OPA1 Processing Reconstituted in Yeast Depends on the Subunit Composition of the m-AAA Protease in Mitochondria (original) (raw)
Abstract
The morphology of mitochondria in mammalian cells is regulated by proteolytic cleavage of OPA1, a dynamin-like GTPase of the mitochondrial inner membrane. The mitochondrial rhomboid protease PARL, and paraplegin, a subunit of the ATP-dependent _m_-AAA protease, were proposed to be involved in this process. Here, we characterized individual OPA1 isoforms by mass spectrometry, and we reconstituted their processing in yeast to identify proteases involved in OPA1 cleavage. The yeast homologue of OPA1, Mgm1, was processed both by PARL and its yeast homologue Pcp1. Neither of these rhomboid proteases cleaved OPA1. The formation of small OPA1 isoforms was impaired in yeast cells lacking the _m_-AAA protease subunits Yta10 and Yta12 and was restored upon expression of murine or human _m_-AAA proteases. OPA1 processing depended on the subunit composition of mammalian _m_-AAA proteases. Homo-oligomeric _m_-AAA protease complexes composed of murine Afg3l1, Afg3l2, or human AFG3L2 subunits cleaved OPA1 with higher efficiency than paraplegin-containing _m_-AAA proteases. OPA1 processing proceeded normally in murine cell lines lacking paraplegin or PARL. Our results provide evidence for different substrate specificities of _m_-AAA proteases composed of different subunits and reveal a striking evolutionary switch of proteases involved in the proteolytic processing of dynamin-like GTPases in mitochondria.
INTRODUCTION
Mitochondria form large networks of interconnected tubules that are maintained by balanced fission and fusion events (Nunnari et al., 1997; Okamoto and Shaw, 2005). The morphology and ultrastructure of mitochondria depend on the tissue, on the physiological condition of the cell, and, in particular, on the functional status of mitochondria. Dynamic processes associated with mitochondria are apparently crucial for the cell, e.g., in apoptosis (Frank et al., 2001; Karbowski et al., 2002; Lee et al., 2004; Jagasia et al., 2005). Likewise, formation of dendritic spines and synapses (Li et al., 2004; Verstreken et al., 2005) and functional complementation of mitochondrial DNA (mtDNA) mutations by content mixing (Nakada et al., 2001; Ono et al., 2001) depend on dynamics of mitochondria. In contrast, vast morphological alterations of mitochondria have been reported to occur in human disorders. Impairment of mitochondrial fusion is causative of neurodegenerative diseases such as Charcot-Marie-Tooth disease type 2A and 4A, and optic atrophy type 1 (Alexander et al., 2000; Delettre et al., 2000; Zuchner et al., 2004; Niemann et al., 2005).
A key player in regulating mitochondrial fusion is the dynamin-like GTPase OPA1 (Olichon et al., 2003; Cipolat et al., 2004). Mutations in the OPA1 gene cause autosomal dominant optic atrophy type I, a prevalent hereditary neuropathy of the optic nerve (Alexander et al., 2000; Delettre et al., 2000). Down-regulation of OPA1 leads to fragmentation of mitochondria, mitochondrial dysfunction, altered maintenance of mtDNA, altered mitochondrial inner membrane morphology, and increased propensity for apoptosis (Olichon et al., 2003; Griparic et al., 2004; Lee et al., 2004; Arnoult et al., 2005; Chen et al., 2005). Eight alternatively spliced mRNAs transcribed from the OPA1 gene were reported (Delettre et al., 2001; Olichon et al., 2002; Satoh et al., 2003), resulting in the accumulation of five apparent isoforms of the OPA1 protein (Delettre et al., 2001; Olichon et al., 2003, 2006; Duvezin-Caubet et al., 2006; Ishihara et al., 2006). Several lines of evidence suggest that OPA1 undergoes limited proteolysis. Dissipation of the mitochondrial membrane potential induces a fast and specific proteolytic conversion of larger OPA1 isoforms into smaller isoforms accompanied by a simultaneous fragmentation of mitochondria (Duvezin-Caubet et al., 2006; Ishihara et al., 2006). Proteolysis of OPA1 is observed in patients and in various model systems of human disorders associated with mitochondrial dysfunction (Duvezin-Caubet et al., 2006). We further showed that mitochondrial dysfunction leads to OPA1 processing, inhibition of mitochondrial fusion, and therefore to segregation of damaged mitochondria from the network of intact mitochondria (Duvezin-Caubet et al., 2006).
Because OPA1 processing has a key role in regulating mitochondrial morphology, it is of major interest to identify this protease. Apparently contradicting results have been reported in this respect, because PARL, a mitochondrial rhomboid protease, and paraplegin, a subunit of the _m_-AAA protease, were proposed to be involved in cleaving OPA1 (Cipolat et al., 2006; Ishihara et al., 2006). PARL seems to be an obvious candidate, because its orthologue, Pcp1, was shown to process the orthologue of OPA1, Mgm1, in baker's yeast (Herlan et al., 2003, 2004; McQuibban et al., 2003; Sesaki et al., 2003). Deletion of Parl in Drosophila led to fragmentation of mitochondria (McQuibban et al., 2006). Moreover, PARL is a critical regulator of OPA1-dependent cristae remodeling during apoptosis, a process that is accompanied by the accumulation of small amounts of a soluble form of OPA1 in the intermembrane space (Cipolat et al., 2006; Frezza et al., 2006). Other observations, however, challenged the requirement of PARL for OPA1 processing. Deletion of Parl in mouse did not have an obvious effect on mitochondrial morphology (Cipolat et al., 2006). Further, cleavage of OPA1 has recently been linked to the hetero-oligomeric _m_-AAA protease (Ishihara et al., 2006), an ATP-dependent metalloprotease in the inner membrane of mitochondria (Atorino et al., 2003). Ishihara and colleagues observed a modestly impaired OPA1 processing in human cells upon down-regulation of paraplegin, a subunit of the _m_-AAA protease, but not when PARL was down-regulated. Notably, deletion of paraplegin is causative for axonal degeneration in hereditary spastic paraplegia (Casari et al., 1998), and it leads to the accumulation of aberrant mitochondria in a paraplegin-deficient mouse model (Ferreirinha et al., 2004). Impaired processing of OPA1 may therefore be of pathogenic relevance for this disease.
