PSF Acts through the Human Immunodeficiency Virus Type 1 mRNA Instability Elements To Regulate Virus Expression (original) (raw)
Abstract
Human immunodeficiency virus type 1 (HIV) gag/pol and env mRNAs contain _cis_-acting regulatory elements (INS) that impair stability, nucleocytoplasmic transport, and translation by unknown mechanisms. This downregulation can be counteracted by the viral Rev protein, resulting in efficient export and expression of these mRNAs. Here, we show that the INS region in HIV-1 gag mRNA is a high-affinity ligand of p54nrb/PSF, a heterodimeric transcription/splicing factor. Both subunits bound INS RNA in vitro with similar affinity and specificity. Using an INS-containing subgenomic gag mRNA, we show that it specifically associated with p54nrb in vivo and that PSF inhibited its expression, acting via INS. Studying the authentic HIV-1 mRNAs produced from an infectious molecular clone, we found that PSF affected specifically the INS-containing, Rev-dependent transcripts encoding Gag-Pol and Env. Both subunits contained nuclear export and nuclear retention signals, whereas p54nrb was continuously exported from the nucleus and associated with INS-containing mRNA in the cytoplasm, suggesting its additional role at late steps of mRNA metabolism. Thus, p54nrb and PSF have properties of key factors mediating INS function and likely define a novel mRNA regulatory pathway that is hijacked by HIV-1.
Many eukaryotic mRNAs are subject to regulated turnover via _cis_-acting signals which are recognized by _trans_-acting factors. The best-studied example is the cytoplasmic mRNA decay mediated by _cis_-acting AU-rich RNA elements and _trans_-acting ARE-binding proteins. Another important mechanism is nonsense-mediated mRNA decay, which serves to eliminate the transcripts that contain premature stop codons (for recent reviews, see references 17, 34, and 40).
In contrast to cytoplasmic pathways, nuclear mRNA turnover is less well understood. Among the mRNAs that are subject to nuclear downregulation are the Rev-responsive element (RRE)-containing mRNAs of human immunodeficiency virus type 1 (HIV-1) (11, 22, 25, 26, 32, 33, 42, 48, 54; reviewed in references 13, 21, 27, and 47). These unspliced and nonterminally spliced transcripts need to be exported from the nucleus before completion of splicing to produce the Gag-Pol and Env proteins. This regulatory step is accomplished by the viral Rev protein, which binds to the RRE and links these transcripts to the CRM1 export receptor. In the absence of Rev, these mRNAs are further spliced to completion or degraded. However, even when devoid of splice sites, the unspliced or nonterminally spliced mRNAs are poorly expressed due to the presence of _cis_-acting instability elements (INS/CRS) that are scattered throughout the gag/pol and env mRNAs (11, 25, 32, 42, 50, 51, 54). These elements act at several steps, impairing mRNA stability, nucleocytoplasmic transport, and translation (5, 15, 22, 29, 33, 51, 54), whereas Rev counteracts these defects, resulting in efficient expression. However, Rev is unable to export nonterminally spliced mRNAs that do not contain a functional INS (51), and hence, INS are an integral part of Rev regulation.
Functionally analogous elements are found in all lentiviruses and viruses of the human T-cell lymphotropic virus family, indicating their biological relevance. Several mRNA-binding proteins, including PTB/hnRNP I, hnRNP A1, and PABP1, were shown to bind specifically to such elements in vitro (2, 3, 9, 10, 41). It is thought that the INS-binding factors may divert these mRNAs from the splicing route and promote their association with Rev, enabling their export and expression. Because these factors are abundant, ubiquitous components of general mRNPs, the functional relevance of such interactions was difficult to address.
Here, we further investigated the mechanism of INS-mediated mRNA regulation, using the best-characterized INS within the 5′ portion of HIV-1 gag (p37_gag_). This INS spans more than 1 kb and can be inactivated by silent point mutations in eight regions, resulting in a mutant gene termed INS(−)M1-10 (50, 51, 54) (Fig. 1). Although AU-rich, these regions are functionally and structurally unrelated to AU-rich RNA elements (37, 50, 51, 54) and do not show homology to other known cellular mRNA instability elements.
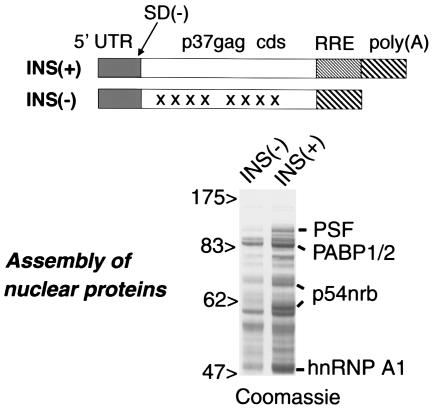
FIG. 1.
Assembly of nuclear proteins onto INS-RNA. The in vitro-transcribed INS(+) and INS(−) RNAs are shown schematically, indicating the 5′ untranslated region (UTR), p37_gag_ cds, RRE, and poly(A) site and the deletion destroying the major HIV-1 5′ splice site [SD(−)]. In INS(−) RNA, the regions containing inactivating mutations (50) are shown by X. The INS(+) and INS(−) RNAs were immobilized on streptavidin beads and incubated with HeLa nuclear extracts. The assembled proteins were separated by SDS-PAGE and stained with Coomassie, and the major bands in the INS(+)-bound fraction were identified by microsequencing, as shown to the right. The sizes of marker proteins are shown (in kilodaltons).
In this study, we found that INS assembles specifically with nuclear factors PSF and p54nrb in nuclear extracts. Human p54nrb and PSF are nucleic acid-binding proteins that form a heterodimer (45, 61), and p54nrb is 71% identical to the C-terminal portion of PSF (16, 24, 44, 61). PSF participates in both early and late steps of splicing (reference 45 and references therein), and p54nrb is also involved in splicing (16). PSF associates with polypyrimidine tract-binding protein (44) and with the small nuclear ribonucleoprotein (snRNP)-free form of U1A protein (31), whereas both p54nrb and PSF bind specifically to U5 snRNA in vitro (45), but their exact role in splicing remains unclear.
Mammalian PSF and p54nrb have also been implicated in transcriptional regulation (reviewed in reference 56). Apparently, p54nrb/PSF has the potential to bind pre-mRNA cotranscriptionally, because both subunits bind avidly to the carboxy-terminal domain of the largest subunit of RNA polymerase II (19). Both p54nrb and PSF were found in the nuclear subdomains known as splicing speckles (18), and p54nrb also localizes to paraspeckles, a distinct nuclear domain that additionally contains a p54nrb-related PSP1 protein (23). PSF, p54nrb, PSP1 as well as PSP2/CoAA, another paraspeckle-specific protein, were shown to associate with the nucleolus upon inhibition of transcription (4), indicating that they are dynamically localized. The Chironomus tentans p54nrb-related protein hrp65 has been found in filaments attached to some nuclear mRNPs, suggesting a role in mRNA trafficking (39). Hence, p54nrb and PSF are multifunctional factors that are involved in a variety of nuclear processes (for a recent review, see reference 56).
In addition, p54nrb was recently shown to bind specifically to hyperedited, inosine-containing RNAs (I-RNA), leading to their nuclear retention. The I-RNA-binding complex also includes PSF and a nuclear matrix protein, matrin 3 (62). These data predicted that p54nrb/PSF could act similarly on other high-affinity RNA ligands, such as INS-containing mRNAs (INS-mRNA). Here, we show that p54nrb and PSF have properties of trans_-acting factors that bind directly to the INS located in p37_gag of HIV-1. We further found that PSF specifically downregulates INS-containing mRNA produced by an HIV molecular clone, resulting in loss of _gag_-pol and env expression and hence no virus production. Therefore, PSF is one of the key factors mediating the posttranscriptional regulation of HIV-1. Thus, this study provides insight into a new control of HIV-1 replication and suggests that PSF is likely part of a novel mRNA regulatory mechanism.
MATERIALS AND METHODS
Transcription in vitro.
As DNA templates for in vitro transcription, PCR fragments that were produced with sense primers that contained T7 promoter sequences were used. For polyadenylated transcripts, the antisense primers also included a 98-nucleotide oligo(dT) sequence. The capped, polyadenylated RNAs were produced with Message Machine kits (Ambion). For uncapped transcripts, Megascript kits (Ambion) were used. For biotinylated RNAs, transcription mixes were supplemented with biotin-16-UTP (Roche), at 1/100 of the total UTP concentration. Radioactively labeled transcripts were produced by standard protocols with an equimolar mix of [32P]αATP and [32P]αGTP.
Protein-RNA assembly in nuclear extracts.
