Ler and H-NS, Regulators Controlling Expression of the Long Polar Fimbriae of Escherichia coli O157:H7 (original) (raw)
Abstract
Enterohemorrhagic Escherichia coli (EHEC) serotype O157:H7 colonizes the human intestine and is responsible for diarrheal outbreaks worldwide. Previously we showed that EHEC produces long polar fimbriae (LPF) and that maximum expression is observed during the exponential phase of growth at 37°C and pH 6.5. In this study, we analyzed the roles of several regulators in the expression of LPF using the β-galactosidase reporter system, and we found that H-NS functions as a transcriptional silencer while Ler functions as an antisilencer of LPF expression. Interestingly, deletion of the hns and ler genes in EHEC caused constitutive expression of the fusion reporter protein. Semiquantitative reverse transcription (RT)-PCR was also used to analyze LPF expression in the EHEC ler or hns mutant strain. The hns mutant exhibited an increase in lpf mRNA expression, while expression in the ler mutant was decreased, compared to that in the wild-type strain. Using primer extension analysis, we identified two potential transcriptional start sites within the regulatory region of lpf and located consensus hexamers of −10 (CAAGAT) and −35 (TTCAAA), which are commonly found in σ70-dependent promoters. Further, we determined whether H-NS and Ler interact directly with the lpf promoter region by using purified His-tagged proteins and electrophoretic mobility shift assays. Our data are the first to show direct binding interactions between the H-NS and Ler proteins within the regulatory sequence of the lpf operon. Based on the electrophoretic mobility shift assay, RT-PCR, primer extension, and β-galactosidase assay results, we concluded that the E. coli O157:H7 lpf operon possesses a promoter dependent on σ70, that H-NS binds to the regulatory sequence of lpfA and “silences” the transcription of lpf, and that Ler binds to the regulatory sequence and inhibits the action of the H-NS protein.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a serious food-borne pathogen causing diarrhea that is often bloody and accompanied by severe abdominal cramps and can result in a life-threatening condition known as the hemolytic-uremic syndrome (reviewed in reference 33). The organism can be found living in the intestines of healthy cattle, and eating contaminated meat, especially ground beef, has resulted in multiple outbreaks worldwide (23). Recent E. coli O157:H7 outbreaks have drawn attention to food-borne illnesses, and though official sources were saying that the overall number of cases is on the decline in the United States, consumption of produce, particularly leafy vegetables, is becoming increasingly associated with human infections. This alternate source of infection has set up new challenges for the scientific community in trying to identify novel determinants and regulatory mechanisms implicated in the colonization, survival, and/or pathogenic processes (9, 10, 40).
During the infectious process, EHEC adheres to the intestinal epithelium, where it produces Shiga toxins responsible for the hemorrhagic symptoms. Adhesion of E. coli O157:H7 to enterocytes induces the formation of the attaching-and-effacing (A/E) lesion (reviewed in reference (49). The A/E phenotype is mainly conferred by the locus of enterocyte effacement (LEE), which encompasses 41 genes, encoding structural components of a type III secretion apparatus, including translocator and secreted effector proteins, an adhesin (intimin), and the intimin receptor, Tir (reviewed in reference 47). Previous studies with EHEC and enteropathogenic E. coli (EPEC) have shown that the expression of LEE-encoded virulence factors is regulated by a complex assortment of environmental cues and a variety of regulatory elements encoded inside and outside the LEE (reviewed in reference 29). Various regulators outside the LEE have been characterized for EHEC and EPEC strains, including H-NS, a histone-like protein acting as a transcriptional silencer; IHF, the integrated host factor acting positively; Fis and Hha, two other nucleoproteins acting positively and negatively, respectively; and QseA and QseD, activating the expression of the LEE in response to quorum sensing (4, 21, 35, 37, 42). While the regulatory networks controlling expression of LEE-encoded proteins have been quickly elucidated, the roles of these regulators in the expression of other E. coli O157:H7 factors associated with the colonization process are still unclear.
Histone-like DNA binding proteins, such as H-NS and IHF, in association with topoisomerases, play important roles in the maintenance of bacterial nucleoid organization (13). H-NS, a small (136 amino acids), relatively neutral protein, functions as a homodimer in binding DNA, showing preference for curved double-stranded DNA and becoming nucleation sites, where the protein polymerizes along DNA (reviewed in references 14 and 15). H-NS has been implicated as a transcriptional repressor for a diverse array of genes, particularly those involved in environmental adaptation or virulence, via a preferential interaction with intrinsically curved DNA (reviewed in references 14 and 15). In the case of EHEC and EPEC strains, H-NS plays an important role in the “silencing” of genes located in the different operons of the LEE (LEE1 to -5), and it is responsive to multiple environmental signals and regulatory proteins (29). It has been shown that LEE1, encoding the regulatory protein Ler, is inhibited by H-NS at 27°C and activated at 37°C (50). Then, in a cascade fashion, Ler activates transcription at the other LEE operons (LEE2, LEE3, LEE4, and LEE5) (29). It has also been shown that H-NS binds directly to the LEE1, LEE2, and LEE3 regulatory regions (50), whereas Ler binds directly to the LEE2 and LEE5 regulatory regions (22, 43). Thus, both of these regulatory proteins act directly on the LEE.
It is evident that certain E. coli strains encode additional H-NS-like proteins, such as Ler in the cases of EPEC and EHEC (30). The Ler protein is particularly interesting, since it acts as an anti-H-NS factor to activate the expression of other genes on the LEE island (6, 30). The predicted 15.1-kDa Ler protein exhibits amino acid sequence similarity with the H-NS family of DNA binding proteins and shows greater similarity to the C terminus of H-NS, predicted to be a DNA binding domain (43). As indicated above, H-NS silences the expression of several LEE genes, while Ler induces the expression of these genes by counteracting the H-NS-mediated repression (6, 22, 50). In addition, it has been reported that Ler regulates the expression of proteins encoded outside the LEE and which are not essential for A/E lesion formation, including EspC in EPEC or “long fine” fimbriae in EHEC (17).
Very little is known about fimbria expression and regulation in EHEC strains. E. coli O157:H7 contains two nonidentical lpf loci homologous to the long polar fimbriae of Salmonella enterica serovar Typhimurium (reviewed in reference 49). Expression of the E. coli O157:H7 lpf operon 1 (lpf1) in E. coli K-12 has been linked to increase adherence to tissue culture cells and has been associated with the appearance of peritrichous long fimbriae (46). Further, E. coli O157:H7 strains harboring mutations in one or both of the lpf loci have diminished colonization abilities in swine and sheep animal models (24), and these mutations also influence E. coli O157:H7 human intestinal tissue tropism (18). Recently we have investigated the environmental cues that promote expression of lpf1 genes and the role of E. coli O157:H7 long polar fimbriae (LPF) in intestinal colonization of lambs. Our results indicated that expression of lpf1 is regulated in response to growth phase, temperature, and pH, while in vivo data support the impact that LPF have on the ability of E. coli O157:H7 to persist in the intestines of infected animals (48). However, clarification of the connection between regulatory pathways acting on the lpf1 loci in response to environmental cues is important as a prerequisite to fully understanding the pathogenesis of EHEC O157:H7. In the current study, we investigated whether E. coli O157:H7 LPF 1 expression is controlled positively or negatively by global regulators, which then can act directly or as coregulators, in response to environmental conditions of growth.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Strains were routinely grown in Luria-Bertani broth (27) or Dulbecco's minimal Eagle medium (D-MEM) (Gibco/Invitrogen) at 37°C. Additional conditions used to grow bacteria in D-MEM were as follows: temperature (37°C) and pH (adjusting at pH 6.5 by buffering with morpolinepropanesulfonic acid MOPS at a concentration of 0.1 M). Antibiotics were added to media at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; streptomycin, 100 μg/ml; tetracycline, 12.5 μg/ml; and nalidixic acid, 15 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli strains | ||
| 86-24 | E. coli O157:H7, Smr Nalr | A. G. Torres stock collection; 44 |
| EDL933 | Prototype E. coli O157:H7 | A. G. Torres stock collection |
| AC425 | EDL933 hns::Tn_10_, Tcr | J. B. Kaper |
| CB49 | EDL933 ler::kan, Kmr | J. B. Kaper |
| SDP01 | EDL933 hns::Tn_10 ler_::kan, Tcr Kmr | This study |
| MC4100 | E. coli K-12 | 7 |
| K5185 | MC4100 ihfA::Tn_10_, Tcr | 20 |
| CSH56 | MC4100 hns::kan, Kmr | 8, 51 |
| JMH73 | MC4100 fis::kan Kmr | 8 |
| DL844 | MC4100 lrp::Tn_10_, Tcr | 11 |
| BL21/pLys21 | F−ompT (lon) _hsdS_B(rB− mB−) gal dcm (λDE3), Cmr | Invitrogen |
| Plasmids | ||
| pRS551 | Protein fusion vector, Apr Kmr | 39 |
| pPLPFA | pRS551 with EcoRI/BamHI 1,164-bp lpf promoter region | 46 |
| pT6Ler | pMPM-T6 derivative expressing Ler-His6 under control of arabinose-inducible promoter, Tcr | 1 |
| pT6HNS | pMPM-T6 derivative expressing H-NS-His6 under control of arabinose-inducible promoter, Tcr | 1 |
| pCVD442/ler::kan | ler::kan in suicide vector pCVD442, Apr Kmr, sucrose sensitive | This study |
Recombinant DNA techniques.