To identify proteases capable of cleaving OPA1, we have reconstituted OPA1 processing in yeast, and, in parallel, we have analyzed this process in PARL and paraplegin-deficient mammalian cell lines. Our results demonstrate that PARL can functionally substitute for the yeast rhomboid Pcp1, consistent with a previous report (McQuibban et al., 2003), but it does not affect OPA1 processing. Rather, yeast and mammalian _m_-AAA proteases mediate OPA1 cleavage when expressed in yeast. Different efficiencies of OPA1 cleavage by homo-oligomeric and hetero-oligomeric, paraplegin-containing _m_-AAA proteases point to different substrate specificities of these proteolytic complexes and rationalize efficient OPA1 processing in paraplegin-deficient cell lines.
MATERIALS AND METHODS
Plasmids
OPA1 splice variants 4, 7, and 8 were amplified from human cDNA and cloned into pYES2 (Invitrogen, Carlsbad, CA). All sequences were verified by DNA sequencing. OPA1 splice variant 8 encoded the reported A210V polymorphism (Yao et al., 2006). PARL splice variant 1 was amplified from human cDNA and cloned into pES425#1. Human AFG3L2 together with the ADH1 promoter was subcloned from pRS316-hAFG3L2 (Atorino et al., 2003) into pRS314 (Sikorski and Hieter, 1989). PCP1 was amplified from genomic DNA from Saccharomyces cerevisiae and cloned in pYES2 (Invitrogen). Other plasmids are described in Supplemental Table 1.
Yeast Strains and Growth Conditions
For complementation analysis, the PCP1/Δ_pcp1_ strain (EUROSCARF, Frankfurt, Germany) was transformed with pYES2-PCP1 or pES425-PARL. After sporulation and dissection of tetrads, haploid strains that retained mitochondrial DNA were used for further analysis. Screening of proteases was performed using deletion strains and corresponding wild-type (WT) cells (BY4742) from BioCat (Open Biosystems, Huntsville, AL) and transformed with pYES2-OPA1 plasmids. Saccharomyces Genome Database (SGD) nomenclature is used throughout. Haploid W303 was used as wild-type control elsewhere. All strains used in this study are described in Supplemental Table 2. Cells were grown under standard conditions (Guthrie and Fink, 1991). Strains expressing OPA1 were cultured using 2% galactose and 0.5% glucose unless indicated differently.
Mammalian Cell Culture
HeLa cells, immortalized mouse embryonic fibroblasts from control (Parl+/+) and knockout mice (_Parl_−/−) were used for this study (Cipolat et al., 2006). In addition, primary dermal fibroblasts were isolated from newborn wild-type and _Spg7_−/− mice (Ferreirinha et al., 2004). Mammalian cells were cultured in Dulbecco modified Eagle's medium (DMEM) containing 4.5 g/l glucose and 2 mM l-glutamine supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. Cell culture reagents were obtained from PAA Laboratories (Cölbe, Germany), and carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was purchased from Sigma Chemie (Deisenhofen, Germany). Mitochondria were prepared from HeLa cells by differential centrifugation as described previously (Duvezin-Caubet et al., 2006). Transient transfections of HeLa cells were performed using FuGENE HD (Roche Diagnostics, Basel, Switzerland).
Antibodies
Anti-OPA1 antibodies were raised against a C-terminal peptide of OPA1 as described in Duvezin-Caubet et al. (2006) and from BD Biosciences (San Jose, CA). Anti-Pcp1 antibodies were affinity purified from a rabbit polyclonal serum raised against the C terminus of Pcp1 by using the synthetic peptide CEKQRQRRLQAAGRWF (Pineda Antikörper-Service, Berlin, Germany).
Immunoprecipitation of OPA1
Cells were lysed in lysis buffer (0.5% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 5 mM EDTA, supplemented with complete protease inhibitor cocktail; Roche Diagnostics), and then they were subjected to standard immunoprecipitation with OPA1 antibodies coupled to protein A-Sepharose CL-4B beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Mass Spectrometry
Cut gel slices from SDS-polyacrylamide gel electrophoresis (PAGE) were washed two times with water and two times with 40 mM ammonium bicarbonate for 30 min each. After 2 × 5-min treatment with 50% acetonitrile, trypsin (sequencing grade modified; Promega, Madison, WI) was added, and proteins were digested overnight in 40 mM ammonium bicarbonate at 37°C while shaking (650 rpm). Peptides were directly analyzed by nano-electrospray ionization (ESI)-liquid chromatography (LC)-tandem mass spectrometry (MS/MS) for which they were separated on a C18 reversed phase column (75 μm i.d. × 15 cm, packed with C18 PepMap, 3 μm, 100 Å; LC Packings, Sunnyvale, CA) via a linear acetonitrile gradient; MS and MS/MS spectra were recorded on a QSTAR XL mass spectrometer (Applied Biosystems, Foster City, CA) and analyzed via the Mascot Software (Matrix Science, London, United Kingdom) by using the National Center for Biotechnology Information non-redundant database.
