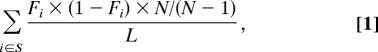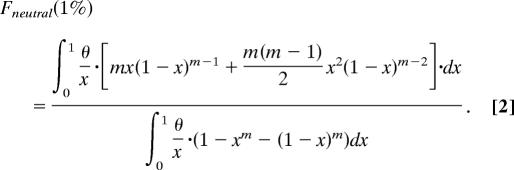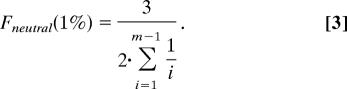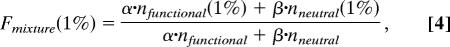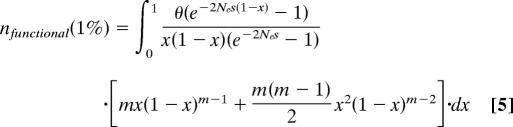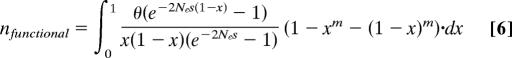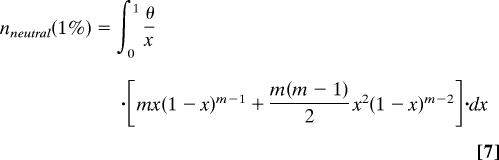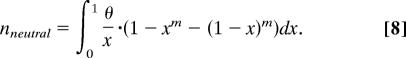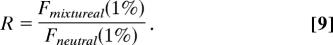Widely distributed noncoding purifying selection in the human genome (original) (raw)
Abstract
It is widely assumed that human noncoding sequences comprise a substantial reservoir for functional variants impacting gene regulation and other chromosomal processes. Evolutionarily conserved noncoding sequences (CNSs) in the human genome have attracted considerable attention for their potential to simplify the search for functional elements and phenotypically important human alleles. A major outstanding question is whether functionally significant human noncoding variation is concentrated in CNSs or distributed more broadly across the genome. Here, we combine wholegenome sequence data from four nonhuman species (chimp, dog, mouse, and rat) with recently available comprehensive human polymorphism data to analyze selection at single-nucleotide resolution. We show that a substantial fraction of active purifying selection in human noncoding sequences occurs outside of CNSs and is diffusely distributed across the genome. This finding suggests the existence of a large complement of human noncoding variants that may impact gene expression and phenotypic traits, the majority of which will escape detection with current approaches to genome analysis.
Higher eukaryotes are believed to carry a large burden of “junk DNA” in their genomes. Although >98% of the human genome comprises nonprotein-coding DNA (1), the true density and distribution of functional nucleotides in these regions is currently unknown. Comparison of the human genome with those of other species has revealed the existence of a large number of nonprotein-coding sequences that appear to have been conserved through purifying natural selection (2). Such conserved noncoding sequences (CNSs) are widely believed to harbor the preponderance of human noncoding nucleotides under active selection (3–6). Frequently cited estimates suggest that only ≈5% of the human genome, including ≈3.5% of its noncoding fraction, consists of regions under purifying natural selection (2, 5). This figure has led to the widespread supposition that most of the human genome landscape comprises a vast evolutionary junkyard, a situation that contrasts sharply with that in lower animals and simple eukaryotes (7–9). Recent estimates suggest that selectively constrained and hence presumably functional nucleotides comprise 43% of the yeast genome (7). Likewise, a substantial fraction (40–50%) of noncoding DNA in the genome of Drosophila melanogaster appears to be under selection. This estimate is based on the findings that intronic and intergenic sequences are evolving more slowly than 4-fold degenerate (synonymous) sites in coding regions (8, 9). These findings are the opposite of what is observed in the human genome, where noncoding regions appear to be evolving more quickly than 4-fold degenerate sites (after correcting for hypermutable CpG dinucleotides) (10). Whether selected nucleotides in lower genomes are confined to specific regions or dispersed throughout noncoding regions is unknown.
A major unanswered question is whether CNSs in the human genome accurately capture the distribution of functionally significant noncoding nucleotides, or, conversely, to what extent noncoding sites outside of CNSs are functionally significant in modern humans. Comparison of sequence divergence between species with population polymorphism provides a powerful approach for analyzing selective forces acting on genomic sequences (8, 11, 12).
