Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis (original) (raw)
Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating motor neuron degenerative disease whose etiology and pathogenesis remain poorly understood. Most cases of ALS (≈90%) are sporadic (SALS), occurring in the absence of genetic associations. Approximately 20% of familial ALS (FALS) cases are due to known mutations in the copper, zinc superoxide dismutase (SOD1) gene. Molecular evidence for a common pathogenesis of SALS and FALS has remained elusive. Here we use covalent chemical modification to reveal an attribute of spinal cord SOD1 common to both SOD1-linked FALS and SALS, but not present in normal or disease-affected tissues from other neurodegenerative diseases, including Alzheimer's, Parkinson's, and Huntington's diseases and spinal muscular atrophy, a non-ALS motor neuron disease. Biotinylation reveals a 32-kDa, covalently cross-linked SOD1-containing protein species produced not only in FALS caused by SOD1 mutation, but also in SALS. These studies use chemical modification as a novel tool for the detection of a disease-associated biomarker. Our results identify a shared molecular event involving a known target gene and suggest a common step in the pathogenesis between SALS and FALS.
Keywords: copper, zinc superoxide dismutase; motor neuron; neurodegeneration
Amyotrophic lateral sclerosis (ALS) pathology and symptoms in copper, zinc superoxide dismutase (SOD1)-linked familial ALS (FALS) patients closely resemble those of sporadic ALS (SALS) patients. In light of the linkage of the SOD1 gene to FALS (1), it has long been speculated that there may be a common molecular basis for both forms of the disease, but no such feature has been identified. A plethora of evidence has demonstrated that mutant SOD1 causes motor neuron death by gain of toxic properties (2, 3). However, mechanisms of mutant SOD1-mediated toxicity to motor neurons remain elusive. Many studies have investigated the possibility of structural differences in SOD1 caused by mutations as a basis for such toxic effects (4, 5). Although several common features of mutant SOD1 proteins such as instability, higher sensitivity to denaturing conditions, and propensity to form aggregates are found to be associated with mutant SOD1-linked FALS disease course and severity (6–11), no single biochemical feature has been found to be shared by all identified mutant SOD1s, nor has there been a single molecular feature of SOD1 identified to be associated with SALS.
We propose that SOD1 is more conformationally heterogeneous in vivo and that these “conformers” are relevant to ALS but difficult to distinguish by conventional biochemical analyses. To reveal such conformational differences especially those intrinsic to minor subspecies of SOD1 we applied a new approach involving biotinylation of accessible lysine residues in proteins, followed by SDS/PAGE and immunoblot analysis for SOD1. Covalent chemical modification of amino acid side groups has the advantage of rapidly proceeding to completion, being readily quenched, and capable of potentially affecting antibody immunoreactivity (12). The extent of a chemical modification reaction is strongly affected by the degree of the accessibility of a side group to a modifying agent. This, in turn, is influenced by the overall conformational structure of the protein, thereby providing a basis by which different conformers react differently to a covalent modifying agent (13). Thus, a conformational difference in a small proportion of molecules or a subtle difference in structure may be magnified. Using this approach, we now provide direct evidence for a common underlying pathogenesis of SALS and FALS through the identification of a unique, crosslinked protein species of SOD1 that is common to both.
Results
A 32-kDa SOD1-Immunoreactive (IR) Protein Species Revealed by Biotinylation of Spinal Cord Extracts of ALS Patients.
Conventional immunoblotting after protein denaturation under strongly reducing conditions of spinal cord tissue extracts from ALS patients or normal samples revealed only the expected 16-kDa SOD1 polypeptide recognized by an antibody generated to a peptide from human SOD1 (Fig. 1B, “−” lanes). After testing 12 commercially available chemical compounds that can modify side-chains of amino acids of lysine, cysteine, aspartic acid, glutamic acid, and arginine [see supporting information (SI) Table 1], side-chain modification by covalent reaction with _N_-hydroxysulfosuccinimide-long chain-biotin (Sulfo-NHS-LC-Biotin), which covalently adds biotin to conformationally accessible lysine side chains (Fig. 1A), revealed a new 32-kDa species in the SOD1-linked FALS sample [from a patient with a dominant alanine to valine mutation in SOD1 at position 4 (SOD1A4V)], but not a normal sample (Fig. 1B). Surprisingly, a similar biotinylation-dependent 32-kDa species was also seen in a SALS sample (Fig. 1B).
