Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein (original) (raw)
Abstract
Sequestration of the transcriptional coactivator CREB-binding protein (CBP), a histone acetyltransferase, has been implicated in the pathogenesis of polyglutamine expansion neurodegenerative disease. We used a Drosophila model to demonstrate that polyglutamine-induced neurodegeneration is accompanied by a defect in histone acetylation and a substantial alteration in the transcription profile. Furthermore, we demonstrate complete functional and morphological rescue by up-regulation of endogenous Drosophila CBP (dCBP). Rescue of the degenerative phenotype is associated with eradication of polyglutamine aggregates, recovery of histone acetylation, and normalization of the transcription profile. These findings suggest that histone acetylation is an early target of polyglutamine toxicity and indicate that transcriptional dysregulation is an important part of the pathogenesis of polyglutamine-induced neurodegeneration.
Keywords: Polyglutamine, Drosophila, CREB-binding protein (CBP), histone acetylation, gene expression analysis, retinal degeneration
Huntington's disease, spinobulbar muscular atrophy, dentatorubro-pallidoluysian atrophy, and six forms of spinocerebellar ataxia are caused by expansion of CAG trinucleotide repeats, resulting in pathological polyglutamine expansion in the disease proteins (Taylor et al. 2002). These diseases likely share a common mechanism involving a toxic gain of function by the expanded polyglutamine tract. Evidence indicates that the nucleus is an important site of polyglutamine toxicity and that transcriptional dysregulation may be a primary pathogenic process in polyglutamine disease (Klement et al. 1998; Saudou et al. 1998; Lin et al. 2000; Luthi-Carther et al. 2000). Mutant proteins with expanded polyglutamine have been shown to bind and sequester the transcriptional coactivator CREB-binding protein (CBP) in cell culture, animal models, and tissue derived from patients with these diseases (McCampbell et al. 2000; Steffan et al. 2000; Nucifora et al. 2001). Physical interaction between CBP and mutant protein has been found to depend on the acetyltransferase domain of the former and the polyglutamine tract of the latter (Steffan et al. 2001). Sequestration of CBP results in reduced acetyltransferase activity and loss of CBP-mediated coactivation in cell culture models of polyglutamine disease (McCampbell et al. 2000; Nucifora et al. 2001; Steffan et al. 2001). Live-cell dynamic imaging showed that coexpression of GFP-CBP with various proteins with polyglutamine expansions in cell cultures results in not merely colocalization, but functional sequestration of CBP in polyglutamine inclusions (Chai et al. 2002). CBP is an important cofactor in transcriptional activation mediated by CREB (Chrivia et al. 1993). A recent report demonstrates that disruption of CREB function leads to progressive neurodegeneration in a pattern similar to that observed in transgenic mouse models of polyglutamine disease (Mantamadiotis et al. 2002). CBP also serves as a cofactor for other transcription factors in addition to CREB, but this finding indicates that disruption of the CREB pathway alone is sufficient to lead to neurodegeneration and further implicates CBP as a target of polyglutamine toxicity.
Results and Discussion
Morphological and functional rescue of polyglutamine-mediated toxicity
To investigate the role of histone acetylation in polyglutamine disease, we used a Drosophila model of polyglutamine toxicity (Jackson et al. 1998; Warrick et al. 1998; Fernandez-Funez et al. 2000; Kazemi-Esfarjani and Benzer 2000). Polyglutamine-induced neurodegeneration is recapitulated in Drosophila with a GAL4/UAS system and a transgene expressing 127 consecutive glutamines fused with an HA tag (127Q). A genetic cross with a line that expresses GAL4 under the control of the glass mul-timer repeat (GMR) promoter resulted in expression of 127Q in all cells of the retina and led to eye-specific degeneration. As seen in Figure 1B, external eye pigmentation was lost, and the regular array of ommatidia was disrupted. In contrast, retinal expression of 20Q did not induce degeneration, demonstrating that polyglutamine toxicity is length-dependent (data not shown). To evaluate the role of CBP in this process, we crossed the GMRGAL4/UAS-127Q flies with line EP1179, which carries a P(EP)-element insertion 3 kb upstream of the gene nejire, encoding the Drosophila ortholog of CBP (dCBP). The P(EP) insertion results in increased dCBP expression in the presence of the GAL4 transcription factor (Fig. 1G; Marek et al. 2000). This up-regulation of dCBP provided complete morphological rescue of polyglutamine-mediated degeneration with restoration of eye pigmentation and normalization of the ommatidial array (Fig. 1C). Identical rescue was found in a genetic cross with an independent P(EP) line, EP1149, which contains a P(EP) insertion 3 kb upstream of nejire and also up-regulates dCBP in response to GAL4 (data not shown). The importance of dCBP activity in modifying polyglutamine toxicity