Nuclear extracts were prepared (30) from HeLa cells which were grown and fractionated as described previously (7). The extracts were used without nuclease pretreatment to ensure strong competitive binding conditions. Prior to use, the extracts were adjusted to 200 mM NaCl, 0.1% Triton X-100, and 2 mg of tRNA per ml and cleared with empty streptavidin-containing magnetic beads (M-280; Dynal). The biotin-labeled capped, polyadenylated p37_gag_ RNAs were immobilized on M-280 beads according to the manufacturers' protocol and incubated with cleared extracts for 1 h at 30°C. The assembled proteins were eluted by micrococcal nuclease digestion, separated on sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE), and stained with Coomassie.
RNA binding and UV cross-linking in vitro.
Binding reactions contained p54nrb and PSF proteins at ≈1 μM and 32P-labeled RNA probes at 1 μM (oligoribonucleotides) or 20 nM (full-length p37_gag_ and luc RNAs). Binding was performed in 15 mM HEPES (pH 7.7)-50 mM KCl-200 mM NaCl-0.2 mM EDTA-0.5% Triton X-100 for 15 min at room temperature, followed by UV-irradiation in a Stratalinker (15 min at time mode 4C). When using uniformly labeled RNAs, the reactions were treated with 1 mg of RNase A per ml for 15 min at 37°C after irradiation. The products were separated on SDS-10% PAGE and quantified with a phosphorimager.
For competition experiments, the 50% inhibitory concentrations (IC50) were calculated with linear interpolation. Synthetic oligoribonucleotide probes that were used to map the interaction sites within p37_gag_ INS were 32 nucleotides in length and spanned the previously published INS subregions (50, 51). Such probes represented the wild-type or INS(−) HIV-1 gag sequences. As an additional control, the wild-type sequences were randomized while preserving the nucleotide content.
Recombinant DNA.
The eukaryotic expression plasmids for INS(+)-RRE and INS(−)M1-10 (50), green fluorescent protein (GFP)-PSF (18), GFP-p54nrb (45), the Escherichia coli expression plasmids for p54nrb (16) and PSF (44), and the codon-optimized HIV-1 gag gene (28) have been described. The eukaryotic expression plasmid for p54nrb-GFP was constructed by PCR amplification of p54nrb-cDNA (16) and cloning in-frame with the GFP coding sequence (cds) into the _Nhe_I site of the pF25 vector (58). The HIV-1 proviral plasmid pNL4-3 (1), rev expression plasmid pBsrev (14), env cDNA expression plasmids pNL1.5E (53) and pNL1.5EΔSS (42), and nef cDNA expression plasmid pNL1.5.7 (52) were described. The 1.5E INS(−) plasmid was constructed by replacing the env cds of pNL1.5EΔSS (_Eco_RI-_Xho_I) with the mutant INS(−) env cds (_Eco_RI-_Xho_I) derived from HIV-1 isolate 6101, which also lacks the splice sites (Rosati et al., unpublished data).
Cell culture, transfections, shuttling assay, and cell fractionation.
Transfections in human 293 or HeLa-derived HLtat cells, HIV-1 p24_gag_, luciferase, and GFP measurements were performed (65). Nucleocytoplasmic shuttling assays were performed as described previously (6). Preparation of subcellular extracts of 293 cells was described previously (64).
Recombinant proteins.
Recombinant p54nrb and His6-tagged PSF were produced in E. coli as soluble proteins and isolated on Ni-nitrilotriacetic acid-agarose following standard protocols. For RNA-binding studies, the proteins were further purified by affinity chromatography on a biotinylated ribonucleotide (ACAAGAUUUAAACACCAUGCUAAACACAGUG) that was immobilized on streptavidin-coated Dynabeads. The proteins were bound at 80 mM NaCl in 20 mM phosphate buffer, pH 7.4, and eluted stepwise with 80 to 400 mM NaCl. The fractions that were retained at >250 mM were pooled and stored at −20°C.
Antibodies, immunoblots, Northern blots, and immunoprecipitation.
Rabbit anti-p54nrb serum was described previously (60). HIV-1 patient serum (Scripps) and monoclonal antibodies to p54nrb (Transduction Labs) were used according to the manufacturers' instructions. The B92 monoclonal antibodies (55) were obtained from D. Zipori or purchased from Sigma. The HIV-1 rabbit anti-gp120 antiserum was a gift from L. Arthur. Northern blot analyses of HIV-1 mRNAs were performed (25) with a probe that is complementary to all HIV-1 transcripts, and the luciferase mRNA was detected with a full-length cds probe. Western blots of HIV-1 proteins (25) and analyses of mRNP complexes (46, 64) were performed as described previously.
RESULTS
Identification of proteins that assemble with p37_gag_ mRNA in vitro.
To identify the proteins that recognize the INS in p37_gag_ mRNA, we assembled the complexes from nuclear HeLa extracts onto in vitro-transcribed, immobilized RNA. To ensure that the INS is recognized in its authentic context, we used a full-length transcript that was 7mGpppG-capped, contained the HIV-1 5′ untranslated region, wild-type p37_gag_ cds, the RRE element, and a 98-nucleotide poly(A) tail [Fig. 1, INS(+)]. As control, the matching INS(−)M1-10 RNA was used [Fig. 1, INS(-)]. To reduce the assembly of spliceosomal components, both RNAs had a deletion destroying the major HIV-1 5′ splice site [Fig. 1, SD(−)]. The bound proteins were eluted by micrococcal nuclease digestion to exclude the RNA-independent binders and separated on SDS-PAGE.
Figure 1 shows that several proteins associated preferentially with the INS(+) RNA. The major bands were microsequenced, and all of them produced high-confidence matches to known human proteins. Three groups of proteins were identified: (i) PSF and p54nrb; (ii) polyadenylate-binding protein PABP1 or/and its homologs (the peptides did not discriminate between several closely related proteins); and (iii) heterogenous nuclear RNP (hnRNP) A/B family proteins. The identification of PABP1 and hnRNP A1 is in agreement with previous findings (2, 3, 9, 10, 41) and confirms the validity of our assembly conditions. We focused on the novel INS binders p54nrb and PSF.
p54nrb/PSF is sufficient for INS recognition in vitro.
Since p54nrb and PSF form a heterodimer (45, 61), it is possible that one or both subunits bound to INS directly (Fig. 1). Alternatively, additional factors could have facilitated the binding. We therefore asked whether p54nrb/PSF could recognize the INS directly in the absence of nuclear extract. For in vitro RNA binding studies, we used the p54nrb/PSF dimer that was purified from human cells (59) (Fig. 2A, h-p54nrb/PSF). The INS(+) RNA contained the wild-type p37_gag_ coding sequence, whereas in the matching INS(−) RNA, the INS was destroyed by codon swapping (28), and both RNAs lacked the cap structure, the 5′ untranslated region, the RRE element, and the poly(A) site. As an additional nonspecific control, we used an RNA comprising the complete luciferase (luc) cds, which is of similar size. The probes were double labeled at A and G residues to reduce the composition biases, adjusted to the same specific radioactivity, and used at equimolar amounts. All RNAs ran as single bands of the expected size on denaturing PAGE (not shown). Since these RNAs were more than 1 kb in size, we used UV cross-linking to analyze protein binding.
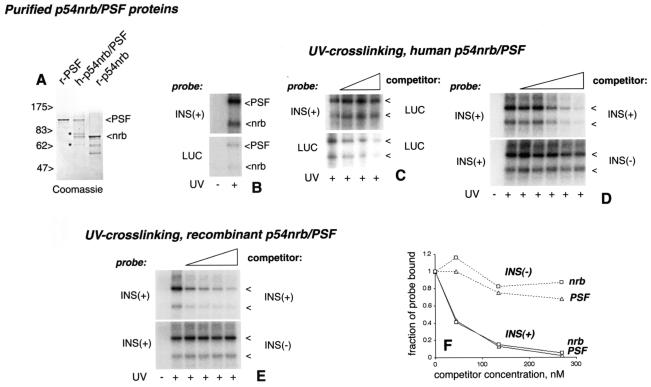
FIG. 2.
p54nrb and PSF bind specifically to INS-RNA in vitro. (A) SDS-PAGE analysis of purified p54nrb/PSF proteins. r-PSF, recombinant His-tagged PSF; h-p54nrb/PSF, purified human p54nrb/PSF dimer; r-p54nrb, recombinant p54nrb. The sizes of marker proteins are shown (in kilodaltons). The positions of p54nrb and PSF are indicated to the right. Asterisks indicate the bands of contaminating proteins in the h-p54nrb/PSF fraction. (B to E) RNA binding/UV cross-linking with purified human (B, C, and D) and recombinant (E) p54nrb/PSF dimer. The radioactive RNA probes (indicated on the left) and the competitor RNAs (indicated on the right) are indicated. The competitors were present at final concentrations of 15, 45, and 135 nM (C); 5, 15, 45, 135, and 270 nM (panel D); and 77, 156, 311, and 622 nM (E), as indicated by triangles. As a control, some reactions were not UV-irradiated, as indicated at the bottom. The cross-linked p54nrb and PSF proteins (indicated by arrowheads) were separated by SDS-PAGE and detected by autoradiography. In panel C, a stronger exposure was used to visualize the luc probe cross-links. (F) Quantification of the cross-links from panel D. The radioactivity values were determined with a phosphorimager, normalized to those obtained without competition (fraction of probe bound, y axis), and were plotted against the competitor concentrations (x axis).