Fluorescence Microscopy
After transformation with the plasmid pVT100U-mtGFP expressing mitochondrially targeted green fluorescent protein (GFP) (Westermann and Neupert, 2000), strains were analyzed by standard fluorescence microscopy on an Axioplan 2 microscope (Carl Zeiss MicroImaging, Jena, Germany) with a numerical aperture 1.3 oil immersion objective (100×; model Plan-Neofluar; Carl Zeiss MicroImaging,) and a charge-coupled device camera 1.1.0 (Diagnostic Instruments, Sterling Heights, MI) at room temperature by using MetaView 3.6a software (Molecular Devices, Sunnyvale, CA). Quantification of cells with different morphology phenotypes was performed without knowing the identity of the strain by counting 100 cells each time of minimum three samples of each strain.
RESULTS
Characterization of OPA1 Isoforms in HeLa Cells
At least five distinct OPA1 isoforms are present in HeLa cells: the two high-molecular-weight OPA1 isoforms L1 and L2 and the three isoforms of lower molecular mass S3, S4, and S5 (Duvezin-Caubet et al., 2006). To examine to which extent they are generated by alternative splicing or limited proteolysis, we isolated mitochondria from HeLa cells, and we purified the different OPA1 species by immunoprecipitation (Figure 1, A and B). The antibodies used were directed against a C-terminal peptide of OPA1 present in all OPA1 isoforms. The various species were resolved by SDS-PAGE and analyzed by LC-MS/MS spectrometry (Figure 1C). The most N-terminal tryptic peptide found in L1 and L2 was located a few amino acid residues C-terminal to the cleavage site of the mitochondrial processing peptidase (MPP) between N87 and F88 (Ishihara et al., 2006). Specifically for L1 and L2 a number of peptides were found located C-terminally to the MPP cleavage site (Figure 1C). These peptides were derived from exon 3 and from alternatively spliced exons 4 and 5b but not from exon 4b. Small OPA1 isoforms (S3–S5) were lacking increasingly more of these peptides from the N terminus (Figure 1C). All S-forms lacked the transmembrane segment TM1. Besides that, numerous peptides corresponding to exon 6–29, which are common to all splice variants, were identified in all isoforms (Figure 1C; data not shown).
Figure 1.
Characterization of OPA1 isoforms by mass spectrometry. (A) Immunoprecipitation of OPA1 from isolated HeLa mitochondria (15 mg) by using anti-OPA1 antibodies. Equal fractions (0.25%) of total mitochondria (T), supernatant (S), and pellet (P) of solubilized mitochondria after clarifying spin, flow through (FT) and 1% of the elution fraction (E) were analyzed by SDS-PAGE and immunoblotting with anti-OPA1 antibodies. (B) Coomassie-stained bands of immunoprecipitated OPA1 isoforms. Each band was separately cut and used for ESI-LC-MS/MS. (C) Alignment of N termini of the eight OPA1 splice variants. MPP cleavage site N-terminal to F88 (Ishihara et al., 2006) is indicated. Vertical dotted lines indicate the exon boundaries. Gray highlighted areas represent hydrophobic stretches called TM1, TM2a, and TM2b (Herlan et al., 2004). Boxes represent peptides found by ESI-LC-MS/MS analysis of any of the different immunoprecipitated OPA1 isoforms. The absence (−) or presence (+) of each peptide in different OPA1 isoforms (L1–S5) is indicated below the alignment. The confidence of peptide identification is indicated by +++ (excellent), ++ (very good), or + (good) based on the number of identifications in separate ESI-LC-MS/MS runs and on the ion score. (D) OPA1 splice variant 7 (Sp.7) was expressed in wild-type yeast cells (left) or was overexpressed in HeLa cells (right), and total cell extracts thereof or mitochondria isolated from HeLa cells (M) were analyzed by Western blotting with anti-OPA1 antibodies. Endogenous OPA1 isoforms (endo) in HeLa cell extracts are shown for comparison. Bands are labeled according to apparent corresponding size of OPA1 isoforms in HeLa mitochondria (L1, L2, S3, S4, and S5). Precursor protein (p) and a degradation product (d) are indicated.
To assign the isoforms to individual splice variants, we took into account the presence or absence of peptides in the purified isoforms that cover or overlap exons affected by alternative splicing. For L1, we obtained peptides that all could be derived from splice variant 7 but not from a different splice variant alone (Figure 1). The calculated molecular weights of each splice variant (Sp.) when cleaved by MPP are as follows: 101.4 kDa (Sp.1), 97.3 kDa (Sp.2), 99.2 kDa (Sp.3), 101.6 kDa (Sp.4), 103.3 kDa (Sp.5), 103.4 kDa (Sp.6), 105.7 kDa (Sp.7), and 107.5 kDa (Sp.8). Thus, because L2 contained the same nine N-terminal peptides as L1, but it was smaller (Figure 1, A and B), it is unlikely to be derived from splice variant 7 or 8. Furthermore, the peptide pattern found for L2 was consistent with splice variant 1 except for one peptide (GLLGELILLQQQIQEHEEEAR) that could theoretically be derived from splice variants 4, 6, 7, or 8 (Figure 1C). However, only the predicted size of MPP-cleaved splice variant 4 is nearly identical to the size of splice variant 1. Therefore, in HeLa cells, L2 most likely represents a mixture of two isoforms derived from splice variants 1 and 4, whereas L1 is derived from splice variant 7 (Figure 1C). These results are consistent with the high level of expression of these variants in HeLa cells (Satoh et al., 2003) and the number and size of bands obtained when splice variants 1, 4, and 7 are expressed in mammalian cells (Ishihara et al., 2006; Olichon et al., 2006). S3 contains peptides representing splice variant 7, whereas S4 contains peptides derived from splice variants 4, 6, 7, or 8, and S5 contains only peptides common to all eight splice variants. In conclusion, these results suggest that S-forms are generated by proteolysis of larger OPA1 isoforms, possibly derived from different splice variants. Still, the expression of more than one splice variant of OPA1 might be required for OPA1 function because expression of single splice variants 4, 7, or 8 was not sufficient to rescue the deletion of mgm1 in baker's yeast (data not shown).