Here, we apply human polymorphism data and divergence data from multiple species toward analyses of conservation and selection at nucleotide resolution in noncoding regions. The results suggest that a substantial fraction of the human noncoding genome is under active negative selection in modern populations, with much of the effect arising outside of CNSs.
Results and Discussion
The hallmark of active negative selection on human sequences is a shift in the allele frequency spectrum toward rarer alleles and a reduction in nucleotide diversity (13) (heterozygosity per nucleotide; π). Such analyses are optimally conducted in the context of data sets that comprehensively ascertain polymorphisms by resequencing multiple individuals. Resequencing permits accurate estimation of nucleotide diversity and correct appreciation of skew in the allele frequency spectrum. We therefore focused our analysis on 13.1 megabases (≈0.5% of the human genome) comprising 567 diverse human gene loci that were resequenced in 90–95 individuals (180–190 chromosomes) from a multiethnic population (14). We analyzed a total of 78,472 polymorphisms (coding and noncoding) and computed both nucleotide diversity and allele frequency spectrum in several classes of genomic regions (Fig. 1). 5′ and 3′ UTRs were excluded from the analysis because of insufficient SNP data. We also excluded sex chromosomes because of their lower average effective population size and lower mutation rate, which will cause them to harbor less polymorphism than autosomes, confounding comparison between chromosomes.
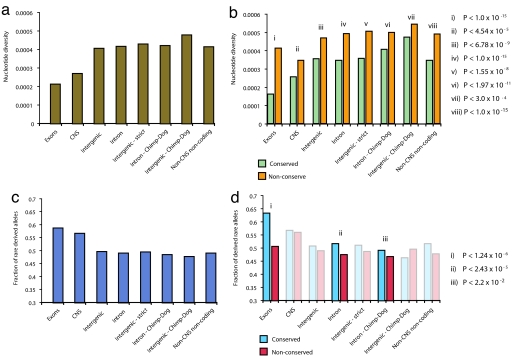
Fig. 1.
Selection at conserved vs. nonconserved nucleotide positions (EGP). Effect of partitioning genomic sequence features into conserved and nonconserved positions on nucleotide diversity and allele frequency distributions. Only EGP SNPs at non-CpG sites were considered. (a) Nucleotide diversity (π; vertical axis) in exons, CNSs defined by PhastCons, introns, introns aligning between chimp and dog but not between chimp and rodents, intergenic sequences, intergenic sequences aligning between chimp and dog but not between chimp and rodents, and all non-CNS noncoding sequences. (b) Nucleotide diversity at conserved (green) and nonconserved (orange) positions within genomic features shown in a. P values (Fisher exact test) for differences in density of segregating sites between conserved and nonconserved positions at corresponding features are shown. (c) Fraction of SNPs with derived allele <1% (vertical axis) within different genomic sequence features. (d) Fraction of SNPs with derived allele <1% at conserved (blue) and nonconserved (red) positions within features shown in c, with corresponding P values. Semitransparent data indicate features for which the number of SNPs within the EGP data set do not provide sufficient power to detect statistically significant differences in allele frequency distribution.
We applied several widely used definitions of CNSs, including CNSs delineated by hidden Markov model analysis of multispecies alignments using the PhastCons algorithm (15), those based on high-scoring human–mouse alignments of varying length (16), and those defined by different combinations of sequence length and percent human–mouse identity (Fig. 2) (see Methods).
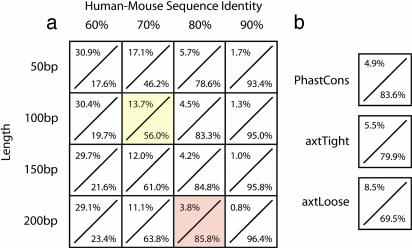
Fig. 2.
Distribution of 4GCBs relative to CNSs. CNSs are typically defined by lengths of human genomic sequence in which the percent human–mouse sequence identity exceeds a threshold value. Shown, for each combination of length (50–200 bp) and percentage of human–mouse sequence identity (60%–90%) (a) or various other CNS definitions (b), is the fraction of the human genome encompassed by that CNS definition (range 30.8% to 0.8%) over the fraction of 4GCBs that fall outside of this definition (range 17.6% to 96.4%). For comparison, parameters used in previous studies of CNSs are highlighted in yellow (34) and red (17). The PhastCons CNS definition is the one used to generate Figs. 1 and 3.