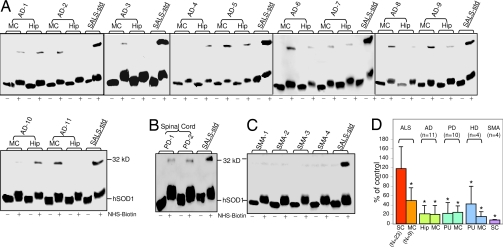
Fig. 1.
A 32-kDa SOD1-IR protein species revealed by biotinylation of spinal cord extracts of ALS patients. (A) Schematic diagram of the detection of a 32-kDa SOD1-IR protein species by immunoblot analysis. Sulfo-NHS-LC-Biotin reacts with structurally available NH2 groups (outside the rings) from the lysine residues in the SOD1 protein resulting in the modification of the protein at the sites of the side-chains (indicated by *). The biotinylated SOD1 proteins were separated on SDS/PAGE gel and immunorecognized by the SOD1 antibody (darker ring). (B–G) Spinal cord cytosolic proteins were incubated with (+) or without (−) Sulfo-NHS-LC-Biotin, separated on SDS/PAGE gel, and immunoblotted with the SOD1 antibody. (B) The FALS sample was from an A4V mutation. Cytosolic proteins from SALS-4 subject were used for C–G. (C) Time course of the reaction. One hundred percent value was taken for the density of the 32 kDa at 150-min reaction time. (D) Sulfo-NHS-LC-Biotin dose responses. One hundred percent value was taken for the density of the 32 kDa at 100 mM biotin concentration. (E) Dose–response of the total amount of proteins used. One hundred percent value was taken at a total amount of 20 μg of proteins. (F) Temperature dependence of the reaction. The optimal temperature for this reaction lies around 25°C. (G) Conformation-specificity of the reaction. NHS-digoxigenin (1 mM) was added before the reaction can block the biotinylation reaction resulting in the disappearance of the 32-kDa protein species (two middle lanes). All error bars represent SD from three independent experiments.
Further characterization of the biotinylation reaction revealed that the intensity of the 32-kDa species depended on time of incubation (Fig. 1C), concentration of Sulfo-NHS-LC-Biotin, the amount of the initial extract and incubation temperature (Fig. 1 D–F). The reaction was highly SOD1 protein conformation-specific, because substrates containing partial or related structural groups to Sulfo-NHS-LC-Biotin including sulfo-NHS, biotin, long chain (LC)-biotin, iodoacetyl-LC-biotin, NHS-Squarin carboxilate (data not shown), and NHS-digoxigenin. (Fig. 1G) did not result in ALS-specific change of SOD1 proteins. Maximum intensity of the 32-kDa SOD1 species was reached after 25 min at an optimal temperature of 25°C in the presence of 10 mM biotin. This condition was used in all efforts below.
The 32-kDa SOD1 Adduct Is Common to Both SALS and FALS.
To determine whether the appearance of the 32-kDa SOD1-IR species in the initial SALS and SOD1-linked FALS samples after reaction with Sulfo-NHS-LC-Biotin was a general feature of FALS and SALS, a series of spinal cord extracts from 19 SALS and four FALS with mutations in the SOD1 gene were examined. After biotinyation, an increased intensity of the 32-kDa protein species IR to SOD1 antiserum was found in all of the ALS samples (Fig. 2A). In 14 of 16 normal samples, this species was absent or at much reduced level, although the expected 16 kDa species representing monomeric SOD1 was found in comparable amounts before and after biotinylation (Fig. 2A). The signal intensities of the 32-kDa species were significantly higher in spinal cord samples from ALS versus normal subjects when determined by either combining FALS and SALS samples (Fig. 2B; 118.0 ± 46.5, n = 23 for all ALS and 25.1 ± 34.2, n = 16, for normal; P < 0.0001) or treating them separately (105.6 ± 37.8, n = 19; P < 0.0001 for SALS and 176.7 ± 81.0, n = 4; P < 0.0001 for FALS). Taken together, there is an ≈5-fold increase in the 32-kDa signal intensity in all ALS compared with normal control (Fig. 2B). In addition, when a sample that contains a mutation in another ALS-linked gene, Senetaxin (14) (also known as ALS4), was examined, the 32-kDa signal was also present (Fig. 2C).