Proteolytic Processing of OPA1 in Yeast
To test whether OPA1 processing can be recapitulated in yeast, we expressed human OPA1 splice variant 7 in yeast. We observed in total five major and a sixth minor OPA1 band (Figure 1D, left). Four bands seem identical in size to OPA1 isoforms L1, S3, S4, and S5 present in HeLa mitochondria. Moreover, we observed an additional degradation product and a large-sized species of very low abundance most likely corresponding to the precursor protein (p). In agreement with our mass spectrometric analysis of OPA1 isoforms in HeLa cells (Figure 1C), L2 did not accumulate in yeast cells expressing the splice variant 7 of OPA1 (Figure 1D, left). Furthermore, upon overexpression of splice variant 7 in HeLa cells, we observed an increase in the formation of L1, S3, and S4 isoforms as well as of a minor band corresponding most likely to the precursor (p) of OPA1 (Figure 1D, right), which is fully consistent with our mass spectrometric analysis and a previous report by Ishihara et al. (2006). Thus, formation of L1, S3, and S4 upon expression of OPA1 splice variant 7 can successfully be reconstituted in yeast mitochondria.
To further corroborate our results, we purified the OPA1 isoforms by immunoprecipitation from total yeast extracts, and we analyzed the different processing products by LC-MS/MS. The peptide patterns obtained were consistent with those obtained for the corresponding isoforms in HeLa cells (data not shown). In particular, the same most N-terminal peptide was found in L1-like isoforms from yeast and HeLa cells, whereas the smaller S-like forms lacked the same N-terminal peptides as the corresponding bands in HeLa cells. Together, these data indicate that OPA1 is processed in a similar manner in yeast and human mitochondria.
Yeast and Mammalian Rhomboid Proteases Are Not Required for OPA1 Processing
To investigate whether rhomboid proteases mediate processing of OPA1 in yeast, we examined OPA1 cleavage in Δ_pcp1_ cells lacking yeast rhomboid and also expressed human PARL in these cells. A heterozygous PCP1/Δ_pcp1_ strain expressing PARL from a plasmid was sporulated, and individual spores were analyzed further. We observed the same pattern of OPA1 isoforms upon expression of OPA1 splice variant 7, irrespective of the presence or absence of Pcp1 or PARL (Figure 2A). This is a puzzling result, because Pcp1 cleaves Mgm1, the yeast homologue of OPA1, and human PARL was shown to restore proteolytic processing of Mgm1 in Δ_pcp1_ cells (McQuibban et al., 2003). Because PARL is not affecting the formation of OPA1 isoforms in yeast cells it is important to provide evidence that PARL is indeed fully functional in the yeast background we used here. We therefore decided to investigate to which extent PARL is taking over the function of Pcp1, and we examined the maintenance of the mitochondrial genome and normal mitochondrial morphology in Δ_pcp1_ cells containing PARL after sporulation. Expression of either PARL or of Pcp1 rescued the growth defects of the Δ_pcp1_ mutant on fermentable and on nonfermentable carbon sources, demonstrating stabilization of mitochondrial DNA (Supplemental Figure 1). The known Pcp1 substrates Mgm1 and Ccp1 (Esser et al., 2002; Herlan et al., 2003; Sesaki et al., 2003) were processed in these cells (Figure 2B), consistent with a previous report (McQuibban et al., 2003). In addition, PARL generated Mgm1 isoforms in the absence of Pcp1 at a ratio compatible with the maintenance of a normal mitochondrial network (Figure 2CD). Thus, PARL can substitute for Pcp1, and it is enzymatically fully active upon expression in yeast. We reasoned that possibly only certain OPA1 splice variants could be cleaved by PARL or Pcp1. Therefore, we also expressed splice variants 4 and 8 of OPA1 in yeast cells containing Pcp1 or PARL. However, the presence or absence of either of the rhomboid proteases did not affect the processing of any of these OPA1 splice variants (Figure 2A). Thus, neither Pcp1 nor PARL affects processing of OPA1 in yeast, suggesting that another protease is responsible for the formation of small isoforms of OPA1.
Figure 2.
Processing of OPA1 in yeast does not depend on rhomboid proteases PARL or Pcp1. (A–D) Functional complementation of Pcp1 by the human mitochondrial rhomboid protease PARL. WT or Δ_pcp1_ (Δ) spores expressing either the human (PARL) or the yeast (PCP1) mitochondrial rhomboid protease were used. (A) Total cell extracts of the indicated strains expressing OPA1 Sp.4, 7, or 8 were analyzed by Western blotting. For splice variant 8, one band was slightly larger than L1 from HeLa mitochondria (data not shown) and was therefore labeled L1′. This is consistent with the larger predicted size of the MPP-cleaved splice variant 8 compared with splice variant 7 forming L1. (B) Processing of the two known substrates of Pcp1 was analyzed by Western blotting of cell extracts of indicated strains. The bands indicated are the large isoform of Mgm1 (l-Mgm1), the small isoform of Mgm1 (s-Mgm1), a degradation fragment of Mgm1 (f; only in Δ_pcp1_), the mature form of Ccp1 (mCcp1), and the intermediate form of Ccp1 (iCcp1; only in Δ_pcp1_). For control, anti-Pcp1 immunodecoration is shown. (C and D) Analysis of mitochondrial morphology of the indicated strains expressing a mitochondrially targeted GFP. (C) Representative images (top, bright field; bottom, fluorescence). (D) Quantification of mitochondrial morphology. Error bars represent the SD (n = 3). (E) Cultured mouse embryonic fibroblasts isolated from Parl+/+ (WT) and _Parl_−/− mice were treated or not with 20 μM CCCP for 30 min. Cell extracts were subjected to Western blotting with anti-OPA1 antibodies. HeLa mitochondria (M) were used as a control.