Standard methods were used to perform plasmid purification, PCR, ligation, restriction digests, transformation, and gel electrophoresis (36). All oligonucleotide primers are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Primer name | Sequence | Use | PCR product size (bp) |
|---|---|---|---|
| LER NCO | 5′-CTTCTTCAGTGTCCTTCACAAG-3′ | Amplification of structural region of Ler | 400 |
| LERXHO | 5′-GAATATGGAAACTAATTCACA-3′ | Amplification of structural region of Ler | 400 |
| LERRDRB | 5′-GTGAGATAACGTTTATCTTAC-3′ | Amplification of regulatory region of Ler | 500 |
| ORF1-A | 5′-CTGGCTGTAGCTTATGTCCG-3′ | Amplification of regulatory region of Ler | 500 |
| 16SKI2232R | 5′-GCCCAGATGGGATTAGCTAAGT-3′ | Amplification of 16S rRNA | 780 |
| 16SL12iORF | 5′-GGAAAGTTCTGTGGATGTCAAG-3′ | Amplification of 16S rRNA | 780 |
| LPFA21R | 5′-TCCGGCAAATTCGAACTTTTTC-3′ | Amplification of lpfA gene | |
| PLPFA262F | 5′-TTTGCGTGGTGAATTCTTTTG-3′ | Amplification of short sequence from lpfA promoter | 262 |
| 520LPFAF | 5′-AGCATTTGTGGATCCATTATCG-3′ | Amplification of long sequence from lpfA promoter | 520 |
| LPFA1256AR | 5′-GTTAAATTTACAGGCGAGATC-3′ | Amplification of full-length lpfA promoter region | 1,335 |
| 1160LPFAF | 5′-GGTCGGTCTGGCGGATGGCGC-3′ | Amplification of full-length lpfA promoter region | 1,335 |
| LPFA1313R | 5′-AATTTAACCGTGCCGTCA-3′ | Primer extension analysis |
Construction of EHEC O157:H7 ler hns double mutant.
The EHEC strain EDL933, defective in ler and hns expression, was constructed by allelic marker exchange as follows. The ler::kan gene from strain CB49 (EDL933 ler::kan) was amplified by PCR with Red Taq polymerase (Sigma) using the primer pair 5ERIC (5′-TTCGCTTACCCCAATCACTTAC-3′) and 3CESAB (5′-TGGCTTTTAGATCTTGCTGGAC-3′) and cloned as a blunted end fragment into the suicide vector pCVD442 (12). pCVD442/ler::kan was introduced into EHEC strain AC425 (EDL933 hns::Tn_10_) by conjugation using the donor strain SM10 (λ pir) (38). Colonies resistant to kanamycin, tetracycline, and sucrose were tested for ampicillin sensitivity. The mutant strain SDP001 (AC425 ler::kan) was further selected as described previously (46), and the presence of the kan cassette within the chromosomal ler gene of SDP001 was confirmed by PCR.
β-Galactosidase assays and statistical analysis.
The cultures were diluted 1:10 in Z buffer (Na2HPO4 [0.06 M], NaH2PO4 [0.04 M], KCl [0.01 M], MgSO4 [0.001 M], and β-mercaptoethanol [0.05 M]) at the different conditions tested and assayed for activity using _o_-nitrophenyl-β-d-galactopyranoside as the substrate as previously described (31). β-Galactosidase specific activity was calculated in accordance with the total proteins of each sample tested. The enzymatic activity units are presented as μg/μl of enzyme/mg of protein. The results of the β-galactosidase assays were analyzed with a paired Student t test.
RNA isolation and RT-PCR.
Bacterial cultures grown in Luria-Bertani broth were diluted in D-MEM and grown at 37°C to either mid- or late exponential phase (optical density at 600 nm [OD600] of 0.6 or 1.2, respectively). Cultures were stabilized with RNAProtect bacteria reagent (QIAGEN, Valencia, CA). Bacteria were harvested by centrifugation at 4°C, resuspended in RNeasy lysis buffer (QIAGEN), and then lysed. RNA was purified using RNeasy columns (QIAGEN) and DNase treated (Ambion, Austin, TX), RNA was quantified and qualitatively analyzed on agarose gels, then 8 μg of total RNA was used for cDNA synthesis using the SuperScript First-Strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The resulting cDNA was utilized for regular PCR with gene-specific primers to amplify lpfA (520 bp; primers 520LPFAF and LPFA1256AR) and 16S (780 bp; primers 16SKI2232R and 16SL12iORF) (Table 2). In addition, a positive control with genomic DNA and a negative control with no reverse transcriptase added were used.
Primer extension and sequencing ladder.
Primer extension analysis was performed as described previously (2, 28). Briefly, primer LPFA1313R, located 69 bp downstream of the lpfA ATG (Table 2), was end labeled using [γ-32P]ATP. A total of 15 μg of RNA from strains EDL933 and 86-24, isolated as described above, was incubated with the end-labeled primer at 90°C for 3 min in 0.2 M NaCl-0.3 M Tris-HCl (pH 8.0), slowly cooling down the reaction to reach 44°C. The mRNA was reverse transcribed using avian myeloblastosis virus reverse transcriptase (Boehringer-Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. The resultant cDNA was treated with RNase H, precipitated, run on an 8% polyacrylamide-7 M urea gel, and visualized by autoradiography. A sequencing ladder was run adjacent to the primer extension reaction. This sequencing ladder was generated with the same primer utilized for the primer extension, following the protocol previously described (28) and using the Sequenase version 2.0 DNA sequencing kit (USB, Cleveland, OH) according to the manufacturer's instructions.
Prediction of lpf promoter and search for motifs.
The bacterial promoter recognition program BPROM (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) was used to predict the location of −10 and −35 hexamers, Shine-Dalgarno sequence, and putative protein binding motifs located within the lpf promoter region.
Expression and purification of His-tagged H-NS and Ler proteins.
