more units, we show that indels often occur in tandem duplicated sequence (i.e., duplet repeats of the motif). We also found that deletions were overrepresented in tandem duplicates. Taken together, these results indicate that small indels might not be distinctly different from microsatellite mutations and may even be the first step in microsatellite genesis. Our results also indicate that similar evolutionary forces act on both SNPs and indels, as their densities are highly correlated.
Acknowledgments
We thank Gane Ka-Shu Wong for useful discussion. Financial support was obtained from the Swedish Research Council.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:** 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson, E., M. T. Webster, N. G. Smith, D. W. Burt and H. Ellegren, 2005. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 15**:** 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27**:** 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangale, T. R., M. J. Rieder, R. J. Livingston and D. A. Nickerson, 2005. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 14**:** 59–69. [DOI] [PubMed] [Google Scholar]
- Bois, P. R., 2003. Hypermutable minisatellites, a human affair? Genomics 81**:** 349–355. [DOI] [PubMed] [Google Scholar]
- Bray, N., and L. Pachter, 2004. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 14**:** 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten, R. J., 2002. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc. Natl. Acad. Sci. USA 99**:** 13633–13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten, R. J., L. Rowen, J. Williams and R. A. Cameron, 2003. Majority of divergence between closely related DNA samples is due to indels. Proc. Natl. Acad. Sci. USA 100**:** 4661–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary, J. V., and L. D. Hurst, 2004. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol. Biol. Evol. 21**:** 1014–1023. [DOI] [PubMed] [Google Scholar]
- Chen, J. M., N. Chuzhanova, P. D. Stenson, C. Ferec and D. N. Cooper, 2005. a Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum. Mutat. 25**:** 207–221. [DOI] [PubMed] [Google Scholar]
- Chen, J. M., P. D. Stenson, D. N. Cooper and C. Ferec, 2005. b A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum. Genet. 117**:** 411–427. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium, 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437**:** 69–87. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M., and M. Kreitman, 2000. The correlation between intron length and recombination in Drosophila: dynamic equilibrium between mutational and selective forces. Genetics 156**:** 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, G. M., M. Brudno, E. A. Stone, I. Dubchak, S. Batzoglou et al., 2004. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. 14**:** 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H., 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5**:** 435–445. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., N. G. Smith and M. T. Webster, 2003. Mutation rate variation in the mammalian genome. Curr. Opin. Genet. Dev. 13**:** 562–568. [DOI] [PubMed] [Google Scholar]
- Fain, M. G., and P. Houde, 2004. Parallel radiations in the primary clades of birds. Evolution 58**:** 2558–2573. [DOI] [PubMed] [Google Scholar]
- Gaffney, D. J., and P. D. Keightley, 2005. The scale of mutational variation in the murid genome. Genome Res. 15**:** 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, T. R., 2004. Insertion-deletion biases and the evolution of genome size. Gene 324**:** 15–34. [DOI] [PubMed] [Google Scholar]
- Gregory, T. R., 2005. Synergy between sequence and size in large-scale genomics. Nat. Rev. Genet. 6**:** 699–708. [DOI] [PubMed] [Google Scholar]
- Hamilton, M. B., J. M. Braverman and D. F. Soria-Hernanz, 2003. Patterns and relative rates of nucleotide and insertion/deletion evolution at six chloroplast intergenic regions in New World species of the Lecythidaceae. Mol. Biol. Evol. 20**:** 1710–1721. [DOI] [PubMed] [Google Scholar]
- Hardison, R. C., K. M. Roskin, S. Yang, M. Diekhans, W. J. Kent et al., 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13**:** 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, I., I. Ebersberger, S. E. Ptak, S. Paabo and M. Przeworski, 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72**:** 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, S, 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6**:** 65–70. [Google Scholar]
- Holmes, I., 2005. Using evolutionary expectation maximization to estimate indel rates. Bioinformatics 21**:** 2294–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. W., R. Friedman, N. Yu, A. Yu and W. H. Li, 2005. How strong is the mutagenicity of recombination in mammals? Mol. Biol. Evol. 22**:** 426–431. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., and M. K. Hughes, 1995. Small genomes for better flyers. Nature 377**:** 391. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., and H. Piontkivska, 2005. DNA repeat arrays in chicken and human genomes and the adaptive evolution of avian genome size. BMC Evol. Biol. 5**:** 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium, 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432**:** 695–716. [DOI] [PubMed] [Google Scholar]
- International Chicken Polymorphism Map Consortium, 2004. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature 432**:** 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K. P., 2004. Deletion bias in avian introns over evolutionary timescales. Mol. Biol. Evol. 21**:** 599–602. [DOI] [PubMed] [Google Scholar]
- Kawakita, A., T. Sota, J. S. Ascher, M. Ito, H. Tanaka et al., 2003. Evolution and phylogenetic utility of alignment gaps within intron sequences of three nuclear genes in bumble bees (Bombus). Mol. Biol. Evol. 20**:** 87–92. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and T. Johnson, 2004. MCALIGN: stochastic alignment of noncoding DNA sequences based on an evolutionary model of sequence evolution. Genome Res. 14**:** 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., and I. B. Rogozin, 2004. Context of deletions and insertions in human coding sequences. Hum. Mutat. 23**:** 177–185. [DOI] [PubMed] [Google Scholar]
- Kozlowski, J., M. Konarzewski and A. T. Gawelczyk, 2003. Cell size as a link between noncoding DNA and metabolic rate scaling. Proc. Natl. Acad. Sci. USA 100**:** 14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher, M. J., and L. D. Hurst, 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18**:** 337–340. [DOI] [PubMed] [Google Scholar]
