NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila (original) (raw)
Abstract
NELF and DSIF collaborate to inhibit elongation by RNA polymerase IIa in extracts from human cells. A multifaceted approach was taken to investigate the potential role of these factors in promoter proximal pausing on the_hsp70_ gene in Drosophila. Immunodepletion of DSIF from a_Drosophila_ nuclear extract reduced the level of polymerase that paused in the promoter proximal region of hsp70. Depletion of one NELF subunit in salivary glands using RNA interference also reduced the level of paused polymerase. In vivo protein–DNA cross-linking showed that NELF and DSIF associate with the promoter region before heat shock. Immunofluorescence analysis of polytene chromosomes corroborated the cross-linking result and showed that NELF, DSIF, and RNA polymerase IIa colocalize at the hsp70 genes, small heat shock genes, and many other chromosomal locations. Finally, following heat shock induction, DSIF and polymerase but not NELF were strongly recruited to chromosomal puffs harboring the hsp70 genes. We propose that NELF and DSIF cause polymerase to pause in the promoter proximal region of hsp70. The transcriptional activator, HSF, might cause NELF to dissociate from the elongation complex. DSIF continues to associate with the elongation complex and could serve a positive role in elongation.
Keywords: NELF, DSIF, transcription, Drosophila, heat shock, elongation
Inhibition of transcriptional elongation is an important step in regulating the expression of numerous genes in eucaryotic cells (Kim et al. 2001). In the case of the hsp70 heat shock gene in Drosophila, RNA polymerase II (Pol II) is stably paused in the promoter proximal region before heat shock induction (Lis 1998). Evidence of promoter proximal pausing has been obtained for a variety of other genes including the c-myc and c-fos genes (Krumm et al. 1992;Plet et al. 1995;Pinaud and Mirkovitch 1998). Law et al. (1998) provided evidence that the RNA polymerase associated with as many as 20% of genes in cultured Drosophila cells might be in a paused state.
Phosphorylation of Pol II appears to be a key step in converting Pol II from an elongation incompetent to elongation competent state (Weeks et al. 1993;O'Brien et al. 1994). During transcription, extensive phosphorylation occurs at the C terminus of the largest subunit—a region called the CTD (Komarnitsky et al. 2000;Cho et al. 2001). The CTD is composed of a 7-amino-acid sequence that is repeated 26 times in yeast, 42 times in Drosophila, and 52 times in human. Pol II associates with the promoter as an unphosphorylated form called Pol IIa (Lu et al. 1991). In vitro, the CTD becomes highly phosphorylated during or shortly after transcriptional initiation (Laybourn and Dahmus 1990). This hyperphosphorylated form of RNA polymerase is called Pol IIo.
In vivo, the Pol II found paused on the hsp70 gene before heat shock induction is in the IIa state (Weeks et al. 1993; O'Brien et al. 1994). Following heat shock, the polymerase detected on the body of the gene is in the hyperphosphorylated Pol IIo state. This suggests that the release of the paused polymerase is coupled to a hyperphosphorylation of the CTD. Consistent with this hypothesis is the finding that expression of a protein consisting of the Gal4 DNA-binding domain fused to the kinase P-TEFb activates transcription of an hsp70 promoter containing binding sites for the Gal4 protein (Lis et al. 2000). Normally, hsp70 is induced during heat shock by HSF. Although no direct association between P-TEFb and HSF is evident, P-TEFb associates with the hsp70 gene during heat shock (Lis et al. 2000). There is mounting evidence that several transcriptional activators function in part by mediating phosphorylation of the CTD either through the kinase P-TEFb or the general transcription factor TFIIH (Blau et al. 1996; Barboric et al. 2001; Eberhardy and Farnham 2001).
An understanding of how phosphorylation of the CTD may control transcriptional elongation has emerged from the biochemical analysis of the sensitivity of transcription toward the nucleoside analog DRB (Yamaguchi et al. 1998). DRB inhibits kinases that are responsible for phosphorylating the CTD, and it causes greater than 50% inhibition of Pol II transcription in human cells (Sehgal et al. 1976;Mancebo et al. 1997;Chao and Price 2001). DRB appears to inhibit elongation as cells are seen to accumulate short transcripts and suppress production of long transcripts (Tamm and Kikuchi 1979;Chodosh et al. 1989). Elongation by purified Pol II is not inhibited by DRB, indicating that DRB targets other proteins that modulate elongation (Kephart et al. 1992). The kinase P-TEFb was first identified as a factor required to reconstitute DRB-sensitive transcription (Marshall and Price 1995). More recently, two other proteins called DSIF and NELF were discovered to be required for DRB inhibition (Wada et al. 1998a;Yamaguchi et al. 1999a). The results of immunodepletion experiments support a model in which DSIF and NELF inhibit elongation by Pol IIa, and P-TEFb relieves this inhibition by phosphorylating the polymerase (Wada et al. 1998b; Yamaguchi et al. 1999a). Additional support for this model comes from the observation that NELF and DSIF only inhibit elongation by Pol IIa and not by Pol IIo in a well-defined elongation reaction (Yamaguchi et al. 1999a).
Chromatin immunoprecipitation analysis has revealed that DSIF is associated with the hsp70 promoter before heat shock induction, so it is reasonable to postulate that this factor is involved in promoter proximal pausing (Andrulis et al. 2000). There is also strong evidence that DSIF stimulates transcriptional elongation, however. Immunofluorescence analysis of polytene chromosomes in_Drosophila_ shows that DSIF associates with strongly transcribed regions of the genome (Andrulis et al. 2000; Kaplan et al. 2000). DSIF stimulates _tat_-dependent elongation in reconstituted transcription reactions apparently by counteracting premature termination and transcriptional arrest (Wu-Baer et al. 1998;Bourgeois et al. 2002). DSIF is also known as Spt4 and Spt5, which correspond to its small and large subunits. Several alleles of spt5 exhibit synthetic lethality when combined with mutations in Pol II or TFIIS, a stimulatory elongation factor (Hartzog et al. 1998). At present, there is no clear explanation for how DSIF functions both as a positive and a negative elongation factor.
We have investigated the potential roles of DSIF and NELF in regulating promoter proximal pausing at the hsp70 promoter in_Drosophila_. We provide both physical and functional data supporting the hypothesis that NELF and DSIF are important for pausing Pol II in the promoter proximal region. Moreover, the mechanism of transcriptional activation appears to involve ejection of NELF from the paused Pol II.
Results
Immunodepletion of DSIF from a Drosophila nuclear extract inhibits promoter proximal pausing in vitro
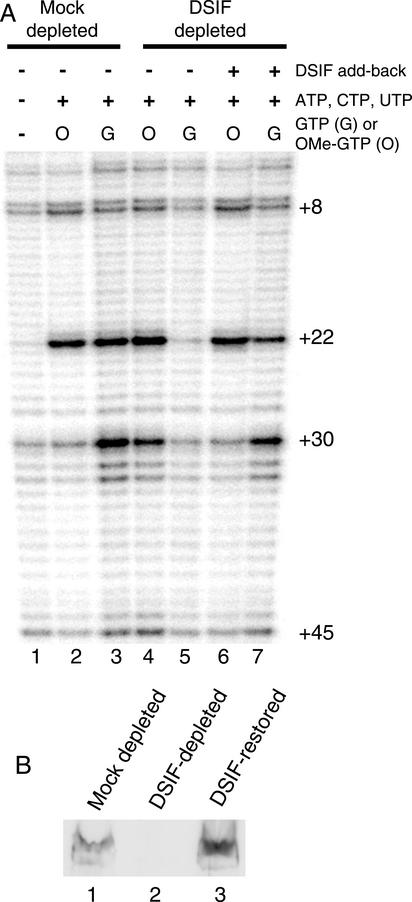
Previously, we showed that promoter proximal pausing could be reconstituted in Drosophila nuclear extracts (Li et al. 1996;Benjamin and Gilmour 1998). The paused polymerase was detected by monitoring the reactivity of the transcribed DNA toward permanganate, which preferentially oxidizes thymines in single-stranded regions. When nuclear extract was incubated with the_hsp70_ promoter in the absence of nucleotides, very little permanganate reactivity was detected in the region 20–40 nucleotides downstream from the transcription start (Fig. 1A, lane 1). When all four nucleotides were present, the permanganate reactivity of the thymines at +22 and +30 was enhanced for a mock-depleted extract, which still contains DSIF (Fig. 1A, lane 3). Immunodepletion of DSIF from the nuclear extract decreased the level of permanganate reactivity detected at +22 and +30 (Fig. 1A, cf. lanes 5 and 3). This decrease could be partially reversed by supplementing the depleted extract with a protein fraction that had been enriched 25-fold for_Drosophila_ DSIF (Fig. 1A, lane 7). As shown by the immunoblot inFigure 1B, the amount of partially purified DSIF was sufficient to return the level of DSIF to normal (cf. lanes 1 and 3).