To substantiate these observations, we examined the requirement of PARL for OPA1 processing in mammalian cells by using murine embryonic fibroblasts derived from a _Parl_−/− knockout mouse (Cipolat et al., 2006). The pattern of OPA1 isoforms was not altered in _Parl_−/− knockout cells compared with wild-type cells (Figure 2E). Thus, PARL is not required for processing OPA1 isoforms during growth of fibroblasts. Moreover, PARL was dispensable for uncoupler-induced processing of OPA1, which occurred both in wild-type and _Parl_−/− cells to a similar extent (Figure 2E).
OPA1 Processing in Yeast Depends on the m-AAA Protease
We screened a series of yeast strains lacking putative or known mitochondrial proteases for the impairment of processing of OPA1 splice variant 7. In agreement with a previous report (Ishihara et al., 2006), small isoforms did not accumulate in cells lacking the yeast _m_-AAA protease subunits Yta10 or Yta12 (Figure 3A). Formation of L1 was not affected consistent with our results that L1 generated upon import into mitochondria by the mitochondrial processing peptidase MPP. In addition, a band most likely representing the precursor form of OPA1 (p) was observed. Similar results were obtained by expressing the OPA1 variants 4 and 8 in these mutants (data not shown). These observations prompted us to examine the role of the human _m_-AAA protease for OPA1 processing in yeast. A complex of human AFG3L2 and paraplegin has been demonstrated to functionally replace the yeast m_-AAA protease (Atorino et al., 2003). We therefore expressed OPA1 splice variant 7 in Δ_yta10_Δ_yta12 cells harboring human AFG3L2 and paraplegin (Figure 3B). In addition to L1 and the precursor form, S3 and S4 accumulated in the presence of these m_-AAA protease subunits. Notably, in contrast to wild-type cells, the OPA1 isoform S5 was not generated in Δ_yta10_Δ_yta12 cells complemented with human AFG3L2 and paraplegin (Figure 3B). Similar-sized bands (L1, S3, and S4) accumulated in HeLa cells upon expression of splice variant 7 (Figure 1D, right) also reported by Ishihara et al. (2006). We also observed a similar dependency on human AFG3L2 and paraplegin for the processing of OPA1 splice variants 4 and 8 (Figure 3B). The three bands (L2, S3, and S4) observed for splice variant 4 in this yeast mutant corresponded to those described by Olichon et al. (2006) when this splice variant was expressed in HeLa cells.
Figure 3.
OPA1 processing depends on the m_-AAA protease. (A) OPA1 splice variant 7 was expressed in yeast strains bearing deletions of putative or known mitochondrial proteases and in WT and analyzed by Western blotting. HeLa mitochondria (M) are shown for comparison. (B) Wild-type and Δ_yta10_Δ_yta12 cells complemented (+) or not (−) with their human orthologues AFG3L2 and paraplegin (L2/PARA) expressing OPA1 Sp.4, 7, or 8. Total cell extracts and HeLa mitochondria (M) were analyzed by Western blotting. (C) OPA1 splice variants 4, 7, and 8 were expressed in Δ_yta10_Δ_yta12_ with (+) and without (−) PARL. Cell lysates were analyzed by Western blotting. (D) Cultured mouse fibroblasts isolated from Spg7+/+ (WT) and _Spg7_−/− mice were treated or not with 20 μM CCCP. Total cell extracts were subjected to Western blotting.
We therefore conclude that the human m_-AAA protease composed of AFG3L2 and paraplegin can restore OPA1 processing in Δ_yta10_Δ_yta12 cells. In contrast, overexpression of PARL in _m_-AAA protease-deficient yeast cells did not lead to the formation of small OPA1 isoforms (Figure 3C), supporting our conclusion that PARL is not required for OPA1 processing.
Processing of OPA1 Can Occur in Paraplegin-deficient Spg7−/− Mice
Our findings are in apparent support of a recent report linking the function of paraplegin to OPA1 cleavage in HeLa cells (Ishihara et al., 2006). However, as only a minor effect of paraplegin down-regulation on OPA1 processing was observed in these experiments, we examined the formation of OPA1 isoforms in fibroblasts isolated from paraplegin-deficient _Spg7_−/− mice (Figure 3D). No difference was observed in the pattern of OPA1 isoforms under steady-state conditions as well as upon dissipation of the mitochondrial membrane potential in mutant cells compared with control fibroblasts (Figure 3D). Thus, processing of OPA1 in mammalian mitochondria can occur independently of paraplegin.
OPA1 Processing by Homo-oligomeric Human m-AAA Protease Complexes in the Absence of Paraplegin
How can these apparent discrepancies be explained? We recently described a homo-oligomeric isozyme of the m_-AAA protease in mammalian mitochondria, which can substitute for the hetero-oligomeric m_-AAA protease composed of paraplegin and AFG3L2 (Koppen et al., 2007). To examine whether homo-oligomeric human AFG3L2 complexes can mediate OPA1 processing, we expressed splice variant 7 of OPA1 in Δ_yta10_Δ_yta12 yeast cells harboring human AFG3L2 or its proteolytically inactive variant AFG3L2E595Q. Processing of OPA1 was restored in Δ_yta10_Δ_yta12 cells by expressing human AFG3L2 (Figure 4) and occurred with similar efficiency as observed upon expression of both human paraplegin and AFG3L2 (Figure 3B). In contrast, expression of AFG3L2E575Q did not promote OPA1 cleavage, demonstrating that processing depends on the proteolytic activity of human AFG3L2 (Figure 4). A similar dependency on human AFG3L2 was also observed for the processing of OPA1 splice variants 4 and 8 (Figure 4). We conclude that homo-oligomeric AFG3L2 complexes are capable of recognizing and cleaving OPA1 in yeast mitochondria in the absence of paraplegin.