We found that both subunits cross-linked efficiently to INS(+) and, to a much lesser extent, to the luc probe (Fig. 2B), indicating preferential binding. Accordingly, luc RNA did not compete with the INS(+) probe, even at the highest concentration tested (135 nM), while it competed efficiently with itself (Fig. 2C). Conversely, the INS(+) RNA competed with the luc probe under the same conditions (not shown). Hence, PSF and p54nrb bound to INS(+) RNA in a structure-specific manner.
To verify that the INS is responsible for specific binding, we compared the ability of INS(+) and INS(−) RNAs to compete for p54nrb/PSF. To assess the competitor efficiencies, their 50% inhibitory concentrations (IC50) were determined by quantification of cross-linked bands. We note that, due to the nature of this assay, such values may represent a semiquantitative estimate. With the INS(+) probe (Fig. 2D and 2F), the INS(+) RNA competed strongly (IC50 ≈40 nM), whereas the INS(−) RNA did not compete even at the highest concentration tested (IC50 >270 nM). Thus, the INS inactivation reduced the binding of p54nrb and PSF by at least sixfold. Under these conditions, both subunits bound to INS with apparent dissociation constants of ≈40 nM. Despite similar constants, the cross-linking efficiencies were reproducibly higher for PSF, suggesting that it makes tighter contacts with RNA. Using luc as the probe also revealed differences between the competitor RNAs INS(+) (IC50 ≈30 nM) and INS(−) (IC50 ≈100 nM) for both subunits (results not shown).
Thus, p54nrb and PSF recognized the INS in the absence of nuclear extracts. Since the human p54nrb/PSF fraction contained only minor contaminants (Fig. 2A) that did not exhibit RNA binding (not shown), p54nrb/PSF was likely sufficient for recognition. To further support this conclusion, we used _E. coli_-produced recombinant proteins that were purified by RNA affinity chromatography (Fig. 2A, r-p54nrb and r-PSF) and were mixed at a 1:1 molar ratio. With INS(+) RNA as the probe, we found that both r-p54nrb and r-PSF discriminated efficiently between the INS(+) and INS(−) competitors: INS(+) IC50 ≈100 nM, and INS(−)IC50, >600 nM (Fig. 2E). When used individually, both the r-p54nrb and r-PSF subunits also showed INS recognition, but they exhibited a lesser degree of discrimination between the INS(+) and INS(−) competitors (not shown). We concluded that p54nrb/PSF can recognize INS by itself. These results also indicated that the INS region is sufficient for the p54nrb/PSF binding that was observed in the initial assembly experiments (Fig. 1).
To study the possible direct recognition sites within INS, we UV-cross-linked h-p54nrb/PSF to synthetic oligoribonucleotides representing the individual regions that are affected by INS-inactivating M1-10 mutations (50). For three of the eight regions tested, p54nrb and PSF cross-linked strongly to the wild-type but not to the mutant or shuffled sequences, suggesting the presence of direct recognition sites, whereas the other probes did not show evidence of sequence-specific binding (data not shown). It is therefore plausible that some of the M1-10 mutations act directly by destroying the p54nrb/PSF recognition motifs, while the others may affect recognition in an allosteric manner.
p54nrb interacts specifically with INS-mRNA in vivo.
To address the binding of p54nrb and PSF to INS in vivo, INS(+)-RRE mRNA was transiently expressed in human 293 cells. As a control, luc was coexpressed, since this mRNA was shown to bind poorly to p54nrb/PSF in vitro (see above, Fig. 2B, C). These cells were fractionated, yielding cytoplasmic, soluble nuclear, and insoluble nuclear fractions (Fig. 3) (38, 64). Western blot analysis showed that p54nrb and PSF were enriched in insoluble nuclear fractions, consistent with their association with the insoluble nuclear components (35, 60). Small amounts of these proteins were also found in the cytoplasmic and soluble nuclear fractions (Fig. 3A). We noted that both subunits contained nuclear export determinants (see below, Fig. 7), and therefore, their presence in the cytoplasm is not likely an artifact.
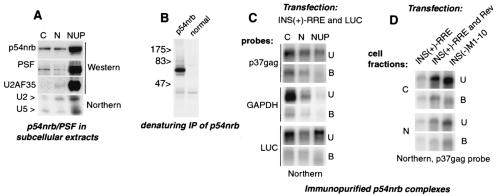
FIG. 3.
p54nrb associates with INS(+) mRNA in vivo. (A) 293 cells were fractionated into cytoplasmic (C), soluble nuclear (N,) and insoluble nuclear (NUP) extracts. Top panels: Western blot analysis with monoclonal antibodies to p54nrb (Transduction Labs) and PSF (Sigma) and with rabbit anti-U2AF35 serum (66), as indicated to the left. Bottom panel: total RNA was extracted from the same fractions, separated on a 15% Tris-borate-EDTA-urea gel, and analyzed on Northern blots for U2 and U5 snRNPs (64). (B) 293 cells were labeled with [35S]methionine and extracted with 0.5× radioimmunoprecipitation assay buffer, and proteins were immunoprecipitated under denaturing conditions with normal or anti-p54nrb rabbit serum (60) as indicated. The sizes of marker proteins are shown (in kilodaltons). (C) 293 cells were cotransfected with 1 μg of INS(+)-RRE and 0.1 μg of luciferase (luc) expression plasmid. At day 2, cell extracts (shown on top) were immunoprecipitated with p54nrb antibodies. The polyadenylated RNA was extracted from immunoprecipitates (bound, B) and from 1:10 aliquots of the supernatants (unbound, U) and analyzed on Northern blots with p37_gag_, luc, and GAPDH probes, as shown to the right. (D) 293 cells were transfected with 1 μg of INS(+)-RRE in the presence or in the absence of 0.05 μg of Rev expression plasmid and with INS(−)M1-10 expression plasmids, as indicated. Cell extraction, immunoprecipitation with p54nrb antibodies, and Northern analysis with p37_gag_ probe were performed as in panel A.
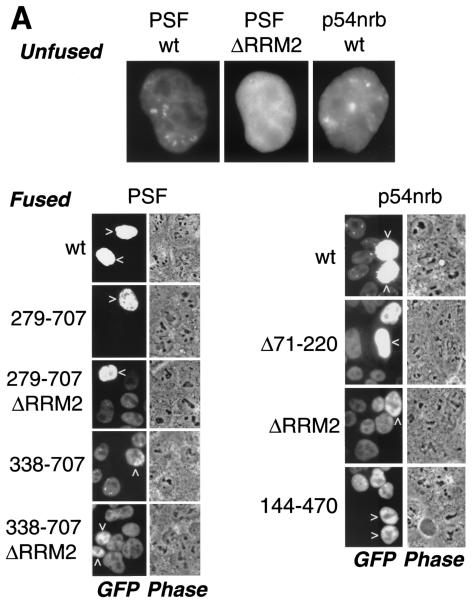
FIG. 7.