Figure 1.
Immunodepletion of DSIF from a Drosophila nuclear extract inhibits promoter proximal pausing. (A) In vitro transcription reactions were performed with extracts that had been subjected to depletions with antibody against DSIF or against an irrelevant protein (Mock depleted). Paused polymerase was detected by permanganate footprinting. Lane 1 shows the results for a transcription reaction performed in the absence of nucleotides. Lanes 3, 5, and 7 were performed in the presence of all four nucleotides. Lanes 2, 4, and 6 were performed with 125 μM 3′ O-methyl GTP substituted for GTP. (B) Immunoblot analysis for DSIF in extracts used in the reconstituted transcription reactions. Lanes 1 and 2 are 2-μL portions of mock-depleted or DSIF-depleted extract. Lane 3 is a mixture of 2 μL of DSIF-depleted extract and 2 μL of the partially purified DSIF, which represents the proportions of proteins used in the transcription reaction used for lanes 6 and 7 in_A_.
The decrease in permanganate reactivity resulting from immunodepletion of DSIF could be caused by either a decrease in pausing or a decrease in initiation. To determine if the depletion of DSIF affected initiation, GTP in the reaction was replaced with 3′ O-methyl GTP. 3′ O-methyl GTP halts elongation when Pol II reaches +15, where the first guanine nucleotide is incorporated into the nascent transcript. The level of permanganate reactivity at +22 was essentially the same for the mock-depleted, DSIF-depleted, and the rescued extracts (Fig. 1A, lanes 2,4,6). Previous analysis demonstrated that our reaction conditions allow only one round of initiation (Benjamin and Gilmour 1998). Therefore, the level of initiation on the hsp70 promoter in the three situations is similar. The decrease in permanganate reactivity at +22 and +30 caused by the depletion of DSIF is therefore attributable to inhibition of promoter proximal pausing.
Identification of NELF-D and NELF-E from Drosophila
We were motivated to investigate Drosophila NELF because biochemical analyses indicated that both DSIF and NELF were required to repress elongation in extracts from human cells (Yamaguchi et al. 1999a). BLAST analyses identified single candidates for NELF-D and NELF-E in_Drosophila_ (data not shown). These genes have been designated CG9984 and CG5994 by the Berkeley Drosophila Genome Project.Drosophila NELF-D has 54% identity over its entire length with human NELF-D. 40% of the amino acids from 1 to 107 and 37% of the amino acids from 140 to 277 of Drosophila NELF-E are identical to human NELF-E. Amino acids 165–236 in Drosophila NELF-E are homologous to the RNA-binding domain of human NELF-E. This region is essential for NELF to inhibit elongation in vitro (Yamaguchi et al. 2002).
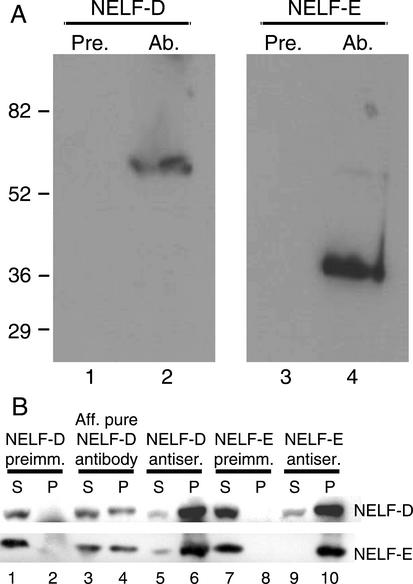
The N-terminal portion of Drosophila NELF-D (amino acids 33–154) and the entirety of NELF-E were expressed and purified from_Escherichia coli_, and each polypeptide was used to raise antibody. A Western blot analysis showed that each antiserum but not the corresponding preimmune serum recognized a polypeptide of the expected size in a_Drosophila_ nuclear extract (Fig. 2A). As further evidence that Drosophila NELF-D and NELF-E are homologs to human NELF subunits, NELF-D and NELF-E were found to associate with each other in Drosophila nuclear extract. Antibodies against either polypeptide coprecipitated the other polypeptide. Affinity-purified anti-NELF-D antibody or crude antiserum against NELF-D precipitated both NELF-D and NELF-E (Fig. 2B, lanes 4,6). Crude antiserum against NELF-E also precipitated both NELF-D and NELF-E (Fig. 2B, lane 10). In contrast, preimmune serum for either NELF-D or NELF-E failed to precipitate either protein (Fig. 2B, lanes 2,8).
Figure 2.
Identification of Drosophila NELF-D and NELF-E. (A) Immunoblot analysis shows that antibodies raised against the_Drosophila_ homologs bind specifically to polypeptides of the sizes predicted for the respective cDNA clones. Nuclear extract was run on SDS-PAGE and blotted to nitrocellulose. Individual nitrocellulose strips harboring nuclear proteins were incubated with preimmune serum for NELF-D or NELF-E (lanes 1,3, respectively) or with antiserum for NELF-D or NELF-E (lanes 2,4, respectively). The NELF-D antiserum specifically binds a polypeptide of ∼60 kD and the NELF-E antiserum specifically binds a polypeptide of ∼36 kD. (B) Coimmunoprecipitation data indicates that Drosophila NELF-D and NELF-E associate with each other. Nuclear extract containing 150 mM KCl was incubated with preimmune antiserum (lanes_1,2_), affinity-purified NELF-D antibody (lanes 3,4), NELF-D antiserum (lanes 5,6), NELF-E preimmune antiserum (lanes_7,8_), or NELF-E antiserum (lanes 9,10). Immune complexes were recovered with protein A sepharose, and the presence of NELF-D and NELF-E was assessed by immunoblot analysis. S, supernatant; P, immunoprecipitated material.
NELF-D, NELF-E, DSIF, and GAGA factor associate with the hsp70 promoter region before heat shock induction
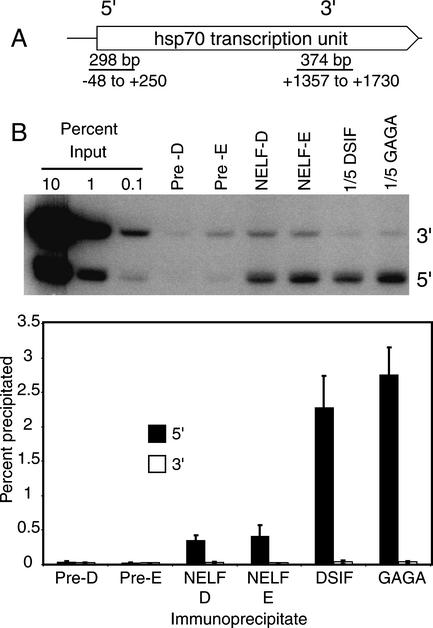
Previously, Andrulis et al. (2000) used chromatin immunoprecipitation to determine that DSIF associates with the hsp70 promoter before heat shock induction. The results inFigure 3 show that NELF-D and NELF-E also associate with the hsp70 promoter. Drosophila cells were treated with formaldehyde to form protein–protein and protein–DNA cross-links, lysed in SDS, sonicated to shear DNA to sizes from 200 to 700 bp, and subjected to immunoprecipitations. DNA recovered from each immunoprecipitate was then analyzed for the presence of DNA corresponding to the promoter region (5′) or the 3′ region of the hsp70 gene (Fig. 3A).
Figure 3.