The H-NS-His6 or Ler-His6 proteins were expressed and purified as described previously (1). Briefly, E. coli BL21/pLys21 harboring the pT6HNS or pT6Ler plasmid, expressing H-NS-His6 or Ler-His6, respectively, was grown to mid-logarithmic phase at 37°C. l-(+)-Arabinose (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 0.1%, and the bacteria were further incubated for 4 h at 30°C. Cells were then pelleted by centrifugation at 4°C, resuspended in urea buffer (pH 8.0), and disrupted by sonication. The suspension was centrifuged at 4°C, and the supernatant was applied to a HiTrap Ni2+-chelating column (ProBond, Invitrogen). Proteins were eluted with a pH gradient (pH 8.0 to 4.5) of urea buffer. Fractions containing purified H-NS-His6 or Ler-His6 were selected based on sodium dodecyl sulfate- polyacrylamide gel electrophoresis analysis. The fractions were loaded into a membrane tubing (molecular weight cutoff, 6,000 to 8,000; Spectrapor; Spectrum Laboratories, Rancho Dominguez, CA), and gradually dialyzed at 4°C in a buffer containing different amounts of urea (4, 1, and 0.2 M), which was changed every hour. The final dialysis was done in storage buffer, and aliquots of the purified proteins were stored at −70°C. Protein concentrations were determined by using a Bradford protein assay (Bio-Rad, Hercules, CA).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as follows. Approximately 500-ng samples of PCR-generated DNA fragments corresponding to the different fragments of the lpf promoter region sequences were mixed with increasing concentrations of purified Ler-His6 or H-NS-His6 protein in a buffer containing 11.7 mM Tris-HCl (pH 7.5), 0.975 mM EDTA, 78 mM NaCl, 9.75 mM 2-mercaptoethanol, 0.975 mM dithiothreitol, and 6.5% glycerol. The reaction mixtures were incubated for 20 min at room temperature and then separated by either electrophoresis in 1.4% agarose gels in 0.5× Tris-borate-EDTA buffer at 4°C or in 6% polyacrylamide gels in 1.0× Tris-borate-EDTA buffer at room temperature. The DNA bands were stained with ethidium bromide and visualized with a UV transilluminator. Fragments containing either the ler structural region or the promoter region from LEE were used as a negative or positive control, respectively, when evaluating H-NS-DNA or Ler-DNA interactions, as previously described (1, 19). 16S RNA was also used as a negative control to evaluate protein-DNA interactions.
RESULTS
The H-NS protein but not other global regulators controls expression of lpf.
We recently demonstrated that the expression of lpf is observed in response to environmental signals, such as temperature (37°C) and pH (pH 6.5), and maximal expression occurs during the late exponential phase of growth (48). However, we did not know what regulatory proteins in E. coli O157:H7 are associated with the transcriptional regulation of these lpf genes. With that question in mind, we selected a group of global regulators known to either control expression of virulence factors in E. coli O157:H7 and other diarrheagenic E. coli strains (IHF, Fis, and H-NS) or to regulate expression of fimbriae in E. coli (Lrp) and tested whether they affect the expression of lpf. Lrp (leucine-responsive regulatory protein) has not been shown to regulate virulence factors in EHEC; however, Lrp is a global transcriptional regulator of metabolism in E. coli which affects the expression of a number of operons involved in amino acid biosynthesis and degradation, nutrient transport, and formation of fimbriae, especially those belonging to the P-regulatory family in other E. coli pathotypes (11). The plasmid pPLPFA (46), containing the fusion lpfAp::lacZ, was introduced into E. coli K-12 strain MC4100 or into the corresponding isogenic ihf, fis, lrp, and hns mutants strains, and the specific β-galactosidase activities were determined at different stages during exponential growth phase. We did not observe differences in β-galactosidase activity while the fusion gene was expressed in the ihf, fis, or lrp mutant compared to the results obtained with the wild-type strain, MC4100 (data not shown). In contrast, β-galactosidase activity was enhanced when the reporter pPLFA plasmid was expressed in the hns mutant strain. While expression of lpfA in the hns mutant was similar to that in the wild-type strain during early phases of growth (e.g., OD600 = 0.3 and 0.6), β-galactosidase activity was induced 3.2-fold over that of the wild type when the hns mutant strain reached the late exponential phase of growth (OD600 = 0.9) (data not shown). Our data indicated that not all the global regulators have an effect on lpf expression and suggested that H-NS is silencing lpf in the E. coli K-12 background.
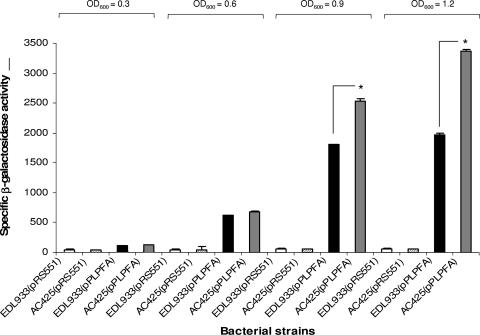
To confirm this initial observation, we introduced our reporter plasmid into the EHEC strains EDL933 (wild type) and AC425 (hns mutant), and the β-galactosidase activity was determined during the exponential phase of growth (Fig. 1). No major differences in expression were observed between the wild-type and hns mutant at early stages of growth; however, expression of lpf was enhanced 1.4- or 1.7-fold when the hns mutant reached an OD600 of 0.9 or 1.2, respectively. These data confirmed that maximum expression of lpf genes occurred at the late exponential phase of E. coli O157:H7 growth and reinforced the observation indicating that H-NS might be acting as a regulator of lpf expression.
FIG. 1.
The H-NS protein “silences” the expression of the lpf operon. (A) The specific β-galactosidase activities were determined using the lpfAp::lacZ fusion in the E. coli O157:H7 strains EDL933 and AC425 (Δ_hns_). The strains were grown with shaking in D-MEM at 37°C, and samples were assayed at OD600s of 0.3, 0.6, 0.9, and 1.2. The asterisks indicate P values of <0.001.
Ler positively regulates expression of lpf in E. coli O157:H7 and acts as an antisilencer.
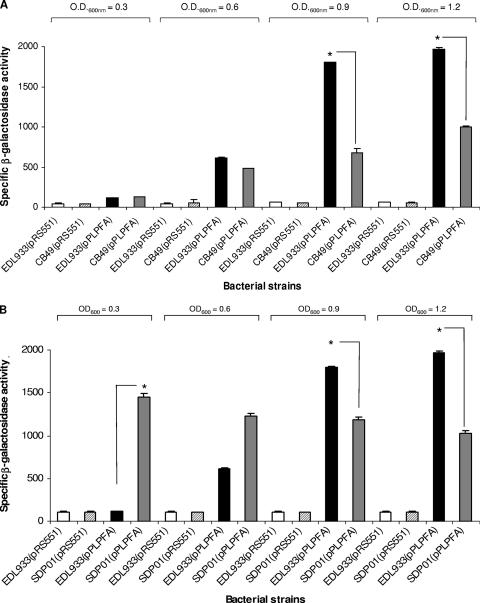
In addition to H-NS, EHEC O157:H7 strains possess Ler, an H-NS-like protein which functions as a positive transcriptional regulator encoded on the LEE pathogenicity island (PAI) (30). Ler is the master regulator of LEE-encoded genes; however, it has been proposed that this protein can regulate the expression of proteins encoded outside the LEE PAI of EHEC and EPEC (17). Because one of the phenotypes previously observed with the EHEC ler mutant involves altered adherence patterns and the expression of long “fine” fimbriae (LFF), we decided to test whether LPF expression is controlled by the Ler protein. Therefore, expression of the lpfAp::lacZ fusion was monitored in EHEC strains EDL933 (wild type) and CB49 (ler mutant). As shown in Fig. 2A, β-galactosidase activity was determined throughout the exponential phase of growth, and while no significant differences in expression between the wild type and the ler mutant were observed at early stages of growth, a significant reduction in enzymatic activity was observed at late stages of growth. β-Galactosidase activity was reduced 2.7- or 2.0-fold for the ler mutant compared with that for the wild-type strain when the cultures reached an OD600 of 0.9 or 1.2, respectively. Overall, our results confirmed again that expression of lpf genes occurs at late exponential phases of growth and suggest that Ler is a positive transcriptional regulator for lpf expression.