We then used whole-genome sequence data from chimp, dog, mouse, and rat to partition the entire human genome sequence at the nucleotide level by identifying bases conserved between the four nonhuman species; these bases may be conserved either because of selective constraint or simply random chance. Such four-genome conserved bases (4GCBs) constitute ≈12% of the genome and are widely distributed in noncoding sequences, with the vast majority (up to 96.4%) occurring outside of CNSs defined by different criteria in both the study regions and across the genome generally (Fig. 2).
To validate the utility of the partition into conserved and nonconserved bases for detecting major features of selective pressure, we first examined protein-coding exons. The evolution of coding regions is driven principally by purifying selection, with significant heterogeny imposed by the structure of the genetic code. We observed marked selective differences between conserved and nonconserved nucleotides within coding regions [Fig. 1 b and d and supporting information (SI) Table 1]. Nucleotide diversity of individually conserved non-CpG sites within coding regions clearly differs from nonconserved sites (0.015% vs. 0.038%; P < 1 × 10−15) and is markedly reduced compared with the average across noncoding regions (0.015% vs. 0.038%; P < 1 × 10−15). As nucleotide diversity is strongly influenced by mutation rate, reduced diversity may be the product of regional mutation rate heterogeneity; however, this reduction is accompanied by a strong shift in the allele frequency distribution toward rare alleles (Fig. 1c), which is significantly more prominent for conserved vs. nonconserved positions (rare allele fraction 0.62 vs. 0.50; P < 1 × 10−5).
Next, we compared polymorphisms within CNSs, coding sequences, and non-CNS noncoding regions. We found that nucleotide diversity within CNSs (0.025%) is markedly reduced compared with other noncoding sequences (0.039%; P < 1 × 10−15), although significantly increased relative to protein-coding regions (0.020%; P < 1 × 10−15), compatible with previous observations (14, 17, 18). The reduction in nucleotide diversity (Fig. 1a) is accompanied by an excess in the proportion of low-frequency variants (Fig. 1c). Despite an overall level of conservation comparable with that of protein-coding sequences, the selective effects in CNSs are markedly weaker than those operating within coding regions.
We then examined differences between conserved vs. nonconserved nucleotides in noncoding regions, both within CNSs and excluding CNSs. In non-CNS noncoding regions, we observed a significant reduction in diversity and an excess of rare alleles at individually conserved positions vs. nonconserved nucleotides (Fig. 1 b and d and SI Table 1). For example, intronic sequences showed an rare allele frequency shift from 0.51 to 0.47 between conserved and nonconserved positions (Fisher exact test; P < 1.2 × 10−5), and a shift in average nucleotide diversity from 0.031% to 0.045%.
To confirm that these estimates are reliable, and that the differences in nucleotide diversity or the fraction of rare alleles between conserved and nonconserved bases are not caused by large coalescent variance, we analyzed independent subsamples of the data set. We subdivided the SNP data set into 25 nonoverlapping subsamples of SNPs pooled according to genomic positions. Using these subsamples we estimated that the standard error of the estimated fractions of rare alleles is 0.0086 for conserved positions and 0.0105 for nonconserved positions. We also applied the nonparametric Friedman test to these subsamples to confirm that the excess of rare alleles in 4GCBs is statistically significant (Friedman test one-sided; P < 0.002).
These differences are of comparable magnitude with those observed when partitioning noncoding sequence into CNSs and non-CNS regions generally. The differences between conserved and nonconserved non-CNS nucleotides are present irrespective of the definition of CNS, even if one excludes the union of all definitions of CNSs. The excess of rare alleles at individually conserved positions cannot be explained either by invoking heterogeneity in mutation rate (19, 20) or the effect of population demographic history. It therefore indicates that a significant fraction of such conserved positions are under active selection in modern human populations.