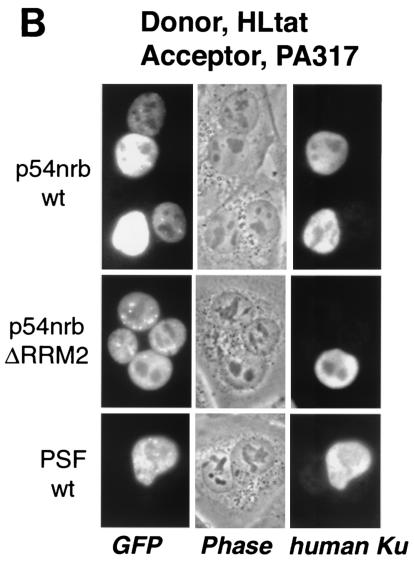
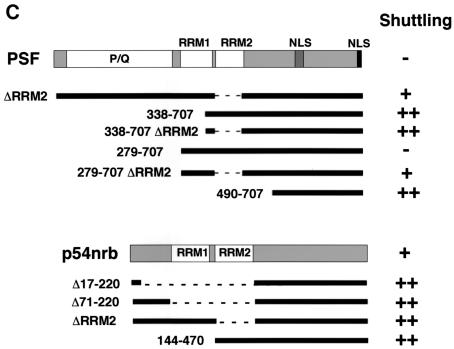
p54nrb and PSF contain nuclear export determinants. (A) HLtat cells were transfected with 1 μg of GFP-tagged PSF and p54nrb expression plasmids and mixed with an excess of untransfected HLtat cells. The next day, cells were fused with polyethylene glycol and fixed with 3.7% formaldehyde 2 h postfusion, and the GFP fluorescence was detected by fluorescent microscopy and a charge-coupled device camera. The top panel shows GFP fluorescence images of cells before fusion. The bottom panels show GFP fluorescence and phase contrast images of the same fields after fusion. Arrowheads indicate donor nuclei. Representative fields are shown, and similar results were obtained in several independent experiments. (B) HLtat cells were transfected, fused, and processed as in panel A but with PA317 cells as the acceptor. The human Ku autoantigen was detected by indirect immunofluorescence with Ku(p80)(Ab-1) monoclonal antibody (Oncogene Research Products) and Alexa 594-conjugated goat anti-mouse immunoglobulin antibody (Molecular Probes). The GFP fluorescence, phase contrast, and indirect immunofluorescence images were captured from the same fields. (C) Summary of GFP-PSF and GFP-p54nrb mutants. In the wild-type proteins, the known domains are indicated (18, 45). P/Q, region rich in proline and glutamine. RRM1 and -2, RNA recognition motifs. NLS, nuclear localization signals. The amino acid residues included in each fusion are shown to the left, with the black bars indicating the corresponding regions of the full-length protein. Dashed lines indicate internal deletions. To the right is a column summarizing the shuttling of fusion proteins: −, undetectable; +, strong; ++, very strong.
To verify the quality of fractionation, we analyzed nuclear markers such as snRNA U2 and U5 and U2AF35 protein. As expected, the bulk of U2 and U5 snRNAs and U2AF35 protein cofractionated with the insoluble nuclear fraction (Fig. 3A), which is enriched in spliceosomal components and pre-mRNA (38, 64). Low levels of these markers were also detectable in the soluble nuclear fraction. While small amounts of U5 snRNA were also found in the cytoplasmic fraction, the U2 snRNA and U2AF35 protein were excluded from this fraction, indicating that there was no general leakage of nuclear components.
To probe the association of INS-mRNA with p54nrb, we performed native immunoprecipitations with monospecific antibodies (Fig. 3B). After immunoprecipitation, the INS-mRNA was analyzed on Northern blots (Fig. 3C, p37_gag_ probe), both in the precipitates (Fig. 3C; bound, B) and the supernatants (Fig. 3C; unbound, U). In the unbound samples, the INS-mRNA was found in all subcellular fractions (Fig. 3C). The anti-p54nrb (Fig. 3C) but not normal rabbit serum (not shown) precipitated INS-mRNAs efficiently from cytoplasmic and soluble nuclear fractions but to a much lesser extent from the insoluble nuclear fraction despite the enrichment of p54nrb in this fraction (Fig. 3A). Thus, the immunoprecipitation likely reflected the interactions prior to cell fractionation rather than reassociation in cell extracts.
Compared to INS-mRNA on the same blots, the coexpressed luc and endogenous glyceraldehyde phosphate dehydrogenase (GAPDH) transcripts were coprecipitated poorly (Fig. 3C), indicating that p54nrb associates with INS-mRNA specifically and is not an abundant component of general mRNP. Quantification of mRNAs revealed that p37_gag_ mRNA was coprecipitated ≈5 times more efficiently than GAPDH. These data demonstrated that p54nrb is part of soluble INS-mRNPs in vivo. Its cytoplasmic association suggested that it may persist during nuclear export, although de novo binding in the cytoplasm could not be excluded.
To verify that INS contributed to the specific association with p54nrb, we transiently expressed the INS(+) RRE and INS(−)M1-10 mRNAs and compared their coprecipitation with p54nrb in soluble nuclear and cytoplasmic fractions. Figure 3D shows that, in both compartments, the INS(−) mRNA accumulated to higher levels than INS(+) mRNA, in agreement with our previous data (50). However, a proportionally smaller amount of INS(−) mRNA was associated with p54nrb in the cytoplasmic complexes.
We next asked whether Rev regulation, known to counteract the effects of INS, affects the p54nrb-INS association. To this end, INS(+)-RRE mRNA was expressed in the presence or in the absence of Rev protein and analyzed as described above. Figure 3D shows that the presence of Rev led to a higher INS(+)-RRE mRNA accumulation but to a lower proportion of p54nrb-associated mRNA in the cytoplasmic complexes. These data demonstrated that the p54nrb-mRNA association was negatively affected by mutational inactivation of INS as well as by the functional counteraction of INS by Rev, further confirming that p54nrb bound via INS. These differential effects were more pronounced in the cytoplasmic than in the nuclear mRNPs, suggesting that p54nrb discriminates INS more stringently in this compartment. It is plausible that p54nrb recognized INS directly, because p54nrb and PSF are sufficient for recognition in vitro (Fig. 2).
To probe the association of the PSF subunit with INS, we performed immunoprecipitations as described above with the monoclonal PSF antibody B92 (55), but INS-mRNA was not coprecipitated detectably. We also noted that in denaturing immunoprecipitation, this antibody performed less efficiently compared to p54nrb antibodies (data not shown), which may explain the lack of coprecipitation. Alternatively, we cannot exclude that p54nrb but not PSF binds to INS-mRNA in vivo. Since both subunits bound to INS in vitro with the same affinity and specificity (Fig. 2), we believe that, in vivo, p54nrb recognizes the INS as part of the p54nrb/PSF heterodimer.
Exogenous PSF acts via INS to inhibit the expression of INS-containing mRNA in human cells.
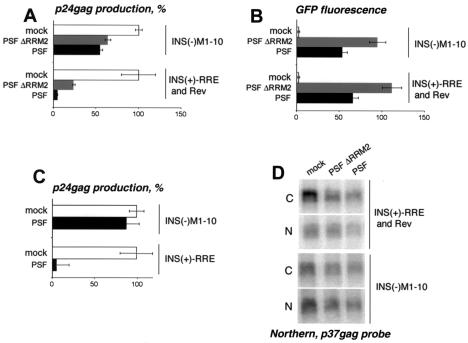
Since p54nrb/PSF interacts with INS, we studied its possible role in INS-mRNA downregulation. First, we tested whether the individual subunits are limiting for INS function in cultured human cells. To this end, we studied the effects of exogenous p54nrb and PSF on INS-mRNA expression in 293 cells (Fig. 4). The INS(+)-RRE and Rev expression plasmids were transfected in the absence or in the presence of the GFP-p54nrb and GFP-PSF plasmids. We found that GFP-PSF strongly inhibited INS-mRNA expression, whereas a PSF mutant lacking one of the RNA recognition motifs (GFP-PSFΔRRM2) was less potent (Fig. 4A). In contrast, GFP-p54nrb reproducibly led to about a 1.5-fold increase of expression. GFP fluorescence measurements of transfected cells confirmed that the different proteins accumulated to similar levels (Fig. 4B and data not shown). Similar results were obtained with various amounts of PSF and p54nrb plasmids (data not shown). We concluded that exogenous PSF inhibited INS-mRNA expression, whereas p54nrb was not limiting. One explanation is that the inhibition was mediated via PSF only. It is also possible that p54nrb can act as an inhibitor but its endogenous levels were saturating and already producing the maximal effect. The RRM2 domain of PSF contributed to inhibition, suggesting its involvement in the interactions with INS. Alternatively, since the ΔRRM2 mutation disrupted the localization of PSF to nuclear foci (18), the effect may be due to mislocalization of the mutant protein. To confirm that PSF acted via INS, we studied its effects on INS(−) mRNA and found that both the wild-type and ΔRRM2 proteins had a much lesser effect on the expression of the INS(−)M1-10 mutant (ranging from zero to 50% inhibition in individual experiments shown in Fig. 4A and C), whereas the presence of the intact INS always led to a ≈10-fold-stronger inhibition in numerous independent transfections. Since both the INS(+) and INS(−) mRNAs were transcribed from the same promoter, we concluded that PSF affected the expression of INS(−) mRNA at the posttranscriptional level.
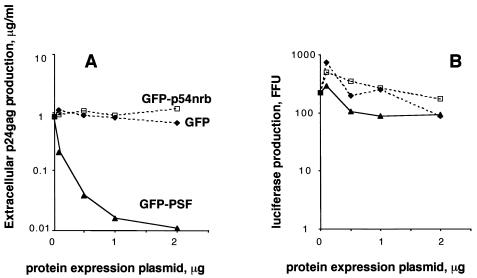
FIG. 4.