Fig. 2.
Prevalence of the 32-kDa SOD1-IR protein species in both SALS and FALS spinal cords after biotinylation. Immunoblot analysis of cytosolic proteins from human spinal cord autopsy samples (A and B) incubated with (+) or without (−) Sulfo-NHS-LC-Biotin. The density of the 32-kDa SOD1-IR signal from SALS-1 spinal cord autopsy sample (indicated as SALS-std) was chosen as a 100%, which was run on all gels as the standard for spinal cord samples. (A) Spinal cords are from normal, SALS, and SOD1-linked FALS autopsy samples. Mutations in SOD1 are G93C, A4V, G127X, D101G for FALS-1,2,3,4, respectively. (B) Bar graphs of intensities of the 32-kDa signal from spinal cords for all ALS (SALS + SOD1-linked FALS, n = 23) and normal (n = 16). Error bars represent SD. *, P < 0.0001. (C) Subject with a mutation in the Senetexin gene (ALS4). (D) The spinal cord samples from one normal and one SALS subject were separated as the white and gray matter before the analysis.
To further determine the area within the spinal cord that gives rise to this 32-kDa signal, we separated the spinal cord into gray and white matter and analyzed them separately. As shown in Fig. 2D, the 32-kDa signal was exclusively associated with the gray matter where motor neuron, glia, and other cells reside in contrast to the white matter containing different axonal tracks, including the cortical-spinal track. Because these assays compare constant amounts of protein before and after biotinylation, the increased immunoreactivity is likely due to enhanced antigen availability to the antiserum as a result of biotinylation.
SOD1 mutations are also observed in SALS, albeit at a very low frequency (<5%) (15–18). To eliminate the possibility that the 32-kDa signals from some SALS subjects could come from subjects that might harbor SOD1 mutations, the coding sequences were determined for the SOD1 genes from spinal cord autopsy samples from SALS-10, -11, -14, -15, and -17, samples which had signal intensities of the 32 kDa that were comparable to those from FALSs. No SOD1 mutations were found (data not shown) in these five samples (Fig. 2A). Although we cannot rule out that there is no SOD1 mutation in the remaining 14 SALS (we have 19 total, minus 5 sequenced) samples analyzed, the 32-kDa signals from the SALS samples represent a typical population of non-SOD1 mutant-linked SALS, rather than SOD1-linked FALS.
The 32-kDa SOD1 Species Is Specific for Disease-Affected Tissues in ALS but Not Those in Alzheimer's Disease (AD), Parkinson's Disease (PD), Huntington's Disease (HD), and Spinal Muscular Atrophy (SMA).
To understand whether the 32 kDa is unique to ALS pathology as opposed to a reflection of a process derived simply from a prolonged agonal state, we further examined this signal in similar samples from three additional non-ALS neurodegenerative diseases, including AD, PD, and HD. Two spinal cord samples were initially examined from PD subjects. The intensities of the 32- kDa signal were comparable to those from normal subjects and were significantly lower than those from ALS (Fig. 3B). As spinal cord is not the primary site of pathology in PD, we also examined affected brain regions in AD, PD, and HD together with the motor cortical area as the common central nervous tissue for all diseases. Hippocampus is one of the areas severely affected in AD. When examined, there was no significant increase of the 32- kDa signal in this brain area compared with the much increased level in ALS spinal cord (P < 0.0001, Fig. 3 A and D). Similar results were found for putamen, an affected brain region in both PD and HD (Fig. 3D, P < 0.0001 and P < 0.02, respectively, compared with ALS spinal cord). For the motor cortexes, the intensities of the 32-kDa signal in ALS, AD, PD, and HD were significantly lower when compared with those from ALS spinal cords (Fig. 3D, P < 0.001, P < 0.0001, P < 0.0001, and P < 0.001, respectively). Furthermore, the difference in the intensity of the 32-kDa signal in the motor cortex between ALS and AD or PD is also significant (Fig. 3D, P < 0.02). This motor cortex value between ALS and HD did not quite reach statistical difference (P = 0.05) because of the large variation and small sample size (n = 4) for HD (Fig. 3D). The higher level of the 32 kDa in the motor cortex of ALS but not AD or PD is consistent with the known upper motor neuron pathology in ALS, demonstrating the selective pathological role of the 32 kDa in ALS. Lastly, to gain some insight on how widely the 32 kDa might be involved in motor neuron damage in other motor neuron diseases, we examined spinal cord tissues from subjects with Type 1 form of SMA. The 32 kDa was not present in the four SMA spinal cords examined (Fig. 3 C and D, P < 0.0001). Motor neuron death in Type I SMA differs from that of ALS by the fact that it is a much accelerated process and devoid of aging factors. Together, these data demonstrate that the 32 kDa appears in the pathologic process of age-related motor neuron degeneration such as that in ALS but not involved in pathology of AD, PD, HD, or SMA.