FIG. 2.
Ler positively regulates the expression of the lpf operon and acts as an antisilencer of H-NS. (A) The specific β-galactosidase activity was determined using the lpfAp::lacZ fusion in the E. coli O157:H7 strains EDL933 and CB49 (ler mutant strain). The strains were grown with shaking in D-MEM at 37°C, and the β-galactosidase activity was determined at different OD600s. The asterisks indicate P values of <0.001. (B) The plasmid pPLPFA, containing the lpfAp::lacZ fusion, was introduced into the EHEC strain EDL933 and the double mutant SDP01 (EDL933 Δler Δhns). The strains were grown with shaking in D-MEM at 37°C, and the specific β-galactosidase activities were determined at OD600s of 0.3, 0.6, 0.9, and 1.2. The asterisks indicate P values of <0.001.
Previous studies have reported that while H-NS commonly acts as a transcriptional silencer in A/E pathogens, Ler can function as an antisilencer and as a positive regulator of transcription (particularly in the case of those LEE genes regulated by both H-NS and Ler) (6, 30, 50). We hypothesize that Ler, in addition to activating lpf expression, functions by antisilencing the action of H-NS. Therefore, we decided to test the expression of the lpfAp::lacZ fusion in an EHEC strain lacking both regulators. The plasmid pPLPFA was introduced in the EHEC strain SDP01 (Δ_ler_ Δ_hns_), and the β-galactosidase activity was compared to that of the wild-type strain EDL933 at different stages during exponential growth. As shown in Fig. 2B, we observed a reduction in expression when the reporter protein was expressed in the double mutant and compared to results for the wild-type strain. The β-galactosidase activity was reduced 1.5- and 1.9-fold for the Δler Δhns double mutant at the later stages of the exponential phase (OD600 = 0.9 and 1.2, respectively) compared to that for the parent strain, a reduction which was similar to the one observed with the EHEC single ler mutant. Interestingly, we noticed that the levels of enzymatic activity in the Δler Δhns double mutant became constitutive throughout the exponential growth phase, a result which was unexpected. This result in combination with further data (see results subsections below) indicates that the silencer H-NS and the antisilencer Ler regulate expression of lpf; however, our data with the Δler Δhns double mutant also suggested that E. coli O157:H7 possesses other regulatory factors that may be controlling expression of lpf in the absence of the Ler and H-NS proteins.
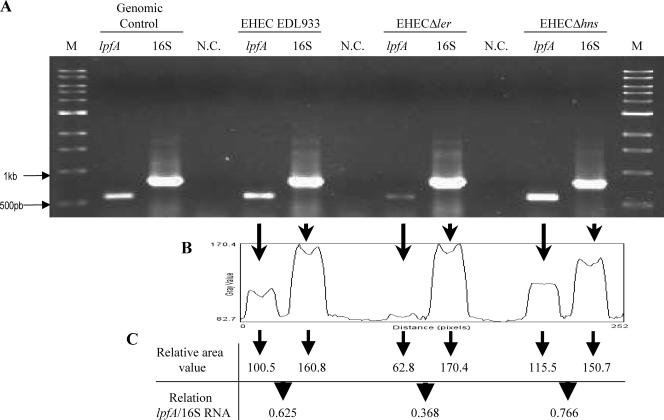
To further confirm that the differences in β-galactosidase activity were due to the regulatory effects of H-NS and Ler on the lpf operon, we performed semiquantitative RT-PCR experiments to determine whether transcription was altered in the different backgrounds. Differences in the lpfA transcriptional levels were observed at late exponential phase, and as shown in Fig. 3, the absence of the Ler protein caused a decrease in the transcript produced compared to the wild-type levels. In contrast, transcription of the lpfA gene was increased in the hns mutant relative to the levels of 16S produced in the wild type and the hns mutant. Overall, the data further support the regulatory role that H-NS and Ler have in the expression of the lpf operon.
FIG. 3.
Semiquantitative RT-PCR to measure transcription of lpf in the different EHEC backgrounds. (A) The level of transcription of the lpfA gene was measured by semiquantitative RT-PCR using EHEC strains EDL933 (wild type), CB49 (Δ_ler_), and AC425 (Δ_hns_). One microgram of RNA obtained at the late exponential phase of growth (OD600 = 1.2) from each of the strains was used for reverse transcription; the lpfA (560 bp) and 16S rRNA (780 bp) genes were amplified to measure transcription in each of the strains tested. The RT-PCR mixture without reverse transcriptase was used as the negative control (N.C.) of the reaction, and the lpfA and 16S rRNA genes amplified from the genomic DNA were used as the positive control of the PCR. M, molecular size marker. (B) The results of RT-PCR were analyzed using the NIH ImageJ program. As shown by the arrows, the areas of intensity were quantified and corresponded to each one of the PCR products. (C) The relation lpfA/16S was calculated from the relative values obtained from each area quantified and represent the transcription level for lpfA and l6S in each strain analyzed.
Primer extension analysis of the lpf TSS and identification of other genetic elements in the lpf promoter region.
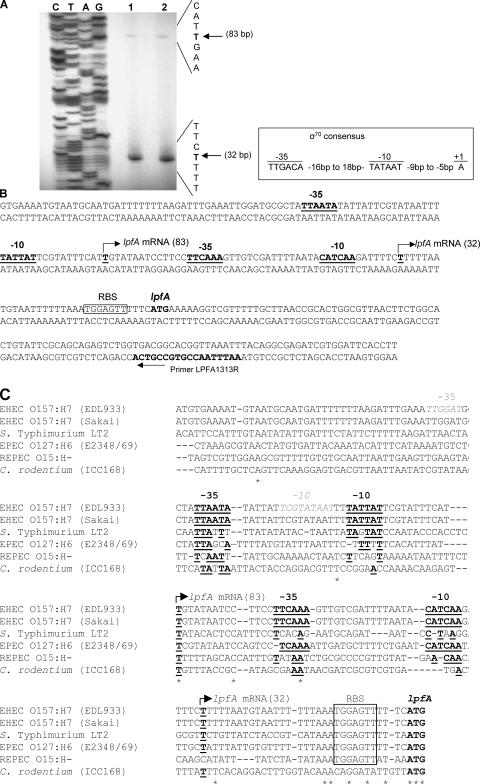
In order to identify the promoter through which H-NS and Ler may act to initiate transcription of lpf, we performed primer extension analysis. We designed a primer located approximately 69 bp downstream from the translational start codon of the lpfA gene, ATG. Primer extension analysis was then performed using RNA isolated from the wild-type strains 86-24 and EDL933, both grown in D-MEM (pH 6.5) at 37°C to an OD600 of 1.0. As observed in Fig. 4A (lanes 1 and 2), a transcriptional start site (TSS) of the lpf operon was mapped to 32 bp upstream of the lpfA translational start site in both wild-type strains. Upstream of this TSS, we identified −10 (3 of 6 bp match the consensus) and −35 (4 of 6 bp match the consensus) hexamers with the σ70-dependent consensus promoter sequence (Fig. 4B). We identified a second TSS which was present in both wild-type strains tested (Fig. 4A, lanes 1 and 2). This second TSS in the lpf promoter region, although with less intensity than the first TSS, was mapped to 83 bp upstream of the lpfA translational start site and contained −10 (5 of 6 bp match the consensus) and −35 (4 bp of 6 bp match the consensus) hexamers, also with the σ70-dependent consensus promoter sequence (Fig. 4B). We then performed a primer extension experiment to determine which of these promoters is regulated by Ler/H-NS, and our preliminary analysis indicates that the TSS located 32 bp upstream of the lpfA translational start site is regulated by the H-NS and Ler proteins (data not shown).
FIG. 4.