Exogenous PSF acts via INS to inhibit INS-mRNA expression. 293 cells were transfected with 1 μg of INS(+)-RRE in the presence or in the absence of 0.05 μg of Rev expression plasmid or with 1 μg of INS(−)M1-10 expression plasmid. (A to C) Transfections were performed in the absence (mock) or in the presence of 1.5 μg of GFP-PSF plasmids. At day 2 posttransfection, p24_gag_ production was measured, and the average values from triplicate plates were normalized to those obtained in the absence of PSF plasmids (100%) and plotted on the x axis (A and C). Panel B shows the normalized fluorescence levels of coexpressed GFP-PSF proteins for the experiment shown in panel A. The average values from triplicate plates are plotted on the x axis as arbitrary units. Bars, standard deviations. Similar data were obtained in several independent transfection experiments. Two representative, independent experiments are shown in panels A and C. (D) Transfections were performed as in panels A and B. At day 2, poly(A)-containing RNA was extracted from the nuclear and the cytoplasmic fractions and analyzed on Northern blots with a p37_gag_ probe.
PSF was able to counteract the Rev regulation (Fig. 4) and hence acted in a dominant manner over Rev-CRM1. To test whether PSF acts before or after Rev binding, we studied its effect on INS(+)-RRE mRNA in the absence of Rev regulation. Figure 4C shows that PSF strongly inhibited the expression of INS(+)-RRE mRNA in the absence of Rev but did not significantly affect INS(−) mRNA expression. Although, as expected, the p24_gag_ values obtained with INS(+)-RRE mRNA in the absence of Rev were relatively low, they permitted unambiguous quantification of PSF's effect (9,000 pg/ml in the absence and 400 pg/ml in the presence of PSF, while the assay's background is 7 pg/ml). We concluded that exogenous PSF can act independently of Rev regulation rather than by affecting the Rev-CRM1 pathway itself. These data further confirmed that PSF's effects are mediated via INS.
To examine the effects of PSF on the mRNA level, we performed transfections like those shown in Fig. 4A and B and analyzed nuclear and cytoplasmic p37_gag_ mRNA on Northern blots. Figure 4D shows that both PSF and PSFΔRRM2 caused a marked decrease in steady-state cytoplasmic INS(+)-RRE mRNA accumulation, while the nuclear levels were not significantly affected. Under the same conditions, the INS(−) mRNA was not affected, in agreement with the lack of expression inhibition.
In summary, these results indicate that PSF participates in posttranscriptional mRNA regulation via INS. It is possible that these effects are mediated via PSF only, although our data do not exclude the contribution of p54nrb.
PSF directly affects Rev-dependent but not Rev-independent mRNAs that are produced from a full-length HIV-1 provirus.
Next, we examined the effects of p54nrb and PSF within the complexity of HIV-1 expression from a full-length NL4-3 proviral plasmid (1), which was transiently transfected in 293 cells. This system faithfully reflects the posttranscriptional regulation of HIV-1 and allows efficient production of infectious virus. To study the effects of p54nrb and PSF on authentic HIV-1 transcripts, we coexpressed NL4-3 with the GFP-tagged p54nrb or PSF and analyzed mRNA accumulation on Northern blots. As an internal control, luc mRNA was coexpressed, which was also transcribed from the HIV-1 long terminal repeat promoter.
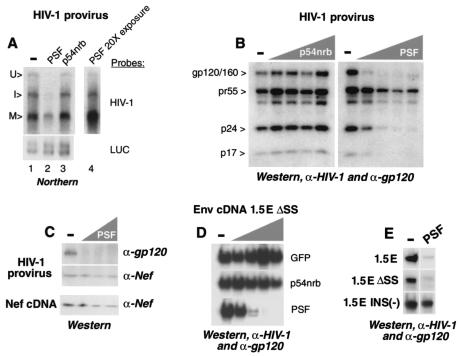
As illustrated in Fig. 5A, this analysis allowed the visualization of three classes of HIV-1 mRNAs: unspliced primary transcripts, intermediate spliced, and multiply spliced. Of these, the unspliced and intermediate spliced mRNAs contained INS elements and were Rev dependent, while the multiply spliced mRNAs did not contain INS and were expressed independently of Rev (25, 54). We found that coexpression of PSF but not p54nrb reduced the levels of all HIV-1 transcripts. A 20-fold-stronger exposure (Fig. 5A, lane 4) revealed that PSF strongly affected the unspliced and the intermediate spliced transcripts and, to much lesser extent, the multiply spliced transcripts. In contrast, neither p54nrb nor PSF affected luc mRNA accumulation significantly (Fig. 5A, lower panel), confirming that PSF affected HIV-1 mRNAs posttranscriptionally.
FIG. 5.
PSF affects Rev-dependent but not Rev-independent HIV-1 expression. (A) 293 cells were cotransfected with 3 μg of pNL4-3 HIV-1 proviral clone and 0.1 μg of L3LUC luciferase expression plasmid in the presence or in the absence (−) of 2 μg of GFP-p54nrb or GFP-PSF expression plasmid, as shown on top. At day 3 posttransfection, the cells were extracted with RNazol, and the poly(A)-containing RNA was isolated with oligo(dT) Dynabeads and analyzed on Northern blots, hybridized with HIV-1 and _luc_-specific radioactive probes, as shown to the right. The mRNAs were visualized by autoradiography. Lanes 1 to 3 represent the same exposure, and lane 4 shows a 20-fold overexposure of lane 2. The bands were quantified with a phosphorimager. U, unspliced; I, intermediate spliced; M, multiply spliced. (B) 293 cells were transfected in parallel to these shown in panel A, with 3 μg of pNL4-3 HIV-1 proviral plasmid in the presence or in the absence (−) of various amounts (0.1, 0.5, 1, and 2 μg) of GFP-p54nrb or GFP-PSF expression plasmid, as shown schematically on top. At day 3 posttransfection, the HIV-1 proteins (shown to the left) were analyzed on Western blots with a mix of patient HIV-1 serum and gp120 antibodies. Similar results were obtained in three independent transfection experiments. (C) Western blot analysis of Env and Nef proteins, as indicated to the right. Top (HIV-1 provirus): same extracts as in panel B. Bottom (nef cDNA): 293 cells were cotransfected with 1 μg of Nef expression plasmid pNL1.5.7 in the absence (−) or in the presence of GFP-PSF plasmid. The amounts of cotransfected PSF plasmid (0.1, 0.5, and 1 μg) are shown schematically. The protein bands were quantified with a Phosphorimager. Similar results were obtained in three independent transfection experiments. (D) 293 cells were cotransfected with 1 μg of p1.5EΔSS and 0.1 μg of BsRev expression plasmids in the absence (−) or in the presence of various amounts (0.1, 0.5, 1, and 2 μg; shown schematically on top) of GFP-p54nrb, GFP-PSF, or empty GFP expression plasmids, as shown to the right. At day 3 posttransfection, the Env proteins were detected on Western blots with a mix of HIV-1 patient serum (Scripps) and gp120 antibodies. Similar data were obtained in several independent experiments. (E) 293 cells were transfected with 1 μg of p1.5E, p1.5EΔSS, or p1.5E INS(−) plasmid, as shown to the left, in the absence (−) or in the presence of 1 μg of GFP-PSF plasmid, as shown on top. The p1.5E and p1.5EΔSS transfections also contained 0.1 μg of BsRev expression plasmid. The Env protein (gp120/160) was detected on Western blots, as described above.
Since all HIV-1 mRNAs were affected (Fig. 5A, compare lanes 1 and 2), PSF likely caused some degradation of the primary transcript prior to splicing. To test this, we used an HIV-1 Rev(−) molecular clone, NL4-3fB (20, 25, 57), whose primary transcripts are spliced to completion, and therefore, the accumulation of splicing products directly reflects the input level of the precursor. By Northern blot analysis, we found that coexpression of PSF significantly reduced the levels of multiply spliced mRNAs produced from NL4-3fB (data not shown). Taken together, these results suggest that exogenous PSF promotes the degradation of primary HIV-1 transcripts, acting upstream of Rev regulation.
To further address the specificity of PSF towards different HIV-1 mRNAs, we studied its effects on protein expression from HIV-1 provirus as well as from cDNA constructs encoding individual HIV-1 transcripts.