Figure 4.
OPA1 processing by homo-oligomeric human m_-AAA protease complexes in the absence of paraplegin. Human OPA1 Sp.4, 7, or 8 were expressed in WT and in Δ_yta10_Δ_yta12 cells harboring the human AFG3L2 or the proteolytically inactive variant AFG3L2E575Q as indicated. Total cell extracts were analyzed by Western blotting. HeLa total cell extract was used as reference.
OPA1 Processing by Homo- and Hetero-oligomeric Murine m-AAA Proteases
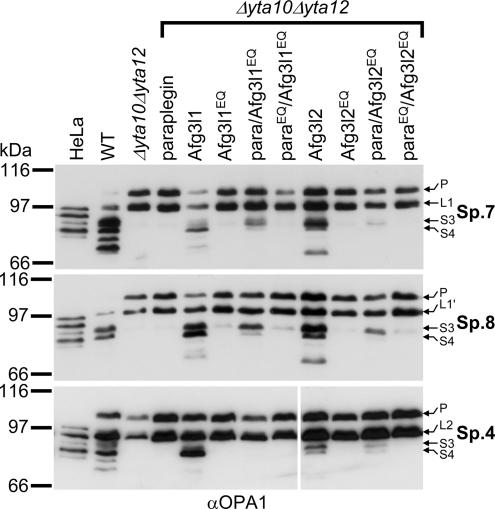
To obtain further evidence in support of the involvement of specific _m_-AAA protease isozymes in OPA1 processing, we analyzed the generation of OPA1 isoforms by murine m_-AAA protease complexes differing in their subunit composition. In addition to homologues of human AFG3L2 and paraplegin, a further subunit, Afg3l1, is expressed in mice (Kremmidiotis et al., 2001). This leads to an even higher number of possible m_-AAA protease complexes with different subunit composition in the inner membrane of murine mitochondria (Koppen et al., 2007). Both homo-oligomeric Afg3l1 and Afg3l2 complexes as well as hetero-oligomeric assemblies of both proteins with paraplegin were shown to be proteolytically active (Koppen et al., 2007). To assess the activity of these complexes toward OPA1, we first expressed the OPA1 splice variant 7 in Δ_yta10_Δ_yta12 yeast cells harboring murine paraplegin, Afg3l1, or Afg3l2 (Figure 5). We did not observe cleavage of OPA1 upon expression of paraplegin alone (Figure 5) consistent with the previous notion that Afg3l1 and Afg3l2 but not paraplegin can form homo-oligomeric, proteolytically active complexes (Koppen et al., 2007). In contrast, expression of each of the murine subunits, Afg3l2 or Afg3l1, in Δ_yta10_Δ_yta12 cells promoted OPA1 processing (Figure 5). Cleavage was abolished when point mutations were introduced in the proteolytic center of Afg3l1 or Afg3l2. Interestingly, expression of Afg3l2 preferentially led to the formation of the S3 isoform, whereas the isoform S4 predominantly accumulated in the presence of Afg3l1 (Figure 5). Furthermore, Afg3l1 and Afg3l2 complexes mediated processing of OPA1 splice variants 4 and 8 in a similar manner (Figure 5). These findings demonstrate that OPA1 splice variants 4, 7, and 8 are recognized and cleaved by homo-oligomeric murine Afg3l1 and Afg3l2 complexes in yeast, and they provide the first evidence for different substrate specificities of _m_-AAA protease complexes composed of different subunits.
Figure 5.
OPA1 processing depends on the subunit composition of the murine m_-AAA protease. OPA1 was expressed in WT; Δ_yta10_Δ_yta12 cells; or Δ_yta10_Δ_yta12_ cells harboring either murine paraplegin (para), Afg3l1, Afg3l2, or their mutant variants paraplegin E575Q (paraEQ), Afg3l1E567Q (Afg3l1EQ), or Afg3l2E574Q (Afg3l2EQ) or combinations of them.
Coexpression of paraplegin with either Afg3l1 or Afg3l2 did not significantly affect OPA1 processing compared with expression of Afg3l1 and Afg3l2 alone (data not shown). To distinguish between the activity of homo-oligomeric and hetero-oligomeric _m_-AAA protease complexes and to examine whether also hetero-oligomeric murine _m_-AAA proteases containing paraplegin can cleave OPA1, we exploited the previous observation that hetero-oligomeric m_-AAA proteases containing both proteolytically active and inactive subunits are capable of mediating protein processing (Arlt et al., 1998; Koppen et al., 2007). We expressed paraplegin or its proteolytically inactive variant in Δ_yta10_Δ_yta12 yeast cells harboring the OPA1 splice variants 7 together with either proteolytically inactive Afg3l1E567Q or Afg3l2E574Q. OPA1 processing, in particular formation of S3, was detectable with both the Afg3l2E567Q/paraplegin complex and the Afg3l1E567Q/paraplegin complex (Figure 5). A similar dependency was observed for splice variant 8 (Figure 5). In contrast, splice variant 4 was processed only to a very minor extent by the Afg3l2E567Q/paraplegin complex but not by the Afg3l1E567Q/paraplegin complex (Figure 5). Because small OPA1 isoforms were not detected when both Afg3l1 and paraplegin or Afg3l2 and paraplegin contained point mutations in their proteolytic centers, the low proteolytic activity observed can be attributed to paraplegin. We conclude that hetero-oligomeric Afg3l1E567Q/paraplegin and Afg3l2E567Q/paraplegin complexes are able to cleave OPA1. However, the processing efficiency by paraplegin-containing _m_-AAA protease isozymes seems to be limited, lower than that of homo-oligomeric Afg3l1 or Afg3l2 complexes, and varies for different splice variants. Together, using yeast as a heterologous reconstituted system, we showed that OPA1 can be processed with variable efficiencies by _m_-AAA protease complexes differing in their subunit composition.