Promoter analysis of the E. coli O157:H7 lpf upstream regulatory region. (A) Mapping the lpfA transcriptional start site by primer extension analysis. Total RNA was obtained from culture samples of EHEC O157:H7 strains EDL933 (lane 1) and 86-24 (lane 2) grown in D-MEM (pH 6.5) at 37°C with agitation until an OD600 of 1.0 was reached. The arrows indicate the transcriptional start sites, 83 and 32 bp upstream from the lpfA translational start codon. (B) Nucleotide sequence of the upstream regulatory region of lpfA. The underlined and bold T, with a broken horizontal arrow, indicates the transcriptional start sites (lpfA mRNA). The −35 and −10 hexamer consensus sequences (bold and underlined) were located according to the primer extension analysis. The putative Shine-Dalgarno sequence (RBS) is boxed. The primer within the lpfA structural gene used for primer extension is indicated with an arrow. The σ70 consensus sequence, indicating the conserved −10 and −35 hexamers and the start of transcription, is displayed in the inlet. (C) Nucleotide sequence alignment of the promoter regions found upstream of lpf1 operons from EHEC O157:H7 strains EDL933 and Sakai, serovar Typhimurium LT2, EPEC O127:H6 strain E2348/69, REPEC O15:H−, and Citrobacter rodentium strain ICC168. Similar to panel B, the transcriptional start sites (underlined and bold T, with a broken horizontal arrow and lpfA mRNA), the −35 and −10 hexamers (bold and underlined), and the putative Shine-Dalgarno sequence (RBS, boxed) are indicated. Further, the software-predicted −10 and −35 hexamers are also indicated (gray characters in italics). Asterisks indicate conserved nucleotides.
There is no experimental evidence available regarding the TSS of other lpf1 operons (e.g., in serovar Typhimurium or EPEC O127:H6), and only predictions have been generated, based on computer software designed to find potential promoter regions (3, 16). Because our data are the first to show that transcription of E. coli O157:H7 lpf1 may occur from two different promoters, we decided to perform an alignment with all the lpf1 promoter regions known so far to identify common elements present in these regions. We aligned the lpf1 promoter regions found in the EHEC O157:H7 strains EDL933 and Sakai, serovar Typhimurium LT2, EPEC O127:H6 strain E2348/69, rabbit EPEC (REPEC) O15:H−, and Citrobacter rodentium strain ICC168. As shown in Fig. 4C, the predicted TSS located 32 bp upstream of the EHEC EDL933 lpfA translational start site is conserved in EHEC O157:H7 strain Sakai, serovar Typhimurium, EPEC O127:H6, and C. rodentium; however, the −10 and −35 hexamers upstream of the TSS are found only in both EHEC O157:H7 strains and EPEC O127:H6. Similarly, the second predicted TSS, located 83 bp upstream of the EHEC EDL933 lpfA translational start site, is also conserved in all of the sequences analyzed, but the −10 and −35 hexamers were not. The only consensus sequences were found in the two EHEC O157:H7 isolates. Interestingly, the bacterial promoter recognition program BPROM helped us to predict −10 and −35 hexamers (Fig. 4C, hexamers in gray); however, they did not corresponded to those obtained experimentally. Further, all the sequences analyzed, with the exception of C. rodentium, contained an identical Shine-Dalgarno sequence located at the same distance of the translational start site as in the prototype EHEC O157:H7 strain EDL933. Our comparative analysis indicates that although some elements are present in the different lpf promoter regions, no conservation exists regarding the different hexamers and the spacing between the two TSS (with the exception being the different isolates of EHEC O157:H7), which might suggest that lpf genes may be similarly expressed in the case of EHEC O157:H7 isolates, but this might not be the case in other _lpf_-containing organisms.
EMSAs with purified H-NS.
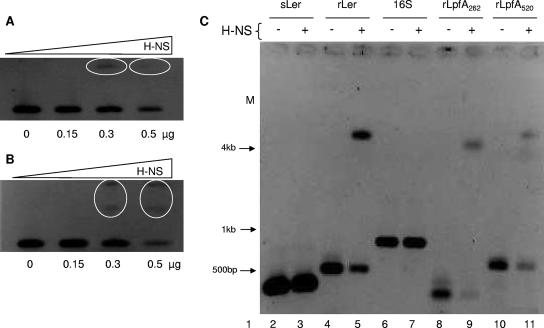
To demonstrate that H-NS was directly binding to the lpf promoter region, EMSAs were performed. Because the pPLPFA plasmid contained a very large DNA fragment including the lpf promoter region (∼1,166 bp, from bp −1047 to +119), we decided to amplify shorter PCR products containing the lpf regulatory region in relation to the first transcriptional start site (32 bp upstream of the ATG) (Fig. 4). Two lpf promoter fragments, a long fragment of 520 bp (−477 to +53; rLpfA520) and a short fragment of 262 bp (−209 to +53; rLpfA262) were amplified by PCR. Both fragments contained the two transcriptional start sites identified in Fig. 4. Initially, our EMSAs were performed using different concentrations of H-NS to determine the appropriate amount of protein required to bind and shift the long and short lpf promoter fragments. As shown in Fig. 5A and B, the addition of 0.3 μg of purified H-NS was sufficient to shift the 520-bp and 262-bp fragments. To validate our initial EMSA's results, we included two negative controls, corresponding to the structural regions of the 16S rRNA gene (16S) (fragment of 780 bp) and the ler gene (sLer) (400 bp). Bustamante et al. (6) found that these two DNA fragments did not have binding sites for H-NS or for Ler proteins. As a positive control of our EMSAs, we included the regulatory region of the ler gene (rLer; 500 bp), where the same authors demonstrate that binding sites exist for both H-NS and Ler (6). An EMSA using the long and short lpf promoter fragments and the appropriate controls showed that H-NS bound and shifted these promoter regions in addition to the ler promoter region. Also, it did not bind or shift the 16S rRNA or ler genes which were used as negative controls (Fig. 5C). These results suggested that the purified HN-S protein is able to bind the lpf regulatory promoter region.
FIG. 5.
H-NS binds to the regulatory region of lpfA. EMSAs were performed to determine the concentration of the H-NS protein needed to shift (A) Five hundred nanograms of the 262-bp PCR product or (B) 500 ng of the 520-bp PCR product containing the regulatory region of lpfA. DNA (first well) and increasing concentrations of H-NS (subsequent wells) were incubated for 20 min and then separated in a 2.5% agarose gel. The ovals denote the DNA-protein complexes. (C) EMSA was performed with 0.3 μg of the H-NS protein and 500 ng of the different PCR products as follows. 1, molecular size markers; lane 2, free DNA containing the structural region of the ler gene (sLer); lane 3, H-NS plus sLer; lane 4, free DNA containing the regulatory region of the ler gene (rLer); lane 5, H-NS plus rLer; lane 6, free DNA containing the 16S gene; lane 7, H-NS plus 16S gene; lane 8, free DNA fragment (262 bp) containing the regulatory region of lpfA (rLpfA262); lane 9, H-NS plus rLpfA262; lane 10, free DNA fragment (520 bp) containing the regulatory region of lpfA (rLpfA520); lane 11, H-NS plus rLpfA520.
EMSAs with purified Ler.
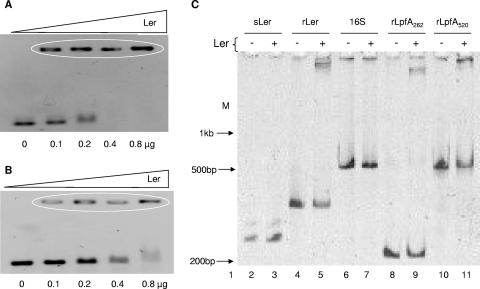
We then performed EMSAs to demonstrate that Ler also binds the lpf regulatory promoter region. Similar to the experiments performed with the purified H-NS protein, we first determined the concentration of purified protein required to bind and shift the long and short lpf promoter fragments. We found that 0.1 μg of purified Ler was sufficient to initiate shifting of the 520-bp (rLpfA520) and 262-bp (rLpfA262) fragments (Fig. 6A and B), and with a concentration of 0.3 μg of Ler, all of the free DNA was completely shifted (data not shown). With that in mind, we performed an EMSA to compare the mobility of the long and short lpf promoter fragments with the appropriate controls. We found that Ler bound and shifted these promoter regions in addition to the ler promoter region and did shift the 16S rRNA or ler (rLer) genes. This result confirmed that Ler binds the lpf regulatory promoter region and further suggested that this protein may be acting as an anti-H-NS-silencer.