In transfections parallel to these shown in Fig. 5A, we coexpressed NL4-3 with different amounts of GFP-tagged p54nrb and PSF proteins or the GFP moiety alone. As an internal control, all transfections included an HIV-1 long terminal repeat promoter-driven luc plasmid. The HIV-1 proteins were analyzed on immunoblots. Figure 5B shows that the expression of PSF led to a striking, dose-dependent inhibition of Env (gp120 and gp160) and Gag (p24CA and p17MA) accumulation, while p54nrb (Fig. 5B) and the GFP moiety (not shown) had no effect. In contrast, PSF inhibited Nef accumulation only up to two- to threefold, as observed in three independent transfection experiments (Fig. 5C, HIV-1 provirus; and data not shown), in agreement with the Northern blot analysis showing an up to threefold reduction of multiply spliced transcripts (Fig. 5A). Therefore, these small effects may be indirect, reflecting the PSF-mediated downregulation of primary transcript (Fig. 5A). We concluded that PSF preferentially affected the expression from the Rev-dependent (gag, env) compared to the Rev-independent (nef) genes.
Because Env production was inhibited very strongly, PSF likely affected env mRNA directly. To test this, we used an env cDNA construct (pNL1.5EΔSS) that produces a transcript which is devoid of splice sites yet still requires Rev regulation due to the presence of INS (42). As a Rev-independent control, we used wild-type nef mRNA expressed from a cDNA construct (pNL1.5.7). Coexpression of pNL1.5EΔSS in the presence of Rev and increasing amounts of GFP-p54nrb, GFP-PSF, or “empty” GFP moiety revealed that PSF but not p54nrb or GFP had a dramatic, dose-dependent effect on Env protein production (Fig. 5D), demonstrating that PSF can act on env mRNA directly and in a manner that is independent of splicing. In contrast, Nef production from the pNL1.5.7 cDNA construct was inhibited only up to threefold (Fig. 5C, nef cDNA), as when produced from pNL4-3 (Fig. 5C, HIV-1 provirus).
To test directly whether PSF acted via the env INS elements, we used a mutant env mRNA in which the INS were inactivated by codon shuffling, resulting in Rev-independent expression [pNL1.5E INS(−)]. We compared the effects of PSF on this INS(−) mRNA and on pNL1.5EΔSS. As another control, we used a wild-type env transcript that contains functional splice sites, which was produced from the pNL1.5E cDNA construct. Figure 5E shows that the presence of functional INS but not the splice sites is required for PSF-mediated inhibition, confirming that PSF acted via INS determinants and independently of splicing. These results are in agreement with our previous findings that the elements causing the Rev dependence of mRNAs are distinct from splice sites (42). Together, these data provide direct, functional evidence that PSF affects env and gag expression, acting via their INS determinants.
The profound effects of exogenous PSF on HIV-1 mRNA and protein expression predicted that virus production will be strongly inhibited. We therefore quantified the effects of PSF on cell-free virus accumulation by measuring p24_gag_ antigen in the cell culture medium in the experiment shown in Fig. 5A. This analysis showed that PSF caused an up to 100-fold inhibition, in a dose-dependent manner, whereas p54nrb and GFP had no effect (Fig. 6A). In contrast, neither of these proteins significantly affected expression of the cotransfected luc gene (Fig. 6B).
FIG. 6.
Exogenous PSF inhibits HIV-1 virus production. Transfections of 293 cells were performed as in Fig. 5A, with a fixed amount of NL4-3 plasmid, in the absence or in the presence of various amounts (plotted on the x axis) of GFP-PSF (triangles), GFP-p54nrb (rectangles), and the GFP moiety alone (diamonds). All transfections included 0.1 μg of L3LUC luciferase expression plasmid. (A) p24_gag_ antigen was measured in the cell culture medium, and the values are plotted on the y axis. (B) In the same transfections, luciferase activity was measured in cell extracts, and the values are plotted on the y axis (firefly units, FFU). Similar data were obtained in three independent experiments.
Taken together, these results led us to conclude that PSF is a key factor acting via HIV-1 INS elements to regulate viral expression.
p54nrb is continuously exported from the nucleus.
Since p54nrb is bound to cytoplasmic INS-mRNA (Fig. 3), we examined the nucleocytoplasmic trafficking of p54nrb and PSF in shuttling assays with GFP-tagged proteins. Briefly, HeLa-derived HLtat cells were transfected and then mixed with an excess of untransfected HLtat or NIH 3T3-derived PA317 cells. The following day, polyethylene glycol fusion was performed in the presence of cycloheximide, and the ability of proteins to translocate to neighboring nuclei within the syncytia was monitored.
Before fusion, both p54nrb and PSF were localized strictly to the nucleus (Fig. 7A), as expected (18). At 1 h after fusion, large syncytia were observed in all plates. p54nrb was found to relocalize efficiently from the donor to the acceptor nuclei, indicating that it is a shuttle protein (Fig. 7A). In contrast, PSF did not shuttle under the same conditions (Fig. 7A).
We repeated the fusion assays with mouse NIH 3T3-derived PA317 cells as the acceptor (Fig. 7B). After fusion, the human nuclei were visualized with species-specific antibodies for a nuclear nonshuttling protein, the Ku autoantigen. Figure 7B shows that wild-type p54nrb relocalized efficiently to the mouse nuclei, whereas the RRM2 deletion further increased the shuttling efficiency, enabling the protein to equilibrate completely between the heterokaryon nuclei. Staining for the Ku autoantigen showed a low or undetectable signal in the acceptor nuclei, verifying that the donor nuclei were intact and the relocalization of p54nrb proteins was due to bona fide nuclear export. The wild-type PSF also did not shuttle in these heterokaryons (Fig. 7B), in agreement with the data obtained in human-human fusions (Fig. 7A).
One possibility was that PSF contains export determinants but nuclear retention is dominant over export in HLtat cells. We therefore used a panel of GFP-PSF deletions to dissect its trafficking signals (Fig. 7C). The deletions affected the previously described N-terminal proline and glutamine-rich region (P/Q, amino acids 10 to 266) and RNA recognition motifs RRM1 (amino acids 297 to 369) and RRM2 (amino acids 371 to 452) (18). All mutants shared the C-terminal portion (amino acids 490 to 707) that contains two putative nuclear localization signals and were localized strictly to the nucleus. As expected from earlier studies (18), the proteins with an intact RRM2 formed subnuclear foci, whereas the lack of this domain led to diffuse localization in the nucleus. Fusion assays showed that deletion of RRM1 or/and RRM2 (Fig. 7; 1-707ΔRRM2, 279-707ΔRRM2, 338-707, 338-707ΔRRM2, and 490-707) caused the proteins to shuttle, whereas a P/Q deletion (Fig. 7; 279-707) had no effect. Thus, there is a nuclear retention signal that is comprised of RRM1 and RRM2 and requires the presence of both domains.
Since RRM1 deletion did not affect focus formation (18) yet led to shuttling (338-707), nuclear retention is not caused solely by focus association. Hence, the RRM deletions revealed a nuclear export determinant(s) in PSF's C-terminal region (amino acids 490 to 707). No further mapping was performed, because any N- and C-terminal truncations led to a detectable cytoplasmic localization (18), which precluded the use of shuttling assays. Since p54nrb is homologous to the region spanning PSF's retention and export determinants, we studied the matching regions in p54nrb with a panel of GFP-tagged mutants. We found that, similar to PSF, deletion of either of p54nrb's RRM domains (Δ17-220, Δ71-220, ΔRRM2, and 144-470) further augmented shuttling efficiency (Fig. 7). To probe the export mechanism, we performed shuttling assays in the presence of actinomycin D or leptomycin B. As expected (18), low doses of actinomycin D led to redistribution of both p54nrb and PSF to perinucleolar clusters. However, the shuttling of both subunits was not affected, suggesting that their nucleocytoplasmic trafficking is independent of transcription inhibition. The leptomycin B treatment also had no effect, indicating that CRM1 is not involved (data not shown).
In summary, we concluded that both p54nrb and PSF contain nuclear export and retention determinants and that p54nrb is continuously exported from the nucleus.
DISCUSSION
Here, we found that the HIV-1 mRNA instability element in the p37_gag_ region is a specific, high-affinity ligand of p54nrb/PSF. Our functional assays showed that the PSF subunit acts posttranscriptionally via INS to inhibit p37_gag_ mRNA expression. We further demonstrated that PSF is able to downregulate expression of the INS-containing, Rev-dependent mRNAs encoding gag and env, whether produced from the full-length molecular clone of HIV-1 or from cDNA expression plasmids.
These assays measured the cumulative effects of mRNA regulation at all posttranscriptional steps, and therefore PSF affected a step(s) which is both crucial and rate-limiting. Hence, PSF has the properties of a key factor mediating INS function. Because INS act at several levels, from nuclear retention to translation (5, 15, 22, 29, 33, 51, 54), additional INS-binding factors may be involved at steps that are not rate-limiting in our assays. Among the proteins that assembled with INS in vitro, our study identified the previously reported INS binders PABP1 and hnRNP A1 (2, 3, 9, 10, 41). We consider it likely that these and/or additional factors may facilitate INS recognition in vivo or play a role at later steps. In fact, hnRNP A1 has been shown to participate in INS function, possibly by direct interactions (9, 41). The possible mechanistic roles of such factors in INS-mRNA regulation remain to be addressed.