Fig. 3.
The 32-kDa SOD1-IR protein species is specific for ALS pathology. Immunoblot analysis of cytosolic proteins from different brain region autopsy samples were incubated with (+) or without (−) Sulfo-NHS-LC-Biotin. The density of the 32-kDa SOD1-IR signal from spinal cord autopsy sample SALS-1 (indicated as SALS-std) was chosen as a 100%, which was run on all gels as the standard for all samples. (A) The anterior hippocampus (Hip) and primary motor cortex (MC) from AD subjects were analyzed. The spinal cord from two PD subjects (B) and four SMA patients (C) was analyzed. (D) Bar graphs of the relative intensities the 32-kDa signal from ALS spinal cords (SC) and MC, AD Hip and MC, PD putamen (PU) and MC, HD PU and MC, and SMA SC.
The 32 kDa Is a Modified SOD1 Protein Species.
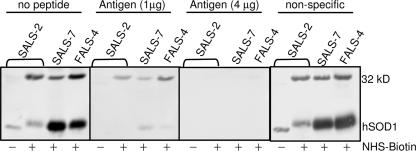
To confirm that the observed 32 kDa is an altered form of SOD1, peptide competition studies were performed. Antigen-specific peptide eliminated reactivity to the 32-kDa (and the 16-kDa SOD1) signal in a dose-dependent manner (Fig. 4). In contrast, a different peptide from SOD1 had no such effect on the SOD1 immunoreactrivity when used at similar a concentration (Fig. 4). To corroborate this conclusion, nanoelectrospray mass spectrometry was used to determine whether the 32 kDa in fact contained SOD1 protein. The gel slice containing the 32 kDa was purified as described (see SI Methods). Peptide sequence analysis based on mass spectra acquired and peptide mapping (see SI Fig. 6 and SI Table 2, respectively) provided 100% coverage of the human SOD1 protein sequence. These results demonstrate that the 32 kDa does indeed contain the human SOD1 protein.
Fig. 4.
Antigen-specific peptide eliminates observed immunoreactivity to SOD1 in the 32 kDa. Biotinylation and immunoblot were done as described. Samples used are cytosolic proteins from muscle (SALS-2) and spinal cords (SALS-7 and FALS-4). Peptides were incubated with the primary antibody for 30 min at room temperature in 500 μl of 5% milk in PBST before incubation with the membranes. Antigen-specific peptide (DDLGKGGNEESTK) was used at 1 μg and 4 μg, respectively. Nonspecific peptide (KESNGPVKVWGSIKGGC) was used at 1 μg.