FIG. 6.
Ler recognizes the regulatory region of lpfA. EMSAs were performed to determine the concentration of the Ler protein needed to shift (A) 500 ng of the 262-bp PCR product or (B) 500 ng of the 520-bp PCR product containing the regulatory region of lpfA. DNA (first well) and increasing concentrations of Ler (subsequent wells) were incubated for 20 min and then separated in a 2.5% agarose gel. The ovals denote the DNA-protein complexes. (C) EMSA was performed with 0.3 μg of the Ler protein and 500 ng of the different PCR products and separated in a 6% polyacrylamide gel as follows, 1, molecular weight marker; lane 2, free DNA containing the structural region of the ler gene (sLer); lane 3, Ler plus sLer; lane 4, free DNA containing the regulatory region of the ler gene (rLer); lane 5, Ler plus rLer; lane 6, free DNA containing the 16S gene; lane 7, Ler plus 16S gene; lane 8, free DNA fragment (262 bp) containing the regulatory region of lpfA (rLpfA262); lane 9, Ler plus rLpfA262; lane 10, free DNA fragment (520 bp) containing the regulatory region of lpfA (rLpfA520); lane 11, Ler plus rLpfA520.
Ler dissociates the complex formed between H-NS and the lpf regulatory promoter region.
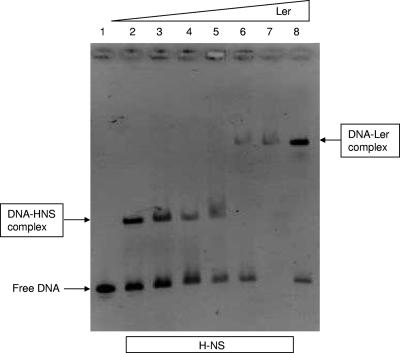
Because our initial data support the role of Ler as an antisilencer protein counteracting the H-NS protein, we designed a competition assay to determine whether the Ler protein can dissociate the complex formed between the H-NS protein and the lpf regulatory promoter region (Fig. 7). Five hundred nanograms of the 262-bp lpf promoter fragment (-rLpfA262) was incubated with a constant amount of the H-NS protein (0.3 μg). We then added increasing amounts of the Ler protein (0 to 0.5 μg). Results shown in Fig. 7 indicated that H-NS binding began to be outcompeted at a concentration of Ler of 0.2 μg and was completely competed out at a concentration of 0.4 μg. These data further support our conclusion that H-NS binds to the regulatory sequence of lpfA and “silences” the transcription of lpf, while Ler binds to the regulatory sequence and inhibits the action of the H-NS protein.
FIG. 7.
Ler and H-NS compete for binding to the regulatory region of lpfA. Competitive EMSA using the 262-bp fragment containing the regulatory region of lpfA (rLpfA262) and the proteins H-NS (constant concentration) and Ler (variable concentration), as follows. Lane 1, free DNA; lane 2, rLpfA262 plus 0.3 μg H-NS; lane 3, rLpfA262 plus H-NS plus 0.1 μg Ler; lane 4, rLpfA262 plus H-NS plus 0.2 μg Ler; lane 5, rLpfA262 plus H-NS plus 0.3 μg Ler; lane 6, rLpfA262 plus H-NS plus 0.4 μg Ler; lane 7, rLpfA262 plus H-NS plus 0.5 μg Ler; lane 8, 262-bp fragment plus 0.4 μg Ler.
DISCUSSION
In the present work, we analyzed the role of several regulators in the expression of LPF and found that H-NS functions as a silencer, while Ler functions as an antisilencer, of the lpf operon. Further, our data are the first to show direct binding of the H-NS and Ler proteins to the regulatory sequence of the lpf operon and confirmed that Ler positively regulates the expression of lpf by counteracting the H-NS-mediated silencing of this promoter. Previously it was shown that H-NS exerts a global regulatory effect on EPEC and EHEC LEE promoters and that Ler acts as an antisilencer counteracting H-NS negative effects (1, 6, 22, 25, 50). In these reports, it was shown that Ler is either competing with H-NS for its binding sites and/or altering the local DNA architecture to hinder H-NS binding, interfering with the formation of a potential repressing nucleoprotein complex that cause silencing of the LEE promoters and facilitating productive RNA polymerase-promoter interactions. In EPEC, for example, H-NS-mediated repression of divergently transcribed LEE operons (LEE2 and LEE3 operons) involved the binding of H-NS to DNA regions known as silencer regulatory sequences 1 and 2 (SRS1 and -2), which are found in the regulatory region between the two EPEC operons. Binding of H-NS to SRS1 and SRS2 favors the formation of a repressor nucleoprotein complex, and the specific binding of Ler to SRS1 destabilizes the complex and releases the expression of LEE2 and LEE3 operons (6). Further, H-NS and Ler can also bind to nonoverlapping sites in the regulatory region of another LEE operon (rorf3-grlRA region) (1). In our case, we have shown that H-NS-mediated silencing can be released by the addition of increased concentrations of Ler. Our data support the role of this protein as a positive regulator of the lpf operon; however, we still do not know whether similar silencer regulatory sequences exist in the lpf regulatory region or whether the mechanism of lpf regulation is similar to any of the LEE operons. Therefore, we are currently pursuing the mapping of the lpf regulatory region to identify in the near future the specific regulatory sequences interacting with Ler and H-NS.
It is clear that up to this date, not much was known about the regulation of colonization factors located outside the LEE pathogenicity island. Therefore, our study is important because it expands our knowledge of the function of two well-established virulence regulators, Ler and H-NS, and helps us to understand how they can control the expression of the LP fimbriae. Previously, Elliott et al. (17) reported that in addition to the effects of Ler on LEE-located genes, Ler regulates the expression of proteins encoded outside the LEE and their associated phenotypes. Their data suggested that Ler regulates the expression of a gene carried on the EHEC pO157 plasmid, tagA (5), as judged by tagA::lacZ fusions in E. coli K-12. TagA, which as been renamed StcE (for secreted protease of C1 esterase inhibitor [C1-INH] from EHEC), is a zinc metalloprotease secreted by the etp type II secretion pathway encoded on pO157, which contributes to intimate adherence of this bacterium to host cells and localizes the inflammatory regulator C1-INH to cell membranes (26). Furthermore, Elliott et al. suggested that Ler also regulates fimbrial expression and adherence phenotypes. A mutation of the ler gene was associated with enhanced adherence to tissue culture monolayers, altered adherence patterns, and expression of LFF (17). Their data suggested that Ler is a repressor of LFF (or perhaps an activator of another repressor) and that these fimbriae mediate the adherence observed in vitro. However, these observations were never further tested or independently confirmed. Our study supports, in part, their findings indicating that Ler regulates expression of EHEC fimbriae. However, in contrast to their findings suggesting that Ler acts as a repressor of LFF, we now clearly show that Ler acts as a positive regulator of the lpf operon, playing a regulatory role similar to that observed in the LEE, where Ler counteracts the silencing effect associated with H-NS regulation. In support of our findings, an independent study by Ogierman et al. (34) showed that Ler increases the level of intimin in Shiga-toxigenic Escherichia coli (STEC) O157. The inability of a STEC O157:H− strain to adhere in a fluorescent actin staining assay (the strain carries eae but did not produce A/E lesions on HEp-2 cells) was shown to be independent of intimin. Thus, the product of ler appears to enhance intimin-independent adherence in STEC O157. Alternatively, the observations of Elliott et al. (17) could indicate that Ler simultaneously promotes the expression of lpf, LEE, and non-LEE genes while another Ler-regulated specific protein(s) represses the expression of gene(s) encoding the LFF. It is plausible to suggest that under intestinal environmental conditions, LPF, LEE, and some non-LEE proteins are required for the pathogenic process of EHEC O157:H7, whereas LFF and other non-LEE-encoded factors are required for the bacterial transit under different environmental conditions. In this way, Ler could function as a critical master regulator for virulence factors in a direct and positive way and become an indirect regulator (antisilencer) when it is interacting with other global or specific regulatory proteins. These and other questions regarding Ler regulation of non-LEE-encoded factors are currently being explored in our laboratory.