Surprisingly, the strength of the observed effect does not depend on the local density of 4GCBs. We computed a local density metric for each 4GCB by counting the number of 4GCBs within nonoverlapping 50-bp windows. We found that the aforementioned selective effect was present even for 4GCBs that fell in low-density windows (<15 4GCBs of 50 bp). Additionally, we confirmed that the selective effect was independent of both regional G+C content and the type of nucleotide substitution (i.e., all combinations of transitions vs. transversions).
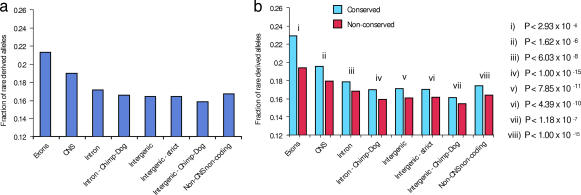
To confirm and extend these findings, we analyzed human polymorphism in two additional settings. First, we examined noncoding regions that are not readily alignable between humans and rodents. In these regions we compared nucleotide diversity and allele frequency spectra between nucleotides that exhibit conservation (identity) between chimp and dog with those that differ between chimp and dog. Chimp/dog conserved sites exhibit lower nucleotide diversity than sites that differ between chimp and dog (0.038% and 0.046%, respectively; P < 1 × 10−12) and a significant shift toward rare alleles (0.48 and 0.46, respectively; _P_ < 0.05). Second, we considered genotype data from the haplotype map of the human genome (HapMap) (21) phase II (Fig. 3 and SI Table 2). The vast majority of SNPs genotyped by the HapMap are common alleles (frequency >10%); however, sufficient numbers of less frequent alleles (<5% frequency) have recently become available. These data recapitulate the allele frequency patterns seen for the resequencing data across all classes of genomic sequence features with much greater statistical significance.
Fig. 3.
Selection at conserved vs. nonconserved nucleotide positions (HapMap). Effect of partitioning genomic sequence features into conserved and nonconserved positions on HapMap allele frequency distributions (non-CpG sites). (a) Fraction of rare derived alleles (frequency <5%; HapMap Yoruba data set) in genomic sequence features (see legend to Fig. 1 for details). (b) Fraction of SNPs with derived allele <5% at conserved (blue) and nonconserved (red) positions within features shown in a, with corresponding P values.
We note that analyses based on 4GCBs are decidedly conservative, and, as suggested by analysis of chimp–dog conservation in nonrodent-aligning regions, our results should extend to other less conserved sequences. We note further that our results and those of other studies (17, 18, 22) do not definitively exclude the possibility that mutation rate heterogeneity has contributed in some way to the genesis and propagation of CNSs in mammalian genomes. A priori, a small proportion of bases under selection in a region with low average mutation rate will be much more likely to be classified as a CNS than a similar situation in a high mutation rate region. Mutation rate is heterogeneous in the human genome and is believed to vary by at least 2-fold between different genomic regions (19, 20); however, the scale over which such fluctuations occur is unknown.
What proportion of noncoding bases in the human genome is under selection? To address this, we attempted to develop a lower-limit estimate by using a modeling approach. We considered a standard infinite number of sites model (23, 24) with heterogeneous mutation rates. We used data from intronic sequences to represent noncoding bases, which showed a rare allele frequency shift from 0.51 to 0.47 between conserved and nonconserved positions and a shift in average heterozygosity from 0.031% to 0.045%. To obtain the most conservative estimate, we allowed for any level of mutational rate differences between conserved and nonconserved sites. We considered two classes of conserved sites, neutral sites and sites under negative selection, the latter all associated with the same selection coefficient. All nonconserved sites are considered neutral. These assumptions yield the most conservative estimate we can make. By varying the selection coefficient, we identified the minimal fraction of functional bases needed to produce the excess of rare alleles observed at 4GCBs (Fig. 4). The curve in Fig. 4 has a minimum because weak selection would produce only a small shift in allele frequencies; strong selection, on the other hand, would produce a larger shift in functional sites, but would also decrease the level of polymorphism there, so that the allele frequency distribution would represent mostly neutral polymorphism (see Methods). This analysis suggested that a minimum of 18.5% of nucleotide positions conserved across four genomes must be under pressure of negative selection to explain the observed shift in the allele frequency spectrum. It is important to emphasize that this fraction represents a lower-limit estimate with respect to selection strength and mutation rate heterogeneity. The only scenario under which our estimate would be rendered nonconservative is if the mutation parameter were considerably higher at functional vs. neutral positions, a situation we consider to be unlikely.