DISCUSSION
Processing of the dynamin-like GTPase OPA1 in the mitochondrial inner membrane is critical for the regulation of mitochondrial morphology (Duvezin-Caubet et al., 2006; Ishihara et al., 2006) and for cristae remodeling during apoptosis (Cipolat et al., 2006; Frezza et al., 2006). In the present study, we have reconstituted OPA1 processing in yeast, to examine the involvement of the rhomboid protease PARL and the paraplegin-containing _m_-AAA protease, which both have been linked to this process in previous studies (Cipolat et al., 2006; Ishihara et al., 2006). The mass spectrometric characterization of OPA1 isoforms not only revealed their formation by alternative splicing and proteolytic processing in HeLa cells but also established yeast as a valid model system for the analysis of OPA1 processing. Using this system, we demonstrate that OPA1 is recognized and cleaved by _m_-AAA protease isozymes but not by PARL in the inner membrane of yeast mitochondria.
OPA1 processing was only modestly affected upon down-regulation of the _m_-AAA protease subunit paraplegin in HeLa cells (Ishihara et al., 2006) and proceeded normally in murine _Spg7_−/− cells. These apparently inconsistent findings can be explained by the activities of homo-oligomeric _m_-AAA protease isozymes in these cells (Koppen et al., 2007). We demonstrate that OPA1 upon expression in yeast is cleaved by homo-oligomeric _m_-AAA protease complexes composed of murine Afg3l1, Afg3l2, or human AFG3L2 subunits. Notably, certain OPA1 processing products are preferentially formed depending on the splice variant analyzed and on the subunit composition of the _m_-AAA protease. In this context, the low activity of paraplegin-containing _m_-AAA protease complexes cannot be neglected, because the expression of _m_-AAA protease subunits varies in different murine tissues (Koppen et al., 2007). Therefore, it is conceivable that hetero-oligomeric forms of _m_-AAA proteases are crucial for OPA1 processing in some tissues and not in others. Tissue-specific differences in the subunit composition of _m_-AAA protease isozymes as well as in the expression of OPA1 isoforms could explain why deficiencies in paraplegin in mouse and human do result in cell type-specific mitochondrial defects. These considerations are relevant for understanding the physiological role of OPA1 cleavage and its potential involvement in the pathogenesis of hereditary spastic paraplegia.
The rhomboid-like protease PARL, in contrast to the _m_-AAA protease, does not cleave OPA1 upon expression in yeast. None of the OPA1 splice variants investigated were converted to smaller isoforms in the presence of proteolytically active PARL, which is able to cleave Mgm1, the yeast orthologue of OPA1 (McQuibban et al., 2003). In agreement with our results in yeast, mitochondrial morphology (Cipolat et al., 2006) and OPA1 processing were unaffected in _Parl_−/− mice. In contrast, a soluble OPA1 isoform generated by PARL cleavage in low amounts was suggested to be responsible for the antiapoptotic effects of OPA1 (Cipolat et al., 2006). The formation of a minor processing product of OPA1 with an antiapoptotic function cannot be ruled out. However, PARL does not control mitochondrial morphology or account for the formation of, at least, the majority of OPA1 isoforms in mammalian cells. Rather, a switch in the protease mediating the processing of OPA1-related GTPases in mitochondria seems to have occurred during evolution.
The intriguing question arises why _m_-AAA proteases rather than PARL mediate OPA1 processing. In view of the different efficiencies of OPA1 processing by _m_-AAA proteases composed of different subunits, it is conceivable that variation in the assembly of _m_-AAA proteases allows adjusting OPA1 processing and thereby mitochondrial dynamics to different needs in different tissues. Moreover, the _m_-AAA protease may play additional roles during OPA1 cleavage that cannot be carried out by PARL. The _m_-AAA protease is known to mediate the ATP-dependent dislocation of proteins from the membrane to allow their complete proteolysis in a hydrophilic environment (Leonhard et al., 2000). Interestingly, the ATP-dependent membrane dislocation of cytochrome c peroxidase by the _m_-AAA protease in yeast was recently found to facilitate maturation by the rhomboid protease Pcp1 (Tatsuta et al., 2007). However, as OPA1 is not recognized and cleaved by PARL in yeast, our results do not favor such a functional interplay between both rhomboid and AAA proteases during OPA1 processing in mammalian mitochondria. In contrast to the situation in yeast, where short forms of the OPA1 orthologue Mgm1 are formed during biogenesis (Herlan et al., 2004), in mammalian cells preexisting large forms of OPA1 can be rapidly converted to small forms, e.g., upon dissipation of the membrane potential across the inner membrane (Duvezin-Caubet et al., 2006). The latter seems to be the consequence of a signaling pathway induced by mitochondrial dysfunction. Possibly, this mechanism has evolved only in higher organisms and depends on the _m_-AAA rather than the rhomboid protease because it allows for fast adaptation of mitochondrial morphology.