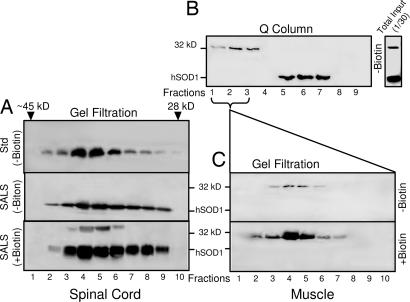
To further demonstrate that the SDS-resistant 32 kDa is a modified SOD1 molecule in complex with itself or a similarly sized protein (19), gel filtration and ion exchange chromatography experiments were performed to characterize the protein(s) that gave rise to the 32-kDa species during biotinylation. As expected, under nonreducing, nondenaturing conditions, most SOD1 polypeptides in spinal cord cytosolic proteins were present in dimers that eluted in gel filtration fractions corresponding to ≈32-kDa molecular mass, a range similar to the elution position of commercially purified human SOD1 protein (Fig. 5A Top and Middle). Biotinylation of gel filtration fractions revealed that the 32-kDa species was observed exclusively in the same molecular mass range (Fig. 5A Bottom), demonstrating the substrate for biotinylation behaved as a 32-kDa protein species.
Fig. 5.
The 32 kDa is a modified SOD1 protein species. (A) The commercial purified hSOD1 protein (Std, 40 μg) and SALS spinal cord cytosolic proteins (SALS-4, 300 μg) were subjected to gel filtration without biotinylation (−Biotin). Molecular mass standards were used during elution, and only the fractions corresponding to ≈32-kDa range (between ≈45 kDa and 28 kDa, as indicated) were shown in fractions 1–10. Fractions from the SALS-4 samples were further subjected to biotinylation (+Biotin). (B) SALS muscle cytosolic proteins (300 μg) were applied to the Q-type ion exchange column, and the flow-through (fractions 1–3) and eluted fractions (4–9) were analyzed. An aliquot of the same sample (total input, 1/30) before the Q-Column is also shown. (C) The Q-column flow-through material was subjected to gel filtrations and analyzed directly (−Biotin) or after biotinylation (+Biotin). All proteins were separated on SDS/PAGE and immunoanalyzed with the SOD1 antibody.
Although biotinylation was required to observe the 32-kDa SOD1 species in spinal cord extracts, it was observable even without biotinylation in muscle extracts from FALS or SALS patients (Fig. 5B). We exploited this property to determine the native characteristics of this 32-kDa SOD1 species without biotinylation and to eliminate the possibility of noncovalent dimeric SOD1 becoming covalently linked during biotinylation. Cytosolic ALS protein extracts were fractionated before biotinylation by Q-column ion exchange chromatography. The normally folded, non-covalently associated SOD1 dimers bound to the matrix and eluted only at higher ionic strength (Fig. 5B, eluted in fractions 5–7). A small amount of IR SOD1 did not bind to the column. Subsequent immunoblotting after denaturation under reducing conditions revealed that this unbound SOD1 exclusively migrated at a position corresponding to 32 kDa (Fig. 5 B, fractions 1–3; C Upper, fractions 3–6). As before, the immunoreactivity of this 32- kDa species with the anti-peptide SOD1 antibody was markedly enhanced after biotinylation (Fig. 5C Lower, fractions 2–7). These findings clearly demonstrate that the 32-kDa protein species is an endogenously modified SOD1 that is covalently linked to another SOD1 polypeptide or to a polypeptide of similar size and that this species has substantially divergent biochemical properties relative to the natural non-covalently linked SOD1 homodimer.
Discussion
One mystery associated with not only ALS, but many other neurodegenerative diseases, is how to reconcile disease mechanisms at the molecular level between involvement of mutations in a few molecules that are responsible for a small percentage of familial forms of the disease and unknown molecules that underlie the majority of the diseases that are of sporadic nature. An assumption frequently made is that there is a common path shared by both familial and sporadic forms of ALS that ultimately leads to a common clinical manifestation of mixed lower and upper motor neuron degeneration. Evidence supporting this view has remained elusive until now. Our study demonstrates at least one component of a shared pathway is an already known molecule linked to ALS: SOD1.