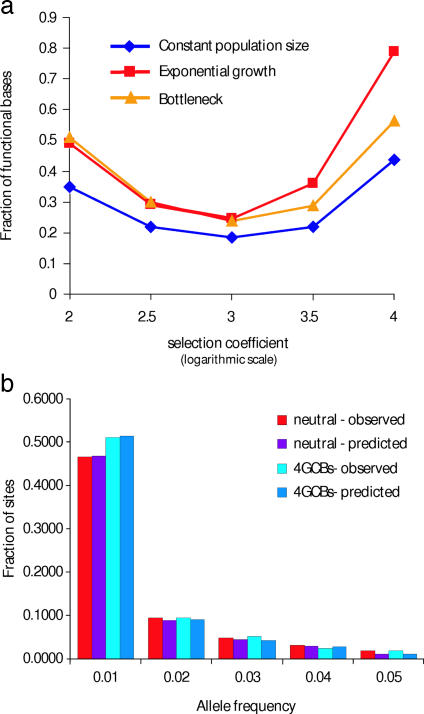
Fig. 4.
Fraction of functionally significant nucleotide positions among 4GCBs. (a) Fraction of functional sites under selection (y axis) sufficient to explain the observed excess of rare alleles in conserved positions, expressed as a function of selection coefficient (x axis; logarithmic scale) for three different population histories. The fraction of functional sites exhibits a minimal value (under any possible strength of selection) needed to explain the observed shift in allele frequency distribution. This minimum provides a lower limit estimate of the fraction of functional sites. (b) Allele frequency distributions for SNPs in non-4GCB (red) and 4GCB intronic positions (light blue) are shown in parallel with theoretical distributions (corresponding to the optima from a) for neutral SNPs (purple) and a mixture of neutral SNPs and functional SNPs (dark blue).
Allele frequency data from multiple populations suggest the complexity of human demographic history (25–27). To quantify the effect of such complexity on our estimate, we performed forward simulations including an expanding population and a population bottleneck followed by population expansion (Fig. 4). Unlike the model of constant population size, the model involving a population bottleneck followed by an expansion is generally consistent with the observed fraction of rare alleles (Fig. 4b). Models involving expansion and bottleneck give slightly higher estimates of the fraction of functionally significant nucleotides (24.5% and 23.6%, respectively).
Selection against deleterious alleles can also be inferred from comparison of polymorphism to divergence (e.g., McDonald–Kreitman test). Under weak purifying selection, reduction of population nucleotide diversity in functionally important sites compared with neutral sites should be lower than the corresponding reduction in divergence between species. This effect was observed earlier in protein-coding human genes (12) and highly conserved noncoding regions of the human genome (18). To test whether our analysis based on allele frequency distribution is in agreement with the polymorphism-to-divergence ratio, we compared human nucleotide diversity to sequence divergence in the human lineage after it split from chimpanzee. To identify substitutions in the human lineage we used the genome sequence of macaque as an outgroup. We note that the macaque genome was not used for defining 4GCBs. A possible disadvantage of using the macaque genome as outgroup is that it is relatively distant and multiple substitutions per site can slightly bias the estimates.
Indeed, divergence in the human lineage in 4GCBs is 38% lower than divergence in non-4GCBs, which is higher than the corresponding difference of 30% for the nucleotide diversity of the human population. As with the analysis of the allele frequency spectrum, we derived a conservative estimate of the fraction of functionally significant 4GCBs. At least 18% of 4GCBs were estimated as selectively constrained, which is in good agreement with the estimate obtained from the allele frequency shift.