Intriguingly, p54nrb was recently shown to mediate RNA retention in the nucleus by directly binding to inosine-rich determinants in hyperedited RNAs (62). The I-RNA-binding complex also includes other nuclear matrix-associated proteins, PSF and matrin 3, suggesting that the RNA is retained via anchoring to insoluble nuclear components. Because both the INS and the I-RNA are directly recognized by the components of this complex, they may share this proposed mechanism. In support of this model, both the I-RNA (62) and the INS elements (our unpublished data) mediate nuclear retention of microinjected RNAs in Xenopus laevis oocytes, and the retention of I-RNA can be counteracted by Rev/RRE regulation (62). In contrast, in human cultured cells, the presence of INS leads to reduced mRNA levels both in the nucleus and in the cytoplasm but does not cause significant nuclear retention (Fig. 3D). This is in agreement with our previous studies, where such effects were attributed to the fast decay of retained INS-mRNA (54). Similarly, the p54nrb-containing INS-mRNPs did not accumulate in the insoluble nuclear fractions which are enriched in p54nrb/PSF.
One explanation is that mRNA in the putative insoluble complexes is subject to fast decay. It is also possible that the retention of INS-mRNPs is not rate-limiting and therefore difficult to detect. In any case, because p54nrb is found in soluble INS-mRNP, both in the nucleus and in the cytoplasm, it may have an additional role downstream of nuclear retention. This is consistent with the view that the INS elements cause mRNA decay in both compartments (54) and can inhibit translation directly (15; our unpublished data). Taken together, our results suggest that p54nrb/PSF participate in INS-mediated regulation both in the nucleus and in the cytoplasm.
In further support of the possible cytoplasmic role of p54nrb/PSF, we found that p54nrb is a shuttling protein and that both subunits contain nuclear export determinants. This link to export is in agreement with the recent identification of PSF as well as PSP2/CoAA and matrin 3 as components of nuclear pore complex proteome, pointing to a probable physical connection to the nuclear pore complex/lamina (12). p54nrb and PSF have also been identified among tyrosine-phosphorylated proteins that are associated with the nuclear envelope, suggesting that they may bind in a regulated manner (43).
Regarding the role of p54nrb/PSF in the regulation of authentic HIV-1 mRNAs, we show that, at elevated concentrations, PSF acts specifically via INS elements in both gag and env regions, causing mRNA downregulation. We also observed a weak effect on expression of the Rev-independent, multiply spliced mRNAs, attributable to the reduced levels of pre-mRNA in the nucleus.
These results are consistent with the previously proposed model in which the INS-binding factors divert the Rev-dependent transcripts from splicing to an alternative compartment where they are committed to export (2, 3, 8, 22, 37, 50, 54). This pathway is only productive in the presence of Rev, and in its absence, all HIV-1 transcripts are spliced to completion. Thus, Rev-dependent transcripts are not withheld from splicing when Rev is absent, suggesting that they interact with INS-binding factors reversibly. We speculate that, at elevated PSF concentrations, such interactions become irreversible, leading to sequestration of INS-containing transcripts from the productive pathways such as splicing and export, which results in fast decay.
Because p54nrb (and possibly PSF) is a component of paraspeckles, a distinct nuclear compartment that is adjacent to splicing speckles but is not enriched in spliceosomal components, it is plausible that p54nrb/PSF diverts the INS-containing transcripts from splicing by targeting them to paraspeckles. Furthermore, both p54nrb and PSF are dynamic factors that shuttle between their nucleoplasmic locations and the nucleolus (18, 23; our unpublished results), a domain that is highly enriched in Rev (22). We found previously that the nucleolus is part of the CRM1 export route and the site of assembly of Rev-CRM1 complexes with CRM1's downstream partners, such as the mobile nucleoporins Nup214 and Nup98 (63). We and others have also shown that the INS-containing HIV-1 mRNAs are enriched in the nucleolus (49) and are accessible to nucleolus-targeted ribozymes (36). Together, these data implicated the nucleolus as a nuclear export compartment for Rev-dependent HIV-1 mRNAs. Hence, p54nrb/PSF also has the potential to deliver the INS-containing transcripts to the sites of Rev-CRM1 export. In support of this idea, we show here that p54nrb is found in specific, soluble complexes with INS-mRNA, which may represent such transport intermediates.
In summary, PSF is one of the key factors mediating the posttranscriptional regulation of HIV-1 and is likely part of a novel mRNA control mechanism that is hijacked by the virus. At elevated concentrations, PSF causes a dramatic, dose-dependent inhibition of virus production in cell culture, pointing to the possibility that PSF can contribute to the control of HIV-1 propagation in vivo. Further work is needed to examine such possible effects in targets relevant for virus replication. Thus, our findings may provide insight into the control of HIV-1 replication in vivo and suggest novel antiviral strategies.
Acknowledgments
We thank L. Arthur, F. Boege, A. Krainer, G. Nabel, and D. Zipori for the generous gifts of reagents. Some of the mutant p54nrb and PSF constructs were made by Billy T. Dye. We gratefully acknowledge the assistance of A. Waddelow, C. Jodrie, A. Gainer, P. Sood, and A. Whitten. We thank Susan Lindtner for help in some experiments and for critically reading the manuscript.
The protein microsequencing was performed at the Beckman Research Institute facility, funded by National Institutes of Health Cancer Center Support grant CA33572.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59**:**284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonina, E., M. Neumann, and G. N. Pavlakis. 1997. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 272**:**2307-2311. [DOI] [PubMed] [Google Scholar]
- 3.Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273**:**13015-13021. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12**:**1-11. [DOI] [PubMed] [Google Scholar]
- 5.Arrigo, S. J., and I. S. Y. Chen. 1991. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 5**:**808-819. [DOI] [PubMed] [Google Scholar]
- 6.Bear, J., W. Tan, A. S. Zolotukhin, C. Tabernero, E. A. Hudson, and B. K. Felber. 1999. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol. 19**:**6306-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergamini, G., T. Preiss, and M. W. Hentze. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6**:**1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berthold, E., and F. Maldarelli. 1996. cis-acting elements in human immunodeficiency virus type 1 RNAs direct viral transcripts to distinct intranuclear locations. J. Virol. 70**:**4667-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black, A. C., J. Luo, S. Chun, A. Bakker, J. K. Fraser, and J. D. Rosenblatt. 1996. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes 12**:**275-285. [DOI] [PubMed] [Google Scholar]
- 10.Black, A. C., J. Luo, C. Watanabe, S. Chun, A. Bakker, J. K. Fraser, J. P. Morgan, and J. D. Rosenblatt. 1995. Polypyrimidine tract-binding protein and heterogeneous nuclear ribonucleoprotein A1 bind to human T-cell leukemia virus type 2 RNA regulatory elements. J. Virol. 69**:**6852-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochrane, A. W., K. S. Jones, S. Beidas, P. J. Dillon, A. M. Skalka, and C. A. Rosen. 1991. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 65**:**5305-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158**:**915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2000. Nuclear RNA export pathways. Mol. Cell. Biol. 20**:**4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Agostino, D. M., V. Ciminale, G. P. Pavlakis, and L. Chieco-Bianchi. 1995. Intracellular trafficking of the human immunodeficiency virus type 1 Rev protein: involvement of continued rRNA synthesis in nuclear retention. AIDS Res. Hum. Retroviruses 11**:**1063-1072. [DOI] [PubMed] [Google Scholar]
- 15.D'Agostino, D. M., B. K. Felber, J. E. Harrison, and G. N. Pavlakis. 1992. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 12**:**1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, B., D. S. Horowitz, R. Kobayashi, and A. R. Krainer. 1993. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 21**:**4085-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3**:**195-205. [DOI] [PubMed] [Google Scholar]
- 18.Dye, B. T., and J. G. Patton. 2001. An RNA recognition motif (RRM) is required for the localization of polypyrimidine tract-binding protein-associated splicing factor (PSF) to subnuclear speckles. Exp. Cell Res. 263**:**131-144. [DOI] [PubMed] [Google Scholar]
- 19.Emili, A., M. Shales, S. McCracken, W. Xie, P. W. Tucker, R. Kobayashi, B. J. Blencowe, and C. J. Ingles. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8**:**1102-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinberg, M. B., R. F. Jarrett, A. Aldovini, R. C. Gallo, and F. Wong-Staal. 1986. human T-cell lymphotropic virus-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell 46**:**807-817. [DOI] [PubMed] [Google Scholar]
- 21.Felber, B. K. 1998. Posttranscriptional control: a general and important regulatory feature of HIV-1 and other retroviruses, p. 101-122. In G. Myers (ed.), Viral regulatory structures and their degeneracies, vol. XXVIII. Addison-Wesley, Boston, Mass.