Like other neurodegenerative diseases, ALS has been considered a disease of protein misfolding (20). Demonstrations that mechanisms of mutant SOD1-mediated toxicity are due to gain of toxic properties (2) suggest that altered conformations of SOD1 proteins because of mutation have different behaviors than the WT SOD1 (SOD1WT) protein. This is fully consistent with the more than 110 mutations in the SOD1 molecule linked to ALS (21) for which a normally folded “WT” state and a misfolded one linked to toxicity have frequently been found. Our observation of a covalently crosslinked, biotinylation-dependent, SOD1-IR species (32 kDa) within the spinal cord demonstrates that SOD1 exists in at least two forms, one of which is disease-associated and common to both FALS and SALS. The increased level of the 32-kDa species in ALS patients must originally come from the normally folded SOD1WT. Our data demonstrate that an aberrant SOD1 species is present (or associated with) human ALS pathology, suggesting a possible involvement in that pathology. This conclusion extends previous results from in vitro cellular and animal models of ALS indicating that SOD1WT or its oxidized form can be toxic in ALS disease mechanisms (22–28). Interestingly, the WT normal α-synuclein proteins, when produced by an additional locus in human, can also cause PD (29).
Our observation that the 32-kDa species is not present in normal and three non-motor neuron diseases including AD, PD and HD, nor in Type I SMA which involves motor neuron death somewhat different from that in ALS, strongly suggests that the 32 kDa is ALS pathology-related. Compared with the vast majority of SOD1 molecules that are almost certainly irrelevant to disease, the low levels of this disease-specific SOD1-IR species (≈1% of total SOD1) are usually below detection limits, but can be revealed after biotinylation. Thus, although chemical modification has been used to study overall differences in protein conformation, the approach we have used here has uncovered a molecular signature common to both familial and sporadic ALS.
SOD1 IR protein species of molecular weight approximately the size of human SOD1 dimer are reported in refs. 30–32. More recently, studies from transgenic mouse models of FALS have suggested that the intermolecular disulfide-linked SOD1 aggregates are associated with disease toxicity (8, 23). One study used immunoprecipitation to conclude an apparent SOD1 species of ≈35 kDa was a complex between SOD1 and Bcl-2 (19), albeit our mass spectrometric analysis offered no support for any linkage to Bcl-2, at least in SALS. The covalently crosslinked, partially misfolded species we have identified here is different from these previously proposed complexes. Its identification suggests a common underlying pathogenic mechanism in inherited and sporadic ALS, a finding that carries a substantial implication for design of therapeutic approaches for sporadic disease.
Materials and Methods
Obtainment of Human Samples.
Human autopsy samples were obtained as described in ref. 24, from The Brain and Tissue Bank for Developmental Disorders of the National Institute of Child Health and Human Development (Baltimore, MD), Forbes Norris MDA/ALS Research Center (San Francisco, CA), and ALS Tissue Bank at University of Pittsburgh School of Medicine (Pittsburgh, PA). Informed consents were obtained from all subjects before sample collections. Handling of all human samples is in accordance with the approved protocols by California Pacific Medical Center Internal Board Review, University of Pittsburgh Internal Board Review, and Committee for Oversight of Research Involving the Dead. Patients' information is listed in SI Table 3.
Subcellular Fractionation and Biotinylation.
Cytosolic proteins from the autopsy samples were prepared as described in ref. 33. Protein concentrations were measured by using the BCA method (Pierce Chemical, Rockford, IL). Biotinylation reaction, unless indicated otherwise, was performed with a total of 10 μg of cytosolic proteins incubated with 10 mM Sulfo-NHS-LC-Biotin (Pierce Chemical) in PBS buffer, pH 7.4, for 25 min at 25°C. The reaction was stopped by adding free lysine-HCl at a final concentration of 20 mM for 20 min at 25°C. Control treatment was carried out in identical procedure except omitting Sulfo-NHS-LC-Biotin in the reaction.
Gel Filtration.
Sephacryl 300 column (Amersham Biosciences, Piscataway, NJ) was equilibrated with PBS buffer, pH 7.4, containing 150 mM sodium chloride. The protein sample was applied onto the column at a ratio of 60 μg of cytosolic proteins per 1 ml of Sephacryl 300. Proteins were eluted under gravity in the same buffer at 4°C, and a volume of 500 μl was collected for each fraction. All fractions were dialyzed against deionizated water for 24 h at 4°C followed by lyopholization. Lyopholized samples were resuspended in PBS for biotinylation and/or immunoblot analysis.
Column Chromatography.