As with other recent work (12) on the analysis of genomewide SNP datasets, our analysis uses a model of completely unlinked sites. However, allele frequency distributions and nucleotide diversity can also be affected by selection in linked sites. Selective sweeps and background selection can reduce nucleotide diversity. Selective sweeps can also increase the fraction of rare alleles. The effect of background selection associated with efficient purifying selection can be modeled as a reduction of effective population size in a locus and will not change our estimate. Weak purifying selection in a small population can change the allele frequency distribution in linked neutral sites (28). Although the action of background selection associated with weak purifying selection has been observed in humans, its effect was “subtle over the majority of the human genome” (29). Further, it has been hypothesized that biased gene conversion can produce disparities in allele frequency distributions. Indeed, we observed that nucleotide diversity is lower, whereas the fraction of rare alleles is higher, in regions with low recombination rate when compared with regions with high recombination rate. We also observed that SNPs resulting from G/C to A/T mutations have lower allele frequency than SNPs resulting from to A/T to G/C mutations, as expected under the hypothesis of biased gene conversion. However, the difference between 4GCBs and non-4GCBs in nucleotide diversity and fraction of low-frequency alleles is almost identical for both categories of substitutions and is independent of recombination rate. Furthermore, the difference in these statistics in adjacent pairs of 4GCB and non-4GCBs (see Methods) remains numerically identical to the difference across all genomic sites, suggesting that the observed effect does not depend on any kind of regional variation.
How do our findings compare with previous estimates of the fraction of noncoding sequence under selection in humans? Estimates have been made both at the nucleotide level and the level of fixed short sequence windows. Per-nucleotide estimates of constraint in humans indicate that 0.6% of bases in the genome are constrained coding positions and 0.8% of bases are constrained positions in CNSs (22). Our results indicate that at a minimum 3.5-fold more noncoding nucleotides (2.8% of nucleotides) are under selection than estimates based on CNSs, and that 71.4% of positions under selection (2% of nucleotides) lie outside CNSs.
Comparisons between the human and mouse genomes have conservatively suggested that 5% of nonoverlapping 50-bp windows are under selection, 1.5% in coding regions and 3.5% in noncoding regions. We find that nearly all (99%) such 50-bp windows in the four-species alignable fraction of the genome contain at least one 4GCB. Depending on the distribution of functional 4GCBs in these windows, between 5.5% and 26% of 50-bp windows in noncoding regions may contain at least one functional nucleotide position under active selection in humans (see SI Table 1). Current analysis does not have sufficient power to identify constrained positions with high specificity but it may become possible given sufficient additional genome sequences (30, 31).
In summary, we have shown that partitioning human noncoding sequence into individually conserved and nonconserved positions using comparative sequence data provides a powerful approach for analyzing the selecting forces shaping the majority of human genome territory. The results suggest that a substantial fraction of the human noncoding genome is under active negative selection in modern populations, with much of the effect arising outside of CNSs. This disparity has significant implications for the discovery of noncoding mutations with functional consequences for human disease and quantitative phenotypes. Systematic discovery of functional variants impacting gene regulation and quantitative phenotypic variation may therefore require prior large-scale definition of functional noncoding elements using experimentally based approaches (32).
Methods
Positions in the human genome were classified according to annotations provided by the University of California, Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu). Coding positions were determined according to the RefSeq track from UCSC. PhastCons CNS elements were taken directly form the UCSC browser. Tight CNS positions were defined by the regions alignable between human (hg17) and mouse (mm5) according to the axtTight alignment set available from UCSC (16). Loose CNS positions were defined by rescoring axtNet alignments (also available from UCSC) using the same criteria used to generate the axtTight set with the subsetAxt program (by Jim Kent, available from UCSC), but with a relaxed score threshold (2,000 instead of 3,400). Windowed CNSs were defined by considering human–mouse alignments (hg17 vs. mm5; from UCSC) and identifying all windows with a given minimum length (50, 100, 150, and 200 bp) and minimum percent identity (60%, 70%, 80%, and 90%) criteria. All exonic positions, plus a 50-bp margin surrounding each exon, were excluded from both CNS sets. Intronic and intergenic positions were also defined according to the RefSeq track from UCSC by excluding exons plus a surrounding 50-bp margin and excluding all tight CNS positions. A strict intergenic set was defined by additionally excluding any positions for which there was an annotated mRNA (according to the UCSC mRNA track).
Genomewide site-specific conservation was computed by using the UCSC multiz8way alignment (33). 4GCBs were those positions alignable between all four genomes showing perfect identity between chimp, mouse, rat, and dog. Four-genome nonconserved bases (4GNB) were those alignable positions that did not show perfect identity. Human sequence was excluded in determining conservation, because the inclusion of derived alleles in the human consensus sequence can produce artificially strong correlation between nonconserved positions and derived allele frequency (DAF). A weaker conservation definition was made on the basis of chimp–dog identity, if chimp and dog were alignable. Any positions that were also alignable with mouse or rat were excluded from this second set, so that the set represented the worst-conserved positions available.