In further support of our findings, Jennifer Smart in James Kaper's laboratory recently performed microarray analysis of EHEC O157:H7 wild-type, ler, and hns mutant strains to identify coregulated genes that may be important for intestinal colonization and survival. They identified more than 1,300 genes significantly activated by Ler and no genes repressed by Ler. In contrast, 443 genes were repressed by H-NS, while 751 genes were activated by H-NS. Upon comparison, they identified 165 genes that were repressed by H-NS and activated by Ler, while 90 genes were activated by both H-NS and Ler. Within the first set of genes, they found that the lpf operon is silenced by H-NS and that the silencing effect is inhibited by Ler (41). Overall, these results strongly support our findings and argue against the role of Ler as a repressor of LFF and whether these fimbriae actually act as a mediator of adherence in vitro. Because Ler is required in the A/E pathogens to activate the LEE genes responsible for the formation of the histological lesion, we propose that this protein also regulates the expression of other virulence factors in the later stages of exponential phase that might help the bacteria to attach closely to the epithelium or form stable microcolonies (46). One of the factors that fulfills that pattern of expression and which is associated with the adherence patterns observed in EHEC is the LPF. Overall, our data suggest that Ler and H-NS are two global regulators controlling the expression of genes acquired by horizontal gene transfer (e.g., LEE genes, the lpf operon, and the plasmid-encoded stcE) and which are required for full EHEC virulence.
Using primer extension analysis, we identified two transcriptional start sites, one mapped to 32 bp and the second one to 83 bp upstream of the same translational start site. Also identified were −10 and −35 hexamers for each of the transcriptional start sites with σ70-dependent consensus promoter sequences (Fig. 4). However, one general question raised by our results concerns the functional significance of these two transcriptional start sites: are they redundant in such a way that the occupancy of one is sufficient to produce full activation of the lpf promoter (83 bp), or do they cooperate and have additive or synergistic effects? We proposed that the second lpf promoter might be present to ensure a basal level of expression of these genes, which may allow the organism to respond quickly in the presence of the correct environmental stimuli, and the first lpf promoter (32 bp) is tightly regulated by the H-NS and Ler proteins. It is also possible to hypothesize that one promoter represents the constitutive expression of the lpf operon (particularly in absence of the Ler and H-NS proteins) and the second one responds to specific environmental stimuli. In support of the regulation by environmental stimulus, our experiments were performed under optimal conditions for lpf expression (48), and such conditions influence the differential expression of the two promoters, favoring the transcriptional start site closer to the ATG.
One additional characteristic associated with the regulation of the lpf operon (and one that makes the LP fimbriae an interesting surface structure to study) is that the genes encoding the fimbrial proteins are present in multiple A/E lesion-forming bacteria and in serovar Typhimurium; however, some of these DNA regions (specifically in EPEC O127:H6 and C. rodentium) which have been shown to be complete and intact are not expressed (45). The absence in expression could be attributed to tight regulation or to the absence of specific promoter elements which are present only in EHEC O157:H7. In the case of C. rodentium, it is evident that its promoter region differs significantly from the one found in EHEC O157:H7 (Fig. 4C). This difference could explain the absence of lpf expression, because this fimbrial operon might be subjected to other regulatory mechanisms that are found only in the mouse intestine. In contrast, the regulatory region of EPEC O127:H6 shared several common elements with the EHEC O157:H7 lpf promoter region, particularly at the TSS located 32 bp upstream of the lpfA translational start site, which makes it more difficult to predict the reason for the lack of expression of this operon. Based on this information, are the differences in the predicted TSS located 83 bp upstream of the lpfA translational start site in EPEC O127:H6 or the absence of regulatory proteins present only in EHEC O157:H7 responsible for the lack of lpf expression? Preliminary studies using the lpfAp::lacZ fusion in an EPEC Δler Δhns strain showed that H-NS also acts as a silencer for the expression of lpf and confirmed the role of Ler as an antisilencer protein (G. N. Lopez-Sanchez and A. G. Torres, unpublished data). Interestingly, the levels of β-galactosidase activity during the exponential growth phase of EPEC were low and not close to the activity observed when the reporter plasmid is expressed in an EHEC background. This reduction in lpf expression in the EPEC background cannot be attributed to a mutation in the lpf operon and might be related more to the absence of EHEC-specific regulatory factors. Finally, are the lpf operons in serovar Typhimurium LT2 and REPEC (which have been shown to express and produce fully functional fimbriae) regulated similarly to regulation of the EHEC O157:H7 lpf operon? Our prediction based on the DNA alignments of the promoters suggested that these regions are quite different and indicated that these fimbrial operons might be subjected to another regulatory mechanism(s) that need further study.
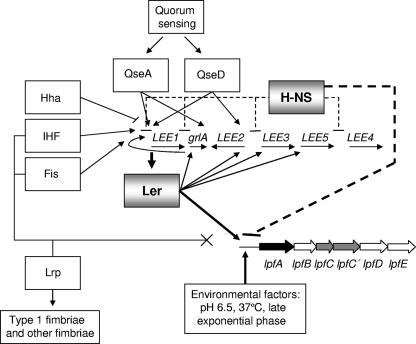
Figure 8 depicts the model for transcriptional regulation of the lpf operon, emphasizing the need of a coordinated regulatory cascade, in conjunction with the LEE pathogenicity island, to achieve expression of the LPF. The model may be dependent on strain backgrounds, because the presence of an intact lpf operon does not guarantee functional fimbriae expression, as we demonstrated for EPEC and C. rodentium, where the lpf operon seems to be strongly repressed and which could suggest that additional factors controlling expression and found only in EHEC O157:H7 are missing from other bacterial backgrounds (45). Under the growth conditions used in the current study and utilizing E. coli O157:H7 EDL933 as the background strain, the expression levels of lpf were induced and the operon was controlled by the H-NS and Ler proteins. No regulatory effects were observed when the lpf reporter plasmid was expressed in the ihf, fis, or lrp mutant strain. In contrast, we demonstrated that H-NS and Ler can bind to the regulatory sequence of lpf, and while H-NS acts as a “silencer” of transcription, Ler inhibits the action of the H-NS protein. An important number of regulators have been shown to modulate expression of the LEE genes in EHEC O157:H7 and other EHEC and EPEC strains (reviewed in reference 29). However, only two of those regulators, Ler and H-NS, were shown for the first time to directly regulate expression of E. coli O157:H7 genes located outside the LEE. Whether additional factors are connected to the regulatory mechanism of the lpf operon (e.g., the formation of H-NS heterodimers with StpA [32] or with other H-NS homologues [14]) and whether this type of regulation occurs only in E. coli O157:H7 have to be further clarified to understand the complexity of the E. coli O157:H7 virulence regulon.
FIG. 8.
LPF expression is controlled by global regulators and LEE-encoded regulators. Transcription of lpf genes occurs in response to environmental signals, and their regulation seems to be linked to expression of LEE-encoded genes. While global and specific regulators control expression of LEE, IHF, Fis, and Lrp did not seem to have any regulatory role on lpf gene expression. In contrast, H-NS binds to the regulatory sequence of lpfA and “silences” the transcription of LPF, while Ler binds to the regulatory sequence and inhibits the action of the H-NS protein.