Fast-flow Q Sepharose anion exchange column was equilibrated with 20 mM triethanolamine buffer, pH 7.8. The protein sample was applied onto the column at a ratio of 320 μg of cytosolic proteins per 1 ml of Q-Sepharose. Proteins were eluted with the same buffer containing 0, 0.025, 0.05, 0.075, 0.1, 0.15, 0.2, 0.25, 0.35, and 0.4 M NaCl (corresponding to fractions 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10, respectively) at a flow rate of 0.5 ml/min. All fractions were dialyzed against deionized water for 24 h followed by concentration in Speed Vac (Savant, Holbrook, NY) at 25°C to a final volume of ≈40 μl.
Immunoblot Analysis.
Proteins were separated on 12% SDS/PAGE gels and transferred onto PVDF membranes. Membranes were incubated with a rabbit polyclonal anti-SOD1 antiserum (34) followed by an HRP-conjugated anti-rabbit IgG secondary antibody before visualization with ECL plus solution (GE Biosciences, Little Chalfont, U.K.). Protein signals were quantified on Typhoon 9410, using Image Quant program (GE Biosciences).
Analysis of Human SOD1 cDNA Sequence.
Sequence analysis of RT-PCR-generated SOD1 cDNAs from the spinal cords of five individuals was performed. Total RNA was extracted by using TRI-Reagent (Sigma–Aldrich, St. Louis, MO). Primers for human SOD1 RT-PCR were synthesized based on GenBank cDNA complete nucleotide sequence (GenBank accession no. AY049787) as follows: forward 5′-ATGGCGACGAAGGCCGTGTGCGTGC-3′ and reverse 5′-TTATTGGGCGATCCCAATTACACCACAAGCC-3′. The RNA was reverse-transcribed and PCR-amplified by using the ImProm-II Reverse Transcription System and Pfu DNA polymerase (Promega, Madison, WI). Synthesis of SOD1 cDNA and PCR were carried in a PTC-100 programmable thermal controller. The PCR products were purified, the sequencing was performed on both strands, and the sequence was determined at the DNA Analysis Unit of Integrated DNA Technologies (Coralville, IA).
Statistical Analysis.
Statistical significance was assessed between groups, using a two-tailed Student's t test. Significance is set at P < 0.05.
Supplementary Material
Supporting Information
Acknowledgments
We thank William J. Hansen, Yelena Mikityansky, William J. Santoni, and David Ovadia for technical support and help with preparation of the article and Jason Mass for coordinating tissue collections at The Forbes Norris MDA/ALS Research Center. This work has been supported by the California Pacific Medical Center Research Institute (V.R.L. and J.L.), The Forbes Norris MDA/ALS Research Center (J.L.), the ALS Association (J.L.), the Center for Excellence in Bioinformatics at University at Buffalo (T.D.W.), and the National Institutes of Health (R.B. and D.W.C.).
Abbreviations
AD
Alzheimer's disease
ALS
amyotrophic lateral sclerosis
FALS
familial amyotrophic lateral sclerosis
HD
Huntington's disease
Hip
anterior hippocampus
IR
immunoreactive
PD
Parkinson's disease
SALS
sporadic amyotrophic lateral sclerosis
SMA
spinal muscular atrophy
SOD1
copper, zinc superoxide dismutase
Sulfo-NHS-LC-Biotin
_N_-hydroxysulfosuccinimide-long chain-biotin.
Footnotes
Conflict of interest statement: V.R.L. is a founder and J.L. a consultant to Prosetta Corporation, an early stage biotechnology company.