To remove biases associated with regional variation in genomic processes, “adjacent pairs” subsets of 4GCB and 4GNB positions were defined by taking all 4GNB positions that were directly followed by a 4GCB position and all 4GCB positions that were directly preceded by a 4GNB position. This partition ensured that both these subsets had the same size and distribution throughout the genome.
SNP allele frequency and position information was retrieved from the Environmental Genome Project (EGP) (14) (http://egp.gs.washington.edu) for all available candidate genes. HapMap phase II genotype data for the Yoruba population was also retrieved from the International HapMap Project (21) (http://hapmap.org) and used to compute SNP allele frequencies. DAF was determined for each SNP where available by using the maximum parsimony method, with the aligned chimpanzee nucleotide serving as the outgroup (UCSC hg17/panTro1 alignment). CpG SNPs were excluded from all allele frequency analysis to prevent the possibility of errors caused by nonparsimony.
Average heterozygosity (π) was computed for EGP data only, because of bias in the HapMap SNP discovery process. π was computed for each data class as:
where F was the frequency of the minor allele, L was the total number of positions in the data class, N was the number of chromosomes in the sample (taken to be 185), and S was the set of all SNPs in the data class. Positions that fell within CpG dinucleotides or that contained a CpG allele were excluded from the data class.
The fraction of rare DAFs was computed for both EGP data and genomewide HapMap data. For EGP data, the fraction of rare DAFs was the fraction of DAFs in a class with frequency ≥0.01; this set encompassed singlets and doublets (i.e., only one or two chromosomes in the sample contained the allele). For HapMap data, discovery bias meant a shifted distribution with a paucity of rare SNPs. Therefore, for these data, the rare fraction was that portion with frequency ≥0.05.
To determine the fraction of sites under selection, we considered a standard infinite number of sites model with constant effective population size. To obtain the most conservative estimate with respect to mutation rate heterogeneity at any scale, we focused solely on differences in allele frequency spectra.
We postulated that individually nonconserved positions outside of CNSs and protein-coding genes evolve neutrally, whereas individually conserved positions represent a mixture of neutral and functionally significant positions. We further assumed that all new mutations in functional positions are associated with the same selection coefficient s and there is no dominance (see below). Thus, the model has two parameters: (i) fraction of functionally significant nucleotide positions among completely conserved bases, and (ii) selection coefficient associated with mutations in functionally significant positions.
The shift in the allele frequency distribution was measured as the ratio of the fractions of SNPs with DAF <1% for individually conserved and nonconserved bases. Alleles with frequencies <1% in the EGP data set were represented once or twice. Therefore, the fraction of neutral SNPs with DAF <1% is given by the sum of the fractions of alleles represented by a single chromosome and alleles represented by two chromosomes. As follows from the diffusion theory approximation of the infinite number of sites model (23, 24):
Here m is the number of sequenced individual chromosomes, x is the allele frequency in the population, and θ is the mutation parameter equal to the product of effective population size and mutation rate multiplied by four. After integration (θ cancels out) this simplifies to:
This is a well known relationship that can be obtained via alternative approaches (13).
If the fraction of functionally significant nucleotides among conserved bases is α and the fraction of neutral bases among conserved bases is β (equal to 1 − α), the fraction of SNPs observed only in one or two individual chromosomes is given by:
Here s is selection coefficient and _N_e is effective population size.
This estimator of the fraction of functionally significant bases has a minimum with respect to selection coefficient (e.g., at _s_0). This implies that the estimate will be the smallest if all sites were associated with selection coefficient _s_0.
Under a more realistic assumption that sites have varying selection coefficients, the contributions of sites with values of s different from _s_0 would increase the estimate. In other words, the most conservative assumption for our purpose is that the distribution of selection coefficients in functional sites is concentrated at a single point, i.e., all sites are associated with the same selection coefficient.