Supplementary Material
[Supplemental Material]
ACKNOWLEDGMENTS
We thank Dr. Bart De Strooper (University of Leuven, Belgium) for cell lines, Drs. Carsten Bornhövd (LMU München, Germany) and Doron Rapaport (University of Tübingen, Germany) for plasmids, and Iris Haag for technical assistance. This work was supported by the DFG/SFB 594 (to A.S.R.), Nationales Genomforschungsnetzwerk I MITOP Subproject III (to A.S.R. and W.N.), the Friedrich-Baur-Stiftung (to A.S.R.), the LMU München/FöFoLe (to J.W. and M.Z.), the DFG/SFB 635 (to T.L.), the Muscular Dystrophy Association (to E.I.R.), the United Mitochondrial Disease Foundation (to E.I.R.), and European Union (to T.L. and E.I.R.).
Footnotes
REFERENCES
- Alexander C., et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Arlt H., Steglich G., Perryman R., Guiard B., Neupert W., Langer T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D., Grodet A., Lee Y. J., Estaquier J., Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J. Biol. Chem. 2005;280:35742–35750. doi: 10.1074/jbc.M505970200. [DOI] [PubMed] [Google Scholar]
- Atorino L., Silvestri L., Koppen M., Cassina L., Ballabio A., Marconi R., Langer T., Casari G. Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol. 2003;163:777–787. doi: 10.1083/jcb.200304112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casari G., et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- Chen H., Chomyn A., Chan D. C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Cipolat S., de Brito O. M., Dal Zilio B., Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S., et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Delettre C., Griffoin J. M., Kaplan J., Dollfus H., Lorenz B., Faivre L., Lenaers G., Belenguer P., Hamel C. P. Mutation spectrum and splicing variants in the OPA1 gene. Hum. Genet. 2001;109:584–591. doi: 10.1007/s00439-001-0633-y. [DOI] [PubMed] [Google Scholar]
- Delettre C., et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S., et al. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 2006;281:37972–37979. doi: 10.1074/jbc.M606059200. [DOI] [PubMed] [Google Scholar]
- Esser K., Tursun B., Ingenhoven M., Michaelis G., E. P. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease pcp1. J. Mol. Biol. 2002;323:835–843. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- Ferreirinha F., et al. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Invest. 2004;113:231–242. doi: 10.1172/JCI20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., Catez F., Smith C. L., Youle R. J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Frezza C., et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Griparic L., van der Wel N. N., Orozco I. J., Peters P. J., van der Bliek A. M. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J. Biol. Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–270. [PubMed] [Google Scholar]
- Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 2004;165:167–173. doi: 10.1083/jcb.200403022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhövd C., Neupert W., Reichert A. S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Lee Y. J., Gaume B., Jeong S. Y., Frank S., Nechushtan A., Santel A., Fuller M., Smith C. L., Youle R. J. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M., Metodiev M. D., Casari G., Rugarli E. I., Langer T. Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia. Mol. Cell. Biol. 2007;27:758–767. doi: 10.1128/MCB.01470-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremmidiotis G., Gardner A. E., Settasatian C., Savoia A., Sutherland G. R., Callen D. F. Molecular and functional analyses of the human and mouse genes encoding AFG3L1, a mitochondrial metalloprotease homologous to the human spastic paraplegia protein. Genomics. 2001;76:58–65. doi: 10.1006/geno.2001.6560. [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Jeong S. Y., Karbowski M., Smith C. L., Youle R. J. Roles of the mammalian mitochondrial fission and fusion mediators fis1, drp1, and opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., Langer T. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell. 2000;5:629–638. doi: 10.1016/s1097-2765(00)80242-7. [DOI] [PubMed] [Google Scholar]
- Li Z., Okamoto K., Hayashi Y., Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- McQuibban G. A., Lee J. R., Zheng L., Juusola M., Freeman M. Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr. Biol. 2006;16:982–989. doi: 10.1016/j.cub.2006.03.062. [DOI] [PubMed] [Google Scholar]
- McQuibban G. A., Saurya S., Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- Nakada K., Inoue K., Ono T., Isobe K., Ogura A., Goto Y. I., Nonaka I., Hayashi J. I. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med. 2001;7:934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- Niemann A., Ruegg M., La Padula V., Schenone A., Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J. Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Marshall W. F., Straight A., Murray A., Sedat J. W., Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Olichon A., Elachouri G., Baricault L., Delettre C., Belenguer P., Lenaers G. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 2006;14:682–692. doi: 10.1038/sj.cdd.4402048. [DOI] [PubMed] [Google Scholar]
- Olichon A., et al. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002;523:171–176. doi: 10.1016/s0014-5793(02)02985-x. [DOI] [PubMed] [Google Scholar]
- Ono T., Isobe K., Nakada K., Hayashi J. I. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- Satoh M., Hamamoto T., Seo N., Kagawa Y., Endo H. Differential sublocalization of the dynamin-related protein OPA1 isoforms in mitochondria. Biochem. Biophys. Res. Commun. 2003;300:482–493. doi: 10.1016/s0006-291x(02)02874-7. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S. M., Hobbs A. E., Jensen R. E. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys Res. Commun. 2003;308:276–283. doi: 10.1016/s0006-291x(03)01348-2. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 2007;26:325–335. doi: 10.1038/sj.emboj.7601514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P., Ly C. V., Venken K. J., Koh T. W., Zhou Y., Bellen H. J. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Westermann B., Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Yao W., Jiao X., Hejtmancik J. F., Leske M. C., Hennis A., Nemesure B. Evaluation of the association between OPA1 polymorphisms and primary open-angle glaucoma in Barbados families. Mol. Vis. 2006;12:649–654. [PubMed] [Google Scholar]
- Zuchner S., et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]