References
- 1.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland DW, Rothstein JD. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 3.Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, Wilcox HM, Flood DG, Beal MF, Brown RH, Jr, et al. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 4.Bruijn LI, Miller TM, Cleveland DW. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 5.Valentine JS, Doucette PA, Potter SZ. Annu Rev Biochem. 2004;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 6.Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH, Jr, Price DL, Sisodia SS, Cleveland DW. Proc Natl Acad Sci USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elam JS, Taylor AB, Strange R, Antonyuk S, Doucette PA, Rodriguez JA, Hasnain SS, Hayward LJ, Valentine JS, Yeates TO, et al. Nat Struct Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa Y, Fu R, Deng HX, Siddique T, O'Halloran T, V Proc Natl Acad Sci USA. 2006;103:7148–7153. doi: 10.1073/pnas.0602048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa Y, O'Halloran TV. J Biol Chem. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 10.Lindberg MJ, Tibell L, Oliveberg M. Proc Natl Acad Sci USA. 2002;99:16607–16612. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Nakanishi T, Yamamoto Y, Andersen PM, Ogawa Y, Fukada K, Zhou Z, Aoike F, Sugai F, Nagano S, et al. Neurology. 2005;65:1954–1957. doi: 10.1212/01.wnl.0000188760.53922.05. [DOI] [PubMed] [Google Scholar]
- 12.Sugo T, Watanabe K, Naraki T, Matsuda M. J Biochem (Tokyo) 1990;108:382–387. doi: 10.1093/oxfordjournals.jbchem.a123210. [DOI] [PubMed] [Google Scholar]
- 13.Lundblad RL, Bradshaw RA. Biotechnol Appl Biochem. 1997;26(Pt 3):143–151. [PubMed] [Google Scholar]
- 14.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battistini S, Giannini F, Greco G, Bibbo G, Ferrera L, Marini V, Causarano R, Casula M, Lando G, Patrosso MC, et al. J Neurol. 2005;252:782–788. doi: 10.1007/s00415-005-0742-y. [DOI] [PubMed] [Google Scholar]
- 16.Corrado L, D'Alfonso S, Bergamaschi L, Testa L, Leone M, Nasuelli N, Momigliano-Richiardi P, Mazzini L. Neuromuscul Disord. 2006;16:800–804. doi: 10.1016/j.nmd.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Redondo A, Bustos F, Juan YSB, Del Hoyo P, Jimenez S, Campos Y, Martin MA, Rubio JC, Canadillas F, Arenas J, et al. Muscle Nerve. 2002;26:274–278. doi: 10.1002/mus.10193. [DOI] [PubMed] [Google Scholar]
- 18.Gellera C, Castellotti B, Riggio MC, Silani V, Morandi L, Testa D, Casali C, Taroni F, Di Donato S, Zeviani M, et al. Neuromuscul Disord. 2001;11:404–410. doi: 10.1016/s0960-8966(00)00215-7. [DOI] [PubMed] [Google Scholar]
- 19.Pasinelli P, Belford ME, Lennon N, Bacskai BJ, Hyman BT, Trotti D, Brown RH., Jr Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 20.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 21.Gros-Louis F, Gaspar C, Rouleau GA. Biochim Biophys Acta. 2006;1762:956–972. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Estevez AG, Crow JP, Sampson JB, Reiter C, Zhuang Y, Richardson GJ, Tarpey MM, Barbeito L, Beckman JS. Science. 1999;286:2498–2500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 23.Deng HX, Shi Y, Furukawa Y, Zhai H, Fu R, Liu E, Gorrie GH, Khan MS, Hung WY, Bigio EH, et al. Proc Natl Acad Sci USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ezzi SA, Urushitani M, Julien JP. J Neurochem. 2007;102:170–178. doi: 10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- 25.Jaarsma D, Haasdijk ED, Grashorn JA, Hawkins R, van Duijn W, Verspaget HW, London J, Holstege JC. Neurobiol Dis. 2000;7:623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson PA, Graffmo KS, Brannstrom T, Nilsson P, Andersen PM, Marklund SL. J Neuropathol Exp Neurol. 2006;65:1126–1136. doi: 10.1097/01.jnen.0000248545.36046.3c. [DOI] [PubMed] [Google Scholar]
- 27.Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 28.Rakhit R, Cunningham P, Furtos-Matei A, Dahan S, Qi XF, Crow JP, Cashman NR, Kondejewski LH, Chakrabartty A. J Biol Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 29.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 30.Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Proc Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonsson PA, Ernhill K, Andersen PM, Bergemalm D, Brannstrom T, Gredal O, Nilsson P, Marklund SL. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Xu G, Borchelt DR. Neurobiol Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Lillo C, Jonsson PA, Vande Velde C, Ward CM, Miller TM, Subramaniam JR, Rothstein JD, Marklund S, Andersen PM, et al. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Pardo CA, Xu Z, Borchelt DR, Price DL, Sisodia SS, Cleveland DW. Proc Natl Acad Sci USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information