Acknowledgments
We thank James B. Kaper, Christine Martin, and Jose L. Puente for providing bacterial strains and plasmids and Cristina Madrid and Antonio Juarez for sharing their protocols and techniques with our laboratory. We also thank Belinda Iles and Christopher Allen for critical reading of the manuscript.
The laboratory of A.G.T. was supported in part by institutional funds from the UTMB John Sealy Memorial Endowment Fund for Biomedical Research, and the laboratory of Y.M.-L. was funded by grant 36477-N from CONACYT. G.N.L-S. and L.M-F. received fellowships from CONACYT, Mexico, and SURP, UTMB.
Footnotes
▿
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Barba, J., V. H. Bustamante, M. A. Flores-Valdez, W. Deng, B. B. Finlay, and J. L. Puente. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187**:**7918-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrios, H., H. M. Fischer, H. Hennecke, and E. Morett. 1995. Overlapping promoters for two different RNA polymerase holoenzymes control Bradyrhizobium japonicum nifA expression. J. Bacteriol. 177**:**1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177**:**2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltrametti, F., A. U. Kresse, and C. A. Guzman. 1999. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181**:**3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26**:**4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39**:**664-678. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104**:**541-555. [DOI] [PubMed] [Google Scholar]
- 8.Castillo, L. 2004. Interacción de IHF y H-NS sobre las regiones reguladoras de bfpA y perA en Escherichia coli Enteropatógena. M.S. thesis. B. Universidad Autonoma de Puebla, Puebla, Mexico.
- 9.CDC. 2006. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. Morb. Mortal. Wkly. Rep. 55**:**1045-1046. [PubMed] [Google Scholar]
- 10.CDC. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 States, United States, 2005. Morb. Mortal. Wkly. Rep. 55**:**392-395. [PubMed] [Google Scholar]
- 11.Crost, C., A. Garrivier, J. Harel, and C. Martin. 2003. Leucine-responsive regulatory protein-mediated repression of clp (encoding CS31A) expression by l-leucine and l-alanine in Escherichia coli. J. Bacteriol. 185**:**1886-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59**:**4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman, C. J. 1995. DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology 141**:**1271-1280. [DOI] [PubMed] [Google Scholar]
- 14.Dorman, C. J. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5**:**157-161. [DOI] [PubMed] [Google Scholar]
- 15.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2**:**391-400. [DOI] [PubMed] [Google Scholar]
- 16.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70**:**6761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68**:**6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzhenry, R., S. Dahan, A. G. Torres, Y. Chong, R. Heuschkel, S. Murch, M. Thomson, J. B. Kaper, G. Frankel, and A. D. Phillips. 2006. Long polar fimbriae and tissue tropism in Escherichia coli O157:H7. Int. J. Med. Microbiol. 296**:**547-552. [DOI] [PubMed] [Google Scholar]
- 19.Flores-Valdez, M. A., J. L. Puente, and E. Calva. 2003. Negative osmoregulation of the Salmonella ompS1 porin gene independently of OmpR in an hns background. J. Bacteriol. 185**:**6497-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friden, P., K. Voelkel, R. Sternglantz, and M. Freundlich. 1984. Reduced expression of the isoleucine and valine enzymes in integration host factor mutants of Escherichia coli. J. Mol. Biol. 172**:**573-579. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41**:**549-559. [DOI] [PubMed] [Google Scholar]
- 22.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71**:**384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussein, H. S., and L. M. Bollinger. 2005. Prevalence of Shiga toxin-producing Escherichia coli in beef cattle. J. Food Prot. 68**:**2224-2241. [DOI] [PubMed] [Google Scholar]
- 24.Jordan, D. M., N. Cornick, A. G. Torres, E. A. Dean-Nystrom, J. B. Kaper, and H. W. Moon. 2004. Long polar fimbriae contribute to colonization by Escherichia coli O157:H7 in vivo. Infect. Immun. 72**:**6168-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laaberki, M. H., N. Janabi, E. Oswald, and F. Repoila. 2006. Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS and RpoS. Int. J. Med. Microbiol. 296**:**197-210. [DOI] [PubMed] [Google Scholar]
- 26.Lathem, W. W., T. E. Grys, S. E. Witowski, A. G. Torres, J. B. Kaper, P. I. Tarr, and R. A. Welch. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45**:**277-288. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Martinez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33**:**153-166. [DOI] [PubMed] [Google Scholar]
- 29.Mellies, J. L., and A. M. S. Barron. 2006. Virulence gene regulation in Escherichia coli. In A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd, and C. L. Squires (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 30.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33**:**296-306. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Muller, C. M., U. Dobrindt, G. Nagy, L. Emody, B. E. Uhlin, and J. Hacker. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188**:**5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11**:**142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogierman, M. A., A. W. Paton, and J. C. Paton. 2000. Up-regulation of both intimin and _eae_-independent adherence of shiga toxigenic Escherichia coli O157 by ler and phenotypic impact of a naturally occurring ler mutation. Infect. Immun. 68**:**5344-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reading, N. C., and V. Sperandio. 2006. Quorum sensing: the many languages of bacteria. FEMS Microbiol. Lett. 254**:**1-11. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sharma, V. K., and R. L. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186**:**7290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1**:**784-791. [Google Scholar]
- 39.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy _lac_-based cloning vectors for protein and operon fusions. Gene 53**:**85-96. [DOI] [PubMed] [Google Scholar]
- 40.Sivapalasingam, S., C. R. Friedman, L. Cohen, and R. V. Tauxe. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67**:**2342-2353. [DOI] [PubMed] [Google Scholar]
- 41.Smart, J. I., and J. B. Kaper. 2006. Comparison of the enterohemorrhagic Escherichia coli O157:H7 LEE-encoded regulator regulon with the H-NS regulon using microarray analysis, p. 105. Abstr. 6th International Symposium on Shiga Toxin (Verocytotoxin) Producing E. coli Infections. Cambridge Publishing, West Leederville, Australia.
- 42.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70**:**3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38**:**781-793. [DOI] [PubMed] [Google Scholar]
- 44.Tarr, P. I., M. A. Neill, C. R. Clausen, J. W. Newland, R. J. Neill, and S. L. Moseley. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J. Infect. Dis. 159**:**344-347. [DOI] [PubMed] [Google Scholar]
- 45.Tatsuno, I., R. Mundy, G. Frankel, Y. Chong, A. D. Phillips, A. G. Torres, and J. B. Kaper. 2006. The lpf gene cluster for long polar fimbriae is not involved in adherence of enteropathogenic Escherichia coli or virulence of Citrobacter rodentium. Infect. Immun. 74**:**265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC_′_DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70**:**5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres, A. G., and J. B. Kaper. 2001. PAIs of intestinal E. coli, p. 31-48. In J. Hacker and J. B. Kaper (ed.), Pathogenicity islands (PAIs) and the evolution of pathogenic microbes, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 48.Torres, A. G., L. Milflores-Flores, J. G. Garcia-Gallegos, S. D. Patel, A. Best, R. M. La Ragione, Y. Martinez-Laguna, and M. J. Woodward. 2007. Environmental regulation and colonization attributes of the long polar fimbriae of Escherichia coli O157:H7. Int. J. Med. Microbiol. 297**:**177-185. [DOI] [PubMed] [Google Scholar]
- 49.Torres, A. G., X. Zhou, and J. B. Kaper. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73**:**18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148**:**2735-2744. [DOI] [PubMed] [Google Scholar]
- 51.Yamada, H., T. Yoshida, K. Tanaka, C. Sasakawa, and T. Mizuno. 1991. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 230**:**332-336. [DOI] [PubMed] [Google Scholar]