The shift in allele frequency distribution was measured by the ratio:
We obtained a conservative estimate of the fraction of functionally significant nucleotides by minimizing α over s while keeping the ratio R constant. The resulting estimate was 18.5%.
This theoretical estimate corresponds to a model of constant size of the human population. A model of constant population size is known to be inconsistent with the data on human genetic variation. Therefore, we analyzed models of population expansion and population bottleneck followed by expansion, which are in much better agreement with observed human allele frequency distributions. These two models of complex demographic history were analyzed by using forward simulations of the Wright–Fisher model, assuming an infinite number of sites. The estimates of fraction of functionally significant sites are 24.5% for the population expansion model and 23.6% for the bottleneck followed by expansion model.
We also estimated the fraction of selectively constrained 4GCBs by using polymorphism-to-divergence comparison (see SI Text). According to this method, the minimal estimate of α over all possible values of s is 0.18, which is in good agreement with the estimate obtained by using the allele frequency spectrum.
Supplementary Material
Supporting Information
Abbreviations
CNS
conserved noncoding sequence
4GCB
four-genome conserved base
4GNB
four-genome nonconserved base
EGP
Environmental Genome Project
HapMap
haplotype map of the human genome
UCSC
University of California, Santa Cruz
DAF
derived allele frequency.
Footnotes
The authors declare no conflict of interest.
References
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Miller W, Makova KD, Nekrutenko A, Hardison RC. Annu Rev Genomics Hum Genet. 2004;5:15–56. doi: 10.1146/annurev.genom.5.061903.180057. [DOI] [PubMed] [Google Scholar]
- 3.Boffelli D, Nobrega MA, Rubin EM. Nat Rev Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- 4.Dermitzakis ET, Reymond A, Antonarakis SE. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 5.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 6.Shabalina SA, Ogurtsov AY, Kondrashov VA, Kondrashov AS. Trends Genet. 2001;17:373–376. doi: 10.1016/s0168-9525(01)02344-7. [DOI] [PubMed] [Google Scholar]
- 7.Doniger SW, Huh J, Fay JC. Genome Res. 2005;15:701–709. doi: 10.1101/gr.3578205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andolfatto P. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 9.Halligan DL, Keightley PD. Genome Res. 2006 doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 11.McDonald JH, Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 12.Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 13.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 14.Livingston RJ, von Niederhausern A, Jegga AG, Crawford DC, Carlson CS, Rieder MJ, Gowrisankar S, Aronow BJ, Weiss RB, Nickerson DA. Genome Res. 2004;14:1821–1831. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, Excoffier L, Attar H, Antonarakis SE, Dermitzakis ET, et al. Nat Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- 18.Kryukov GV, Schmidt S, Sunyaev S. Hum Mol Genet. 2005;14:2221–2229. doi: 10.1093/hmg/ddi226. [DOI] [PubMed] [Google Scholar]
- 19.Silva JC, Kondrashov AS. Trends Genet. 2002;18:544–547. doi: 10.1016/s0168-9525(02)02757-9. [DOI] [PubMed] [Google Scholar]
- 20.Gaffney DJ, Keightley PD. Genome Res. 2005;15:1086–1094. doi: 10.1101/gr.3895005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. Nature. 2005;437:1299–1320. [Google Scholar]
- 22.Keightley PD, Kryukov GV, Sunyaev S, Halligan DL, Gaffney DJ. Genome Res. 2005;15:1373–1378. doi: 10.1101/gr.3942005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura M. Population Genetics, Molecular Evolution, and The Neutral Theory: Selected Papers. Chicago: Univ Chicago Press; 1994. [Google Scholar]
- 24.Sawyer SA, Hartl DL. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marth GT, Czabarka E, Murvai J, Sherry ST. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson SH, Hernandez R, Fledel-Alon A, Zhu L, Nielsen R, Bustamante CD. Proc Natl Acad Sci USA. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlesworth D, Charlesworth B, Morgan MT. Genetics. 1995;141:1619–1632. doi: 10.1093/genetics/141.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed FA, Akey JM, Aquadro CF. Genome Res. 2005;15:1211–1221. doi: 10.1101/gr.3413205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eddy SR. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ENCODE Project Consortium. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nobrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM. Nature. 2004;431:988–993. doi: 10.1038/nature03022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information