- 22.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86**:**1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox, A. H., Y. W. Lam, A. K. Leung, C. E. Lyon, J. Andersen, M. Mann, and A. I. Lamond. 2002. Paraspeckles: a novel nuclear domain. Curr. Biol. 12**:**13-25. [DOI] [PubMed] [Google Scholar]
- 24.Gozani, O., J. G. Patton, and R. Reed. 1994. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13**:**3356-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadzopoulou-Cladaras, M., B. K. Felber, C. Cladaras, A. Athanassopoulos, A. Tse, and G. N. Pavlakis. 1989. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a _cis_-acting sequence in the env region. J. Virol. 63**:**1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammarskjold, M. L., J. Heimer, B. Hammarskjold, I. Sangwan, L. Albert, and D. Rekosh. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 63**:**1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365**:**186-191. [DOI] [PubMed] [Google Scholar]
- 28.Huang, Y., W. P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75**:**4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence, J. B., A. W. Cochrane, C. V. Johnson, A. Perkins, and C. A. Rosen. 1991. The HIV-1 Rev protein: a model system for coupled RNA transport and translation. New Biol. 3**:**1220-1232. [PubMed] [Google Scholar]
- 30.Lee, K. A., A. Bindereif, and M. R. Green. 1988. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Technol. 5**:**22-31. [DOI] [PubMed] [Google Scholar]
- 31.Lutz, C. S., C. Cooke, J. P. O'Connor, R. Kobayashi, and J. C. Alwine. 1998. The snRNP-free U1A (SF-A) complex(es): identification of the largest subunit as PSF, the polypyrimidine-tract binding protein-associated splicing factor. RNA 4**:**1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maldarelli, F., M. A. Martin, and K. Strebel. 1991. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: Novel level of gene regulation. J. Virol. 65**:**5732-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malim, M. H., J. Hauber, S.-Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 _rev trans_-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338**:**254-257. [DOI] [PubMed] [Google Scholar]
- 34.Maquat, L. E., and G. G. Carmichael. 2001. Quality control of mRNA function. Cell 104**:**173-176. [DOI] [PubMed] [Google Scholar]
- 35.Meissner, M., T. Dechat, C. Gerner, R. Grimm, R. Foisner, and G. Sauermann. 2000. Differential nuclear localization and nuclear matrix association of the splicing factors PSF and polypyrimidine tract-binding protein. J. Cell Biochem. 76**:**559-566. [PubMed] [Google Scholar]
- 36.Michienzi, A., L. Cagnon, I. Bahner, and J. J. Rossi. 2000. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. USA 97**:**8955-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikaelian, I., M. Krieg, M. J. Gait, and J. Karn. 1996. Interactions of INS (CRS) elements and the splicing machinery regulate the production of Rev-responsive mRNAs. J. Mol. Biol. 257**:**246-264. [DOI] [PubMed] [Google Scholar]
- 38.Mili, S., H. J. Shu, Y. Zhao, and S. Pinol-Roma. 2001. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell. Biol. 21**:**7307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miralles, F., L. G. Ofverstedt, N. Sabri, Y. Aissouni, U. Hellman, U. Skoglund, and N. Visa. 2000. Electron tomography reveals posttranscriptional binding of pre-mRNPs to specific fibers in the nucleoplasm. J. Cell Biol. 148**:**271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore, M. J. 2002. Nuclear RNA turnover. Cell 108**:**431-434. [DOI] [PubMed] [Google Scholar]
- 41.Najera, I., M. Krieg, and J. Karn. 1999. Synergistic stimulation of HIV-1 rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J. Mol. Biol. 285**:**1951-1964. [DOI] [PubMed] [Google Scholar]
- 42.Nasioulas, G., A. S. Zolotukhin, C. Tabernero, L. Solomin, C. P. Cunningham, G. N. Pavlakis, and B. K. Felber. 1994. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J. Virol. 68**:**2986-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto, H., M. Dreger, L. Bengtsson, and F. Hucho. 2001. Identification of tyrosine-phosphorylated proteins associated with the nuclear envelope. Eur. J. Biochem. 268**:**420-428. [DOI] [PubMed] [Google Scholar]
- 44.Patton, J. G., E. B. Porro, J. Galceran, P. Tempst, and B. Nadal-Ginard. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7**:**393-406. [DOI] [PubMed] [Google Scholar]
- 45.Peng, R., B. T. Dye, I. Perez, D. C. Barnard, A. B. Thompson, and J. G. Patton. 2002. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8**:**1334-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinol-Roma, S., Y. D. Choi, M. J. Matunis, and G. Dreyfuss. 1988. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 2**:**215-227. [DOI] [PubMed] [Google Scholar]
- 47.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52**:**491-532. [DOI] [PubMed] [Google Scholar]
- 48.Pongoski, J., K. Asai, and A. Cochran. 2002. Positive and negative modulation of human immunodeficiency virus type 1 Rev function by cis and trans regulators of viral RNA splicing. J. Virol. 76**:**5108-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romanov, V. I., A. S. Zolotukhin, N. N. Aleksandroff, P. Pinto da Silva, and B. K. Felber. 1997. Posttranscriptional regulation by Rev protein of human immunodeficiency virus type 1 results in nonrandom nuclear localization of gag mRNA. Virology 228**:**360-370. [DOI] [PubMed] [Google Scholar]
- 50.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71**:**4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz, S., M. Campbell, G. Nasioulas, J. Harrison, B. K. Felber, and G. N. Pavlakis. 1992. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 66**:**7176-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyö, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64**:**2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz, S., B. K. Felber, E. M. Fenyö, and G. N. Pavlakis. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 64**:**5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1992. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 66**:**150-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shav-Tal, Y., M. Cohen, S. Lapter, B. Dye, J. G. Patton, J. Vandekerckhove, and D. Zipori. 2001. Nuclear relocalization of the pre-mRNA splicing factor PSF during apoptosis involves hyperphosphorylation, masking of antigenic epitopes, and changes in protein interactions. Mol. Biol. Cell 12**:**2328-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shav-Tal, Y., and D. Zipori. 2002. PSF and p54(nrb)/NonO-multi-functional nuclear proteins. FEBS Lett. 531**:**109-114. [DOI] [PubMed] [Google Scholar]
- 57.Sodroski, J., W. C. Goh, C. Rosen, A. Dayton, E. Terwilliger, and W. Haseltine. 1986. A second post-transcriptional trans-activator gene required for human T-cell lymphotropic virus-III replication. Nature 321**:**412-417. [DOI] [PubMed] [Google Scholar]
- 58.Stauber, R. H., K. Horie, P. Carney, E. A. Hudson, N. I. Tarasova, G. A. Gaitanaris, and G. N. Pavlakis. 1998. Development and applications of enhanced green fluorescent protein mutants. BioTechniques 24:462-. 6**:**468-471. [DOI] [PubMed] [Google Scholar]
- 59.Straub, T., P. Grue, A. Uhse, M. Lisby, B. R. Knudsen, T. O. Tange, O. Westergaard, and F. Boege. 1998. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J. Biol. Chem. 273**:**26261-26264. [DOI] [PubMed] [Google Scholar]
- 60.Traish, A. M., Y. H. Huang, J. Ashba, M. Pronovost, M. Pavao, D. B. McAneny, and R. B. Moreland. 1997. Loss of expression of a 55 kDa nuclear protein (nmt55) in estrogen receptor-negative human breast cancer. Diagn. Mol. Pathol. 6**:**209-221. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, W. W., L. X. Zhang, R. K. Busch, J. Farres, and H. Busch. 1993. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem. J. 290**:**267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Z., and G. G. Carmichael. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106**:**465-475. [DOI] [PubMed] [Google Scholar]
- 63.Zolotukhin, A. S., and B. K. Felber. 1999. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73**:**120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zolotukhin, A. S., W. Tan, J. Bear, S. Smulevitch, and B. K. Felber. 2002. U2AF participates in the binding of TAP (NXF1) to mRNA. J. Biol. Chem. 277**:**3935-3942. [DOI] [PubMed] [Google Scholar]
- 65.Zolotukhin, A. S., A. Valentin, G. N. Pavlakis, and B. K. Felber. 1994. Continuous propagation of RRE(-) and Rev(-)RRE(-) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J. Virol. 68**:**7944-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuo, P., and T. Maniatis. 1996. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10**:**1356-1368. [DOI] [PubMed] [Google Scholar]