Chromatin immunoprecipitation analysis of NELF, DSIF, and GAGA factor.Drosophila Schneider 2 cells were subjected to formaldehyde cross-linking. Sonicated detergent lysates from the cells were immunoprecipited with preimmune serum (Pre-D or Pre-E) or with antiserum against NELF-E, NELF-D, DSIF, or GAGA factor. DNA was purified from each immunoprecipitate, and the presence of DNA from the 5′ and 3′ portions of the hsp70 gene was detected by PCR. Various amounts of material representing the input were amplified to quantify the amount of DNA recovered from the immunoprecipitates. (A) A schematic of the_hsp70_ transcription unit and the regions that were amplified. (B) The results from one of two independent experiments that were done. The bands represent radiolabeled PCR products that were detected after the DNA was run on an acrylamide gel and the gel was subjected to analysis with a PhosphorImager. Because preliminary experiments showed that DSIF and GAGA factor cross-linked more efficiently than NELF-D and NELF-E, 5× less of the DNA recovered from the DSIF and GAGA factor immunoprecipitates were used in the PCR reactions. The graph shows a compilation of the results from two independent experiments. The height of each bar shows the average percent of input material detected in immunoprecipitates from two experiments, and each error bar shows the range of measurements.
Figure 3B provides representative results from an analysis, and the graph quantifies the amounts of DNA from the 5′ and 3′ ends of the hsp70 gene that were recovered from various immunoprecipitates for two independent experiments. The amount of material recovered from all of the preimmune precipitates averaged 0.031% with a standard deviation of 0.015 and a maximum value of 0.068%. This range establishes the background inherent to the technique. In cells, the level of NELF-D, NELF-E, DSIF, and GAGA factor cross-linking to the 5′ region significantly exceeded the level cross-linking to the 3′ region of the gene. Approximately 0.4% of the 5′ region cross-linked to NELF-D and NELF-E, and ∼2.5% of the 5′ region cross-linked to DSIF and GAGA factor. In contrast, the amount of DNA from the 3′ region cross-linking to any of these proteins was in the range of background. Therefore, all four of these proteins appear to preferentially associate with the promoter region before heat shock induction. Previous UV cross-linking and genomic footprinting analyses showed that GAGA factor associates with the promoter region before heat shock (O'Brien et al. 1995;Weber et al. 1997). The detection of DSIF on the promoter region confirms results reported by Andrulis et al. (2000). The results inFigure 3 show for the first time that NELF also associates with the promoter region.
Depletion of NELF-E in salivary glands reduces the level of paused polymerase found on hsp70 in live cells
We tested the hypothesis that NELF is involved in promoter proximal pausing by using RNA interference to deplete NELF-E from salivary glands. Interference RNA against NELF-E was shown recently to cause developmental defects or lethality depending on where the interference RNA was expressed (Enerly et al. 2002). For our experiment, one transgenic fly line, 17A10, contained a transgene encoding an interference RNA against NELF-E that was controlled by Gal4p. A second fly line, 1824, contained a transgene that expressed Gal4p in salivary glands (Brand et al. 1994). The two lines were mated, and salivary glands derived from the progeny and the parental lines were subjected to molecular analysis.
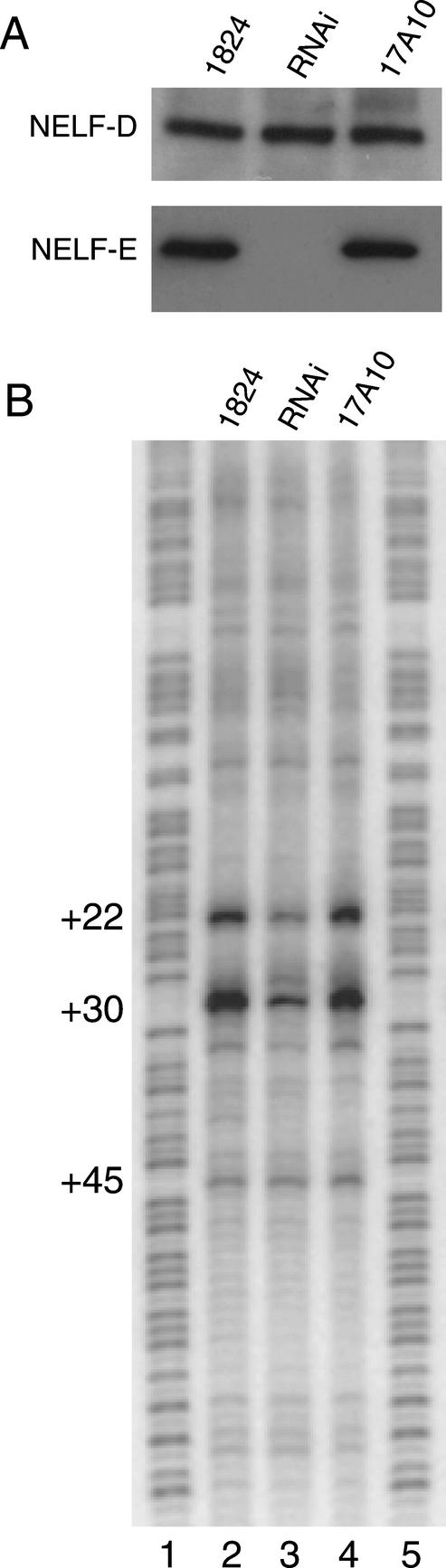
Immunoblot analysis of whole salivary gland lysates revealed that RNAi against NELF-E significantly diminished the amount of NELF-E in the glands (Fig. 4A). Interestingly, the level of NELF-D was unaffected. Genomic footprinting with permanganate was performed to determine if depleting NELF-E caused a decrease in promoter proximal pausing on hsp70. Previously, we showed that the paused Pol II causes thymines at +22 and +30 on the nontranscribed strand to be hyperreactive to permanganate oxidation in salivary glands (Weber et al. 1997). Salivary glands were dissected from third-instar larvae and treated with permanganate. DNA was isolated, and the pattern of permanganate reactivity was analyzed following cleavage at oxidized thymines and ligation-mediated PCR. As shown inFigure 4B, the permanganate reactivity at +22 and +30 was less in the RNAi glands than in glands from either parental line. This affect was observed for three independent RNAi lines (data not shown).
Figure 4.
NELF-E RNA interference inhibits synthesis of NELF-E in salivary glands and inhibits pausing on the hsp70 gene. Fly line 1824 expresses Gal4 in salivary glands. Fly line 17A10 contains a Gal4-activated NELF-E RNAi construct. RNAi is the designation for progeny generated by mating 1824 and 17A10. (A) Immunoblot analysis of NELF-D and NELF-E in whole salivary gland lysates. (B) Permanganate genomic footprinting analysis of_hsp70_ in salivary glands isolated from nonheat-shocked larvae. Lanes_1_ and 5 are derived from genomic DNA partially cleaved at purines. (Lane 2) DNA derived from 1824 salivary glands that were treated with permanganate. (Lane 3) DNA derived from RNAi salivary glands that were treated with permanganate. (Lane 4) DNA derived from 17A10 glands that were treated with permanganate.
Immunofluorescence analysis of NELF-D on polytene chromosomes from Drosophila salivary glands
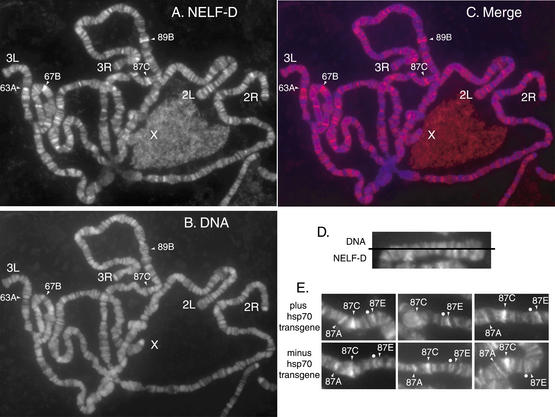
Immunofluorescence analysis of polytene chromosomes was employed to corroborate the results of chromatin immunoprecipitation and to learn more about the genome-wide distribution of NELF. We focused on NELF-D because we were able to affinity-purify NELF-D antibody that stained the polytene chromosome. Affinity-purified antibody against NELF-D broadly stains polytene chromosomes (Fig. 5A). No staining was observed with preimmune serum or with material isolated in a mock affinity purification of antibody (data not shown). Hoechst dye stains DNA and stains polytene chromosomes in a pattern of bands that coincides closely with the pattern of bands on chromosomes typically detected by phase contrast microscopy (Fig. 5B). NELF-D was restricted to the regions known as interbands, which lie between the bands of DNA staining (Fig. 5C,D).
Figure 5.
Immunolocalization of NELF-D on polytene chromosomes from larvae. Polytene chromosome spreads were prepared from salivary glands of third-instar larvae. (A) The chromosomes were incubated with affinity-purified NELF-D antibody and then incubated with Alexa 568 conjugated secondary antibody. (B) DNA was detected by staining with Hoechst. (C) The merged image shows the NELF-staining pattern in red and the DNA staining pattern in blue. (D) A split image of the end of chromosome 2L with the DNA stain on the top half and NELF-D staining on the_bottom_ half. (E) Representative examples of the NELF-D staining pattern in the region encompassing the endogenous hsp70 genes and an hsp70 transgenic promoter that is inserted at 87E. The dot marks a band that serves as a reference for comparing the intensity of staining at 87E where the transgenic promoter resides. The top three pictures are from chromosomes containing the insert and the bottom three pictures are from chromosomes that lackthe insert. Note that the intensity of staining for NELF-D is stronger than the reference in cases containing the transgenic promoter, but not for the cases lacking the transgenic promoter.
Several regions of the polytene chromosomes consistently stained more intensely than other regions of the chromosomes. These regions include 63A, 67B, 87C, and 89B, which are also evident for independent samples shown inFigure 6. The staining at 87C was of interest because at least three copies of the hsp70 gene reside at this locus. Staining at 67B is also noteworthy because four small heat shock genes are located in this region, and three have been shown to harbor paused polymerase (Lis 1998).
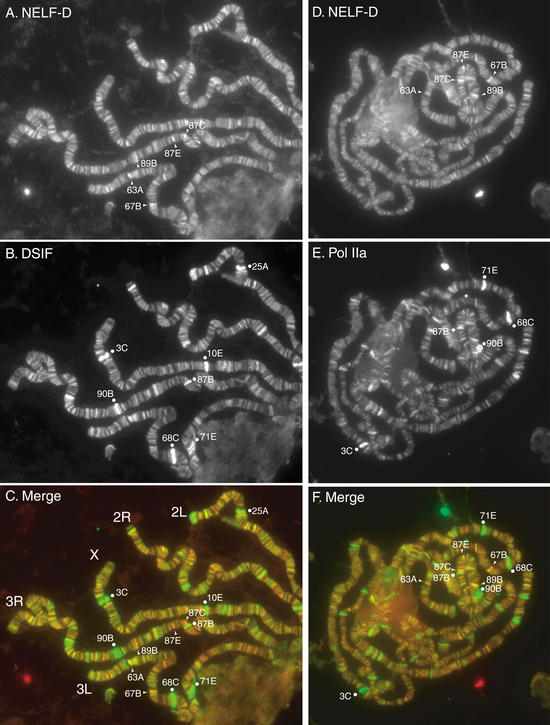
Figure 6.
Immunolocalization of NELF-D, DSIF, and Pol IIa. Chromosome spreads from larvae containing an hsp70 transgenic promoter at 87E were stained with either a combination of affinity-purified rabbit anti-NELF-D antibody and guinea pig anti-DSIF antibody (A–C) or a combination of affinity-purified rabbit anti-NELF-D antibody and a monoclonal antibody (8WG16) against the unphosphorylated CTD of Pol IIa (D–F). The chromosomes were also stained with Hoechst to assist in mapping locations on the chromosomes (data not shown). (A) The NELF-D antibody visualized with Alexa 568 conjugated anti-rabbit antibody. (B) The DSIF antibody visualized with Alexa 488 conjugated anti-guinea pig antibody. (C) A merge with red representing NELF-D and green representing DSIF. (D) The NELF-D antibody visualized with Alexa 568 conjugated anti-rabbit antibody. (E) The Pol IIa monoclonal antibody visualized with Alexa 488 conjugated anti-mouse antibody. (F) A merge with red representing NELF-D and green representing Pol IIa.
A band of fluorescence on a polytene chromosome could encompass many genes and several hundred kilobases of DNA. To test if the fluorescence at 87C could be caused by association between NELF-D and the hsp70 promoter, we compared chromosomes from larvae that had a copy of the hsp70 promoter inserted at 87E to chromosomes lacking this insertion. The_hsp70_ promoter had been inserted as part of a P-element construct, and it consisted of nucleotides -194 to +84 (Wu et al. 2001). Genomic footprinting with permanganate showed that a paused polymerase was present on this promoter (data not shown). Inspection of 20 different chromosomes derived from three different slides for each type of larvae clearly showed that the staining for NELF-D at 87E was enhanced by the presence of the hsp70 promoter. Representative examples are shown inFigure 5E. The band marked with a white dot serves as a reference. NELF-D staining exceeds the intensity of staining at the reference only when the transgenic promoter is present. In addition, NELF-D staining of 87A is evident in all samples where the chromosomes are well spread in this region. Two copies of hsp70 reside at 87A. Taken together, these data corroborate the results of the ChIP analysis showing that NELF associates with the hsp70 promoter before heat shock induction.
Comparison of the distributions of NELF-D and DSIF
We compared the distributions of NELF-D and DSIF because NELF and DSIF collaborate to inhibit transcriptional elongation in vitro (Yamaguchi et al. 1999a). Double-label immunofluorescence reveals that both proteins are broadly distributed among interband regions (Fig. 6A,B). The intensity of staining for DSIF, however, is much more varied than the staining for NELF-D. The sites staining very strongly for DSIF (3C, 10E, 90B, 68C, and 71E) are all sites registered as puffs in third-instar larvae and are regions of high transcriptional activity (Ashburner 1972). Notably, NELF-D does not colocalize with DSIF in these highly transcribed regions.
Outside the regions of intense DSIF staining, there is significant colocalization of NELF-D and DSIF. The relationship between the intensities of fluorescence and the quantities of protein are not known. Therefore, based solely on the intensity of fluorescence, we cannot be certain which regions of the chromosome contain similar amounts of the two proteins. Nevertheless, we observe that the relative intensity of staining for both NELF-D and DSIF at 87A, 87C, and 87E are similar. Moreover, when the intensities of the two colors were adjusted so the merged image appeared yellow at these three loci, 67B also appeared yellow (Fig. 6C). As these loci all harbor paused Pol II, these results are consistent with the hypothesis that these locations contain amounts of NELF-D and DSIF that cause pausing.
Comparison of the distributions of NELF-D and Pol IIa
Pol II paused on the hsp70 promoter before heat shock is in the Pol IIa state (Weeks et al. 1993; O'Brien et al. 1994). The results in Figure 6D and E show that NELF-D and Pol IIa localize to interbands, but as was observed for DSIF, the intensity of staining for Pol IIa is much more varied than the staining for NELF-D. The regions most strongly stained for Pol IIa correspond to highly transcribed regions (71E, 68C, 90B, 3C). Although these regions are known to be enriched in Pol IIo, Weeks et al. (1993) reported previously that antibody against Pol IIa strongly stained highly transcribed regions. Some heptamers constituting the CTD may remain unphosphorylated, or the antibody may weakly bind heptamers phosphorylated on Ser 5 (Cho et al. 2001). Close examination of 87E revealed that Pol IIa staining was enhanced in cases when the hsp70 transgene was present but not in those cases when it was absent (data not shown).
DSIF but not NELF accumulates on the hsp70 gene following heat shock induction
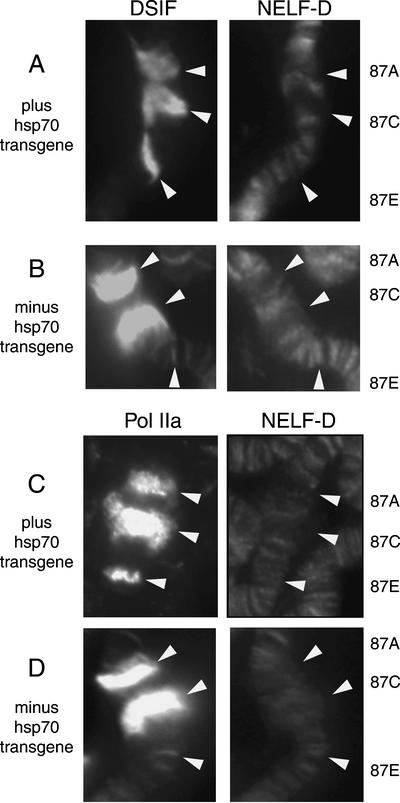
We were intrigued to find that antibody against NELF-D failed to stain the highly transcribed regions on polytene chromosomes that were marked by strong staining against DSIF or Pol IIa (Fig. 6C,F). Perhaps NELF-D dissociates from the Pol II when an activator causes release of paused Pol II. To test this hypothesis, we determined if NELF-D associates with the hsp70 genes during heat shock induction. A 20-min heat shock induces transcription of the heat shock genes and causes 87A and 87C to become puffed (Fig. 7). 87E also becomes puffed in the case where an hsp70 transgene is inserted. The antibodies against DSIF or Pol IIa strongly stained 87A and 87C in accord with previous reports (Andrulis et al. 2000; Kaplan et al. 2000), and 87E when the hsp70 transgenic promoter was present (Fig. 7A,C). In contrast, NELF-D antibody only weakly stained puffed regions. Therefore, NELF-D appears to dissociate from Pol II during active transcriptional elongation.
Figure 7.
DSIF is strongly recruited to the hsp70 gene during heat shock induction but NELF is not. Chromosome spreads were prepared from larvae that had been heat shocked for 20 min at 37°C. (A,B) DSIF and NELF-D staining patterns. (C,D) Pol IIa and NELF-D staining patterns. The samples in A and C were from larvae that have a transgenic_hsp70_ promoter inserted at 87E. 87A and 87C are the locations for the endogenous hsp70 genes.
Discussion
Many proteins have been identified that stimulate transcriptional elongation possibly by alleviating barriers to elongation (Shilatifard 1998). TFIIF, TFIIS, and elongin can stimulate elongation by purified polymerase and function in part to overcome intrinsic features of the nucleic acid sequence that seem to interfere with the movement of Pol II. FACT, elongator, and Swi/Snf seem to function to alleviate barriers to elongation imposed by the chromatin structure. Whether or not these factors are used in gene-specific ways to control gene expression is not known. Evidence for activator-dependent control of elongation has only been provided for Swi/Snf (Brown et al. 1996;Sullivan et al. 2001). Many of these elongation factors could represent part of the repertoire of elongation factors that is called into action at any gene depending on what barriers the polymerase encounters. In contrast, our results provide evidence that DSIF and NELF could be at the heart of a widely used mechanism for implementing gene-specific control of expression.
We have taken a multifaceted approach to provide both physical and functional evidence that NELF and DSIF participate in promoter proximal pausing. Chromatin immunoprecipitation analysis provided physical evidence that NELF and DSIF are associated with the hsp70 promoter region before heat shock. This was corroborated by immunofluorescence analysis of polytene chromosomes. Two types of functional data indicate that NELF and DSIF participate in promoter proximal pausing on hsp70. Immunodepletion of DSIF caused a decrease in the level of paused polymerase that was reconstituted on the hsp70 promoter in nuclear extracts. Attempts to determine if immunodepleting NELF from the nuclear extract affects pausing in vitro have been marginally successful (data not shown). We observed a slight decrease in the permanganate reactivity at +22 after depleting NELF, but Western blot analysis indicated that the NELF depletions were incomplete. We have successfully implemented an RNAi approach to evaluate the contribution of NELF toward pausing on hsp70 in live cells. The permanganate reactivity of thymines in the promoter proximal region of the RNAi salivary glands is twofold less than the reactivity observed for either parent. Note that genomic footprinting with permanganate is a very sensitive way to detect relative levels of polymerase associated with the DNA in salivary glands (Tang et al. 2000). Therefore the decrease in permanganate reactivity caused by the RNAi indicates that there is less paused polymerase. The loss of pausing in RNAi flies was incomplete. A low level of NELF-E persists in spite of the interfering RNA (data not shown). The residual NELF-E may be sufficient for pausing to occur because the paused state appears to be very stable once formed (see below).
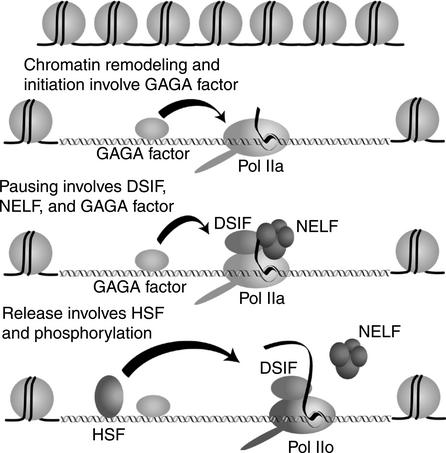
Figure 8 provides a model for the control of promoter proximal pausing on the hsp70 promoter. DSIF and NELF have a central role in the pausing process by associating with Pol IIa during elongation. The generation of the paused polymerase is preceded by steps that participate in chromatin remodeling and initiation. GAGA factor associates with the hsp70 promoter before heat shock induction (Giardina et al. 1992;O'Brien et al. 1995;Weber et al. 1997). GAGA factor interacts with the chromatin remodeling factor, NURF, and could target chromatin remodeling activity to the promoter (Tsukiyama et al. 1994;Xiao et al. 2001;Badenhorst et al. 2002). A preinitiation complex would then assemble. It is not clear what proteins in addition to components of the general transcription machinery participate in assembly of the initiation complex.
Figure 8.
Model for hsp70 transcriptional regulation. GAGA factor participates in chromatin remodeling, initiation, and possibly in pausing. DSIF and NELF associate with Pol IIa shortly after initiation. The nascent transcript associates with NELF when it is 20–40 nucleotides, and this association restricts the forward movement of the polymerase. Heat shock induces HSF to bind DNA. HSF causes the Pol II to resume elongation by triggering the dissociation of NELF.
We propose that promoter proximal pausing occurs when the nascent transcript emerges from the RNA exit channel of the Pol II and is grabbed by the NELF-E subunit. Tethering of the NELF-E to the elongation complex would generate a rigid body that could restrict the movement of the Pol IIa. This model is supported by several observations. The paused polymerase is in the Pol IIa state (O'Brien et al. 1994), and NELF and DSIF only inhibit elongation by Pol IIa (Yamaguchi et al. 1999a). In vitro transcription analysis indicates that the elongation complex is not receptive to inhibition by NELF and DSIF until the nascent transcript is ∼30 nucleotides long (Yamaguchi et al. 1999a). This length coincides approximately to the distance polymerase elongates on hsp70 before it pauses (Rasmussen and Lis 1993). In vitro transcription analyses indicate that DSIF and NELF associate with polymerase shortly after initiation but probably before the polymerase reaches the region of pausing (Ping and Rana 2001; Bourgeois et al. 2002). Finally, NELF-E has an RNA-binding motif that is essential for its inhibitory action (Yamaguchi et al. 2002).
Although NELF and DSIF are sufficient to slow the elongation rate of purified Pol IIa, we suspect additional proteins are involved in stably pausing Pol II on the hsp70 promoter. In cell-free transcription reactions done with other promoters, the pausing caused by DSIF and NELF appears to be transient —the polymerase eventually moves forward if given enough time (Wada et al. 1998a). In contrast, several observations indicate that the Pol II on hsp70 is stably paused. The paused Pol II remains associated with the hsp70 promoter when nuclei are isolated from uninduced cells, and sarkosyl or high salt must accompany addition of nucleotides to cause the Pol II to resume elongation (Rougvie and Lis 1988). In a cell-free system, Pol II remains stably paused on the_hsp70_ promoter for at least 30 min (Li et al. 1996). GAGA factor might be involved in stabilizing the pause because mutations in the GAGA element result in a loss of paused Pol II (Lee et al. 1992).
Heat shock rapidly induces transcription as a result of the association of HSF with sites located upstream from the TATA element (Jedlicka et al. 1997). Our data suggest that HSF may activate transcription in part by causing NELF to dissociate from the Pol II. How HSF might cause the release of NELF is unclear. Phosphorylation of Pol IIa is likely to be an important step as the Pol II found in the body of the gene during heat shock is hyperphosphorylated (Weeks et al. 1993). Phosphorylation of DSIF is another possibility as this has been observed to occur early in elongation in vitro (Bourgeois et al. 2002). It is also unclear which kinase might be responsible for phosphorylating the Pol II. P-TEFb is a candidate because it associates with the hsp70 gene during heat shock induction, and HSF can be bypassed by directing a Gal4/P-TEFb fusion protein to the hsp70 promoter (Lis et al. 2000). No interaction, however, has been detected between P-TEFb and HSF (Lis et al. 2000). Recent results show that HSF associates with the mediator (Park et al. 2001b).Drosophila mediator contains a kinase that phosphorylates the CTD, and phosphorylation can occur synergistically with the TFIIH kinase (Park et al. 2001a). Perhaps HSF recruits the mediator and in turn the mediator releases the paused polymerase by phosphorylating the CTD.
The strong immunofluorescence staining observed for DSIF at heat shock loci during heat shock indicates that DSIF is associated with many of the polymerase molecules transcribing the gene. RNA polymerase initiates at a rate of once every few seconds during heat shock resulting in a train of elongation complexes traversing the gene (Gilmour and Lis 1985). In the absence of NELF, DSIF might act as a positive elongation factor. Shortly after DSIF was discovered, another investigation identified DSIF as a cofactor required for reconstituting tat-dependent transcription (Wu-Baer et al. 1998). In this situation, DSIF appears to be stimulating elongation. DSIF has been found in a complex with another positive elongation factor called Tat-SF1 (Kim et al. 1999; Parada and Roeder 1999). Tat-SF1 was first identified as a stimulatory factor for Tat, but subsequent results indicate that Tat–SF1 may promote elongation on cellular genes (Li and Green 1998). In yeast, DSIF appears to act as either a positive or negative regulator of elongation depending on circumstances (Hartzog et al. 1998). A hypothesis that unites the positive and negative activities of DSIF is to consider this factor an adaptor that connects other modulators to the elongation complex. In this regard, DSIF has been shown to bind on its own to Pol II (Yamaguchi et al. 1999b), whereas the stable association of NELF with Pol II requires the presence of DSIF (Yamaguchi et al. 2002).
NELF and DSIF appear to associate with several hundred interbands in polytene chromosomes. Each interband could contain many genes. The weak staining of interbands by Hoecsht suggests that the DNA in the interbands is in a decondensed state. Residing in these decondensed regions could be genes whose primary control mechanism does not involve a disruption of chromatin structure or even assembly of the initiation complex. Instead, alleviating repression by NELF and DSIF could underlie the mechanism of activation.
Materials and methods
Immunodepletion analysis of nuclear extracts
Drosophila embryo nuclear extract was prepared as described by Biggin and Tjian (1988) except the final ammonium sulfate precipitate was dissolved in 60% the prescribed volume of buffer C modified to contain 0.45 M KCl. The extract was dialyzed against buffer C modified to contain 0.45 M KCl until the conductivity of the extract was equivalent to 0.5 M KCl/HEMG. HEMG is 25 mM HEPES at pH 7.6, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and 0.1 mM PMSF. The extract was clarified by centrifugation and stored at -70°C before depletion. Guinea pig antiserum (200 μL) raised against the largest subunit of DSIF (dSpt5) or an irrelevant protein were covalently coupled to 200 μL of protein A sepharose using dimethylpimelimidate (Harlow and Lane 1999). For each depletion, 600 μL of nuclear extract was incubated at 4°C for ∼3 h with 100 μL of antibody-sepharose. The extract was separated from the sepharose by a brief centrifugation and then incubated with a fresh 100 μL portion of antibody-sepharose. While the second cycle of depletion was occurring, the first portion of antibody-sepharose was washed several times with 0.5 M KCl/HEMG, followed by 0.1 M glycine at pH 2.5, followed by 0.5 M KCl/HEMG. Nuclear extract recovered from the second cycle of depletion was depleted a third time using the regenerated portion of antibody-sepharose. The extract from the third cycle of depletion was dialyzed for 2.5 h against 0.14 M KCl/HEMG. Protein concentrations were determined to be 19 mg/mL for both extracts. Immunoblot analysis indicated that at least 90% of the DSIF was removed from the extract.
Partial purification of DSIF
Nuclear extract from ∼100 grams of embryos was prepared as described above except that the final ammonium sulfate pellet was dissolved in 0.7 M KCl/0.01% NP-40/HEMG and dialyzed against the same solution until the conductivity was equivalent to 0.75 M KCl/0.01% NP-40/HEMG. Then, 14.7 mL of extract was fractionated on a 1.8-L Sephacryl-300 column. Peak fractions of DSIF were identified by immunoblotting, pooled, and dialyzed against 0.1 M KCl/0.01% NP-40/HEMG. The pooled fraction was loaded onto an 8-mL MonoS column and eluted with a 160-mL gradient from 0.1 to 0.3 M KCl. Pooled fractions containing DSIF were loaded onto a 1-mL MonoQ column. The column was washed with 0.2 M KCl/0.01% NP40/HEMG, and DSIF was eluted with 0.3 M KCl/0.01% NP40/HEMG. The total yield of DSIF was ∼4% and the DSIF was enriched ∼25-fold.
Permanganate analysis of paused polymerase in reconstituted transcription reactions
Ten microliters of immunodepleted extract was combined with 10 μL of 0.15 M KCl/0.01% NP-40/HEMGN or 10 μL of partially purified DSIF and the mixture was maintained on ice. Transcription reactions were performed as described previously (Li et al. 1996; Benjamin and Gilmour 1998). After a 30-min incubation at 22°C, samples were subjected to permanganate analysis as described previously (Benjamin and Gilmour 1998).
Cloning and production of antibodies
A BLAST search of the Drosophila genome using the sequence for human NELF-D identified EST clone LD42626. LD42626 DNA was obtained from the Berkeley Drosophila Genome Project, and sequencing showed that the clone contained an open reading frame encoding a 579 amino acid polypeptide with homology to human NELF-D. A primer set, 5′-GCATATCC ATATGCAGGAATGCCTGGAGAAGTTCC-3′ and 5′-GCGG ATCCTTATTCGGCCAGTCGGTAGATTAGC-3′, was used to amplify the region of LD42626 encoding amino acids 33–154. The amplified DNA fragment was digested with _Nde_I and _Bam_HI, and then subcloned into the pET-28-(a) expression vector (Novagen). The resulting 6x-His–tag fusion protein was expressed in BL21(DE3) cells and purified on a Ni-chelated column. Purified recombinant 6x-His–NELF-D protein was injected into rabbits to generate antibody.
A BLAST search identified a non-66Da gene (Saeboe-Larssen et al. 1997) as a possible Drosophila counterpart of human NELF-E. The entire open reading frame encoding a 280-aminoacid protein was amplified using primers 5′-CGCATATGGTT TACATACACTTCCCCAAC-3′ and 5′-CGGGATCCTTAGA GCAAGAAGTCTTCATCGTA-3′, and a Drosophila embryo cDNA library (Novagen) as template. The amplified DNA fragment was digested with_Nde_I and _Bam_HI, and then subcloned into pET-14b. After transformation into BL21(DE3) cells, 6x-His–NELF-E was expressed and purified on a Ni-chelated column. Drosophila NELF-E antibody was raised by injecting recombinant NELF-E protein into rabbits.
Immunoblotting
Nuclear extract was prepared from Drosophila embryos as described by Biggin and Tjian (1988). Proteins were separated on 4%–15% gradient SDS-PAGE and transferred to nitrocellulose membrane. NELF-D and NELF-E were detected by anti-NELF-D (1:5000 dilution) and anti-NELF-E (1:5000 dilution) antiserum, respectively. Preimmune serum (1:5000) served as a negative control.
Immunoprecipitation of NELF from crude nuclear extracts
One hundred microliters of serum was incubated with 20 μL of 50% protein A sepharose beads at 4°C for 1 h. The beads were washed three times with 1 mL 0.15 M KCl/HEMG and most of the liquid was removed after each wash. Ten microliters of Drosophila embryo nuclear extract was mixed with 90 μL of 0.15 M KCl/HEMG, and the diluted extract was incubated with antibody beads for 2 h at 4°C. The antibody beads were collected by brief centrifugation and ∼100 μL of supernatant was transferred to a fresh tube containing 100 μL of 2× SDS-PAGE buffer. The antibody beads were washed three times with 1-mL aliquots of 0.15 M KCl/HEMG. The antibody beads were suspended in 100 μL of 1× SDS-PAGE buffer. Ten microliters of the supernatant and dissolved immunoprecipitate were run on 4%–15% polyacrylamide gradient SDS-PAGE, and NELF-D and NELF-E were detected by immunoblotting.
Chromatin immunoprecipitation analysis
Ten milliters of Drosophila S2 cells were grown to a density of 5 × 106/mL in T-flasks at 25°C in Schneider's Drosophila medium (GIBCO-BRL) supplemented with 10% heat-inactivated fetal bovine serum. Formaldehyde cross-linking was done by adding 675 μL of 37% formaldehyde stock solution directly to the culture medium in the flask giving a final concentration of 1%. Cells were incubated at room temperature for 10 min, dislodged from the surface of the T-flask, transferred to centrifuge tubes on ice, and collected by centrifugation at 700_g_ for 10 min. Subsequent steps were done according to the chromatin immunoprecipitation procedure provided by Upstate Biotechnology. The immunoprecipitated DNA recovered from 5 × 106 cells was dissolved in 20 μL of TE for PCR reactions.
To prepare DNA representing input DNA, 200 μL of the chromatin solution (derived from 1 × 106 cells) was combined with 300 μL of elution buffer. Reversal of formaldehyde cross-links and isolation of the DNA was done as described for DNA eluted from the immunoprecipitates. DNA was resuspended in 20 μL of TE for PCR reactions. Two microliters of this solution corresponded to 20% of the input and other portions were diluted so that 2 μL contained 10%, 1%, or 0.1% of the input material.
Two microliters of each immunoprecipitated DNA or input DNA were subjected to a PCR reaction in a final volume of 50 μL containing 10 pmoles of each primer, 200 μM dNTP, 2 or 4 μCi 32P-dCTP, 2.5 Units of_Taq_ polymerase and buffer (Roche Diagnostics). Each sample was subjected to 20 cycles of: 94°C, 1 min; 60°C, 55 sec; 72°C, 50 sec; and followed by a final extension at 72°C for 10 min. PCR primers were the following: 70 ChIP A.1 spans from -48 to -24 and has the sequence 5′-AAAAGAGCGCCGGAGTATAAATAGA-3′; 70 ChIP A.2 spans from +250 to +229 and has the sequence 5′-GCAGGCAT TGTGTGTGAGTTCT-3′; 70 ChIP B.1 spans from +1357 to +1374 and has the sequence 5′-CTGTGCAGGCCGCTATCC 3′; 70 ChIP B.3 spans from +1730 to +1710 and has the sequence 5′-GGCCTTGCCCGTACTCATCTC-3′. Ten microliters of each PCR reaction was run on a 6% nondenaturing acrylamide. Gels were fixed in 10% acetic acid for 20 min and then dried. Radioactivity was detected in the dried gel using a phosphoimager (Molecular Dynamics). The intensity of each band was quantified by volume analysis using ImageQuant software. The accuracy of the assay was assessed by determining how well the measurements for inputs matched the expected ratio of 0.1:1: 10. In all cases, measurement with an expected value of 0.1 fell between 0.088 and 0.16, and the measurement with an expected value of 10 fell between 8.7 and 10.1.
Permanganate genomic footprinting
The fly line 17A10 containing a NELF-E RNAi construct controlled by the Gal4 UAS has been described previously (Enerly et al. 2002). To direct expression of RNAi in salivary glands, the NELF-E RNAi flies were mated to the Bloomington stock#1824 [genotype: y_1 w*; P(w+mW.hs = GawB)AB1] that expresses Gal4p in salivary glands. Permanganate genomic footprinting was performed on isolated salivary glands as described previously (Weber et al. 1997; Tang et al. 2000). DNA breaks in the nontranscribed strand of the_hsp70 genes at the 87A locus were detected using three overlapping primers that span the region from +165 to +122 (LM1–87A140, 5′-GATTTCAGTAGTTGCAGTTTT TAG-3′; LM2–87A140, 5′-GTAGTTGCAGTTTTTAGTTTAA TTACTTGG-3′; LM3–87A140, 5′-GCAGTTTTTAGTTTAAT TACTTGGTTGTTGG-3′).
Immunofluorescence analysis
Polytene chromosome spreads were prepared as described by Champlin et al. (1991). Antibody against NELF-D was affinity-purified as described by Benyajati et al. (1997) and used at a dilution of 1/20. The antibody against the largest subunit of DSIF (dSpt5) was HMGP11, described by Kaplan et al. (2000), and it was used at a dilution of 1/150. The antibody against Pol IIa was monoclonal antibody 8WG16 obtained from Covance (Richmond CA), and it was used at a dilution of 1/10. All primary antibodies were detected with 1/250 dilutions of Alexa 488 or Alexa 568 conjugated secondary antibodies.
Acknowledgments
We thank Craig Kaplan and Fred Winston for providing antiserum against the largest subunit of DSIF (spt5), and Thomas Kusch and Jerry Workman for use of their fluorescence microscope. Antiserum against NELF-D and NELF-E were produced by Centralized Biolabs at Penn State University. This work was supported by research grant GM47477 from the national Institutes of Health to D.S.G., by a Grant for Research and Development Projects in Cooperation with Academic Institutions from the New Energy and Industrial Technology Development Organization to H.H, and by Norwegian Research Council to A.L. and the Swedish Research Council to J.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1091403.
References
- Andrulis E.D., Guzman, E., Doring, P., Werner, J., and Lis, J.T. 2000. High-resolution localization of drosophila spt5 and spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 14**:** 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. 1972. Puffing patterns in Drosophila melanogaster and related species. In Developmental studies of giant chromosomes (ed. W. Beerman), pp. 101–151. Springer-Verlag, N.Y. [DOI] [PubMed]
- Badenhorst P., Voas, M., Rebay, I., and Wu, C. 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes & Dev. 16**:** 3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M., Nissen, R.M., Kanazawa, S., Jabrane-Ferrat, N., and Peterlin, B.M. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8**:** 327–337. [DOI] [PubMed] [Google Scholar]
- Benjamin L.R. and Gilmour, D.S. 1998. Nucleosomes are not necessary for promoter-proximal pausing in vitro on the Drosophila hsp70 promoter. Nucleic Acids Res. 26**:** 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Mueller, L., Xu, N., Pappano, M., Gao, J., Mosammaparast, M., Conklin, D., Granok, H., Craig, C., and Elgin, S. 1997. Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucleic Acids Res. 25**:** 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M.D. and Tjian, R. 1988. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53**:** 699–711. [DOI] [PubMed] [Google Scholar]
- Blau J., Xiao, H., McCracken, S., O'Hare, P., Greenblatt, J., and Bentley, D. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16**:** 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois C.F., Kim, Y.K., Churcher, M.J., West, M.J., and Karn, J. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22**:** 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H., Manoukian, A.S., and Perrimon, N. 1994. Ectopic expression in Drosophila. Methods Cell. Biol. 44**:** 635–654. [DOI] [PubMed] [Google Scholar]
- Brown S.A., Imbalzano, A.N., and Kingston, R.E. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 10**:** 1479–1490. [DOI] [PubMed] [Google Scholar]
- Champlin D.T., Frasch, M., Saumweber, H., and Lis, J.T. 1991. Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes & Dev. 5**:** 1611–1621. [DOI] [PubMed] [Google Scholar]
- Chao S.H. and Price, D.H. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276**:** 31793–31799. [DOI] [PubMed] [Google Scholar]
- Cho E.J., Kobor, M.S., Kim, M., Greenblatt, J., and Buratowski, S. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 15**:** 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L.A., Fire, A., Samuels, M., and Sharp, P.A. 1989. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 264**:** 2250–2257. [PubMed] [Google Scholar]
- Eberhardy S.R. and Farnham, P.J. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276**:** 48562–48571. [DOI] [PubMed] [Google Scholar]
- Enerly E., Larsson, J., and Lambertsson, A. 2002. Reverse genetics in Drosophila: From sequence to phenotype using UAS-RNAi transgenic flies. Genesis 34**:** 152–155. [DOI] [PubMed] [Google Scholar]
- Giardina C., Perez-Riba, M., and Lis, J.T. 1992. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes & Dev. 6**:** 2190–2200. [DOI] [PubMed] [Google Scholar]
- Gilmour D.S. and Lis, J.T. 1985. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol. Cell. Biol. 5**:** 2009–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E. and Lane, D. 1999. Using antibodies: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Hartzog G.A., Wada, T., Handa, H., and Winston, F. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in_Saccharomyces cerevisiae._ Genes & Dev. 12**:** 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P., Mortin, M.A., and Wu, C. 1997. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 16**:** 2452–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C.D., Morris, J.R., Wu, C., and Winston, F. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes & Dev. 14**:** 2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kephart D.D., Marshall, N.F., and Price, D.H. 1992. Stability of Drosophila RNA polymerase II elongation complexes in vitro. Mol. Cell. Biol. 12**:** 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.K., Yamaguchi, Y., Wada, T., and Handa, H. 2001. The regulation of elongation by eukaryotic RNA polymerase II: a recent view. Mol. Cells 11**:** 267–274. [PubMed] [Google Scholar]
- Kim J.B., Yamaguchi, Y., Wada, T., Handa, H., and Sharp, P.A. 1999. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol. Cell. Biol. 19**:** 5960–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho, E.J., and Buratowski, S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 14**:** 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A., Meulia, T., Brunvand, M., and Groudine, M. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes & Dev. 6**:** 2201–2213. [DOI] [PubMed] [Google Scholar]
- Law A., Hirayoshi, K., O'Brien, T., and Lis, J.T. 1998. Direct cloning of DNA that interacts in vivo with a specific protein: application to RNA polymerase II and sites of pausing in Drosophila. Nucleic Acids Res. 26**:** 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P.J. and Dahmus, M.E. 1990. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J. Biol. Chem. 265**:** 13165–13173. [PubMed] [Google Scholar]
- Lee H., Kraus, K.W., Wolfner, M.F., and Lis, J.T. 1992. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes & Dev. 6**:** 284–295. [DOI] [PubMed] [Google Scholar]
- Li B., Weber, J.A., Chen, Y., Greenleaf, A.L., and Gilmour, D.S. 1996. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol. Cell. Biol. 16**:** 5433–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Y. and Green, M.R. 1998. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes & Dev. 12**:** 2992–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63**:** 347–356. [DOI] [PubMed] [Google Scholar]
- Lis J.T., Mason, P., Peng, J., Price, D.H., and Werner, J. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes & Dev. 14**:** 792–803. [PMC free article] [PubMed] [Google Scholar]
- Lu H., Flores, O., Weinmann, R., and Reinberg, D. 1991. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. 88**:** 10004–10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo H.S., Lee, G., Flygare, J., Tomassini, J., Luu, P., Zhu, Y., Peng, J., Blau, C., Hazuda, D., Price, D., et al. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes & Dev. 11**:** 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N.F. and Price, D.H. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270**:** 12335–12338. [DOI] [PubMed] [Google Scholar]
- O'Brien T., Hardin, S., Greenleaf, A., and Lis, J.T. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370**:** 75–77. [DOI] [PubMed] [Google Scholar]
- O'Brien T., Wilkins, R.C., Giardina, C., and Lis, J.T. 1995. Distribution of GAGA protein on Drosophila genes in vivo. Genes & Dev. 9**:** 1098–1110. [DOI] [PubMed] [Google Scholar]
- Parada C.A. and Roeder, R.G. 1999. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 18**:** 3688–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.M., Gim, B.S., Kim, J.M., Yoon, J.H., Kim, H.S., Kang, J.G., and Kim, Y.J. 2001a. Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol. Cell. Biol. 21**:** 2312–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.M., Werner, J., Kim, J.M., Lis, J.T., and Kim, Y.J. 2001b. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8**:** 9–19. [DOI] [PubMed] [Google Scholar]
- Pinaud S. and Mirkovitch, J. 1998. Regulation of c-fos expression by RNA polymerase elongation competence. J. Mol. Biol. 280**:** 785–798. [DOI] [PubMed] [Google Scholar]
- Ping Y.H. and Rana, T.M. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276**:** 12951–12958. [DOI] [PubMed] [Google Scholar]
- Plet A., Eick, D., and Blanchard, J.M. 1995. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene 10**:** 319–328. [PubMed] [Google Scholar]
- Rasmussen E.B. and Lis, J.T. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. 90**:** 7923–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A.E. and Lis, J.T. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54**:** 795–804. [DOI] [PubMed] [Google Scholar]
- Saeboe-Larssen S., Urbanczyk Mohebi, B., and Lambertsson, A. 1997. The Drosophila ribosomal protein L14-encoding gene, identified by a novel Minute mutation in a dense cluster of previously undescribed genes in cytogenetic region 66D. Mol. Gen. Genet. 255**:** 141–151. [DOI] [PubMed] [Google Scholar]
- Sehgal P.B., Darnell Jr., J.E., and Tamm, I. 1976. The inhibition by DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell 9**:** 473–480. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. 1998. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 12**:** 1437–1446. [DOI] [PubMed] [Google Scholar]
- Sullivan E.K., Weirich, C.S., Guyon, J.R., Sif, S., and Kingston, R.E. 2001. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol. Cell. Biol. 21**:** 5826–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I. and Kikuchi, T. 1979. Early termination of heterogeneous nuclear RNA transcripts in mammalian cells: Accentuation by 5,6-dichloro 1-beta-D-ribofuranosylbenzimidazole. Proc. Natl. Acad. Sci. 76**:** 5750–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Liu, Y., Madabusi, L., and Gilmour, D.S. 2000. Promoter-proximal pausing on the hsp70 promoter in drosophila melanogaster depends on the upstream regulator. Mol. Cell. Biol. 20**:** 2569–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Becker, P.B., and Wu, C. 1994. ATP-dependent nucleosome disruption at a heat shock promoter mediated by binding of GAGA transcription factor. Nature 367**:** 525–532. [DOI] [PubMed] [Google Scholar]
- Wada T., Takagi, T., Yamaguchi, Y., Ferdous, A., Imai, T., Hirose, S., Sugimoto, S., Yano, K., Hartzog, G.A., Winston, F., et al. 1998a. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 12**:** 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Takagi, T., Yamaguchi, Y., Watanabe, D., and Handa, H. 1998b. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17**:** 7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.A., Taxman, D.J., Lu, Q., and Gilmour, D.S. 1997. Molecular architecture of the hsp70 promoter after deleting the TATA box or the upstream regulatory region. Mol. Cell. Biol. 17**:** 3799–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks J.R., Hardin, S.E., Shen, J., Lee, J.M., and Greenleaf, A.L. 1993. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: Correlations with gene activity and transcript processing. Genes & Dev. 7**:** 2329–2344. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Madabusi, L., Nishioka, H., Emanuel, P., Sypes, M., Arkhipova, I., and Gilmour, D.S. 2001. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol. Cell. Biol. 21**:** 1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F., Lane, W.S., and Gaynor, R.B. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol. 277**:** 179–197. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos, R., Wang, H.M., Hamiche, A., Ranallo, R., Lee, K.M., Fu, D., and Wu, C. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8**:** 531–543. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Wada, T., and Handa, H. 1998. Interplay between positive and negative elongation factors: Drawing a new view of DRB. Genes Cells 3**:** 9–15. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Takagi, T., Wada, T., Yano, K., Furuya, A., Sugimoto, S., Hasegawa, J., and Handa, H. 1999a. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97**:** 41–51. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Wada, T., Watanabe, D., Takagi, T., Hasegawa, J., and Handa, H. 1999b. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274**:** 8085–8092. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Inukai, N., Narita, T., Wada, T., and Handa, H. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22**:** 2918–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]