- Levinson, G., and G. A. Gutman, 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4**:** 203–221. [DOI] [PubMed] [Google Scholar]
- Longman-Jacobsen, N., J. F. Williamson, R. L. Dawkins and S. Gaudieri, 2003. In polymorphic genomic regions indels cluster with nucleotide polymorphism: Quantum Genomics. Gene 312**:** 257–261. [DOI] [PubMed] [Google Scholar]
- Ludwig, M. Z., N. H. Patel and M. Kreitman, 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125**:** 949–958. [DOI] [PubMed] [Google Scholar]
- Makova, K. D., S. Yang and F. Chiaromonte, 2004. Insertions and deletions are male biased too: a whole-genome analysis in rodents. Genome Res. 14**:** 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier, W., S. H. Li and C. B. Stewart, 1996. The birth of microsatellites. Nature 381**:** 483. [DOI] [PubMed] [Google Scholar]
- Mills, R. E., C. T. Luttig, C. E. Larkins, A. Beauchamp, C. Tsui et al., 2006. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 16**:** 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:** 520–562. [DOI] [PubMed] [Google Scholar]
- Müller, K., 2006. Incorporating information from length-mutational events into phylogenetic analysis. Mol. Phylogenet. Evol. 38**:** 667–676. [DOI] [PubMed] [Google Scholar]
- Neafsey, D. E., and S. R. Palumbi, 2003. Genome size evolution in pufferfish: a comparative analysis of diodontid and tetraodontid pufferfish genomes. Genome Res. 13**:** 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogurtsov, A. Y., S. Sunyaev and A. S. Kondrashov, 2004. Indel-based evolutionary distance and mouse-human divergence. Genome Res. 14**:** 1610–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto, L., W. Stephan and D. De Lorenzo, 2005. Insertion/deletion and nucleotide polymorphism data reveal constraints in Drosophila melanogaster introns and intergenic regions. Genetics 169**:** 1521–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophir, R., and D. Graur, 1997. Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene 205**:** 191–202. [DOI] [PubMed] [Google Scholar]
- Pereira, S. L., and A. J. Baker, 2006. A molecular timescale for galliform birds accounting for uncertainty in time estimates and heterogeneity of rates of DNA substitutions across lineages and sites. Mol. Phylogenet. Evol. 38**:** 499–509. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., 2002. a DNA loss and evolution of genome size in Drosophila. Genetica 115**:** 81–91. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., 2002. b Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 61**:** 531–544. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., T. A. Sangster, J. S. Johnston, D. L. Hartl and K. L. Shaw, 2000. Evidence for DNA loss as a determinant of genome size. Science 287**:** 1060–1062. [DOI] [PubMed] [Google Scholar]
- Podlaha, O., and J. Zhang, 2003. Positive selection on protein-length in the evolution of a primate sperm ion channel. Proc. Natl. Acad. Sci. USA 100**:** 12241–12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A. L., E. Eskin and P. A. Pevzner, 2004. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 14**:** 2245–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmer, C. R., and H. Ellegren, 1998. Patterns of molecular evolution in avian microsatellites. Mol. Biol. Evol. 15**:** 997–1008. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2006. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Samonte, R. V., and E. E. Eichler, 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3**:** 65–72. [DOI] [PubMed] [Google Scholar]
- Sundström, H., and H. Ellegren, 2004. Reduced variation on the chicken Z chromosome. Genetics 164**:** 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M. S., C. P. Ponting and R. R. Copley, 2004. Occurrence and consequences of coding sequence insertions and deletions in mammalain genomes. Genome Res. 14**:** 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda, T., G. M. Lathrop, A. Sekine, R. Yamada, A. Takahashi et al., 2004. Variation of gene-based SNPs and linkage disequilibrium patterns in the human genome. Hum. Mol. Genet. 13**:** 1623–1632. [DOI] [PubMed] [Google Scholar]
- van Tuinen, M., and G. J. Dyke, 2004. Calibration of galliform molecular clocks using multiple fossils and genetic partitions. Mol. Phylogenet. Evol. 30**:** 74–86. [DOI] [PubMed] [Google Scholar]
- Waltari, E, and S. V. Edwards, 2002. Evolutionary dynamics of intron size, genome size, and physiological correlates in archosaurs. Am. Nat. 160**:** 539–552. [DOI] [PubMed] [Google Scholar]
- Weber, J. L., D. David, J. Heil, Y. Fan, C. Zhao et al., 2002. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 71**:** 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, M. T., E. Axelsson and H. Ellegren, 2006. Strong regional biases in nucleotide substitution in the chicken genome. Mol. Biol. Evol. 23**:** 1203–1216. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A. E., 1997. Nucleotypic effect in homeotherms: body-mass independent resting metabolic rate of passerine birds is related to genome size. Evolution 51**:** 220–225. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A. E., 2002. Growth and decline of introns. Trends Genet. 18**:** 232–236. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A. E., and O. V. Anatskaya, 2006. Genome size and metabolic intensity in tetrapods: a tale of two lines. Proc. Biol. Sci. 273**:** 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X., J. Lu, E. J. Kulbokas, T. R. Golub, V. Mootha et al., 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434**:** 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., A. F. Smit, S. Schwartz, F. Chiaromonte, K. M. Roskin et al., 2004. Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res. 14**:** 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and M. Gerstein, 2003. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 31**:** 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., Y. X. Fu, D. Hewett-Emmett and E. Boerwinkle, 2003. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene 312**:** 207–213. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., J. E. Strassmann and D. C. Queller, 2000. Insertions, substitutions, and the origin of microsatellites. Genet Res. 76**:** 227–236. [DOI] [PubMed] [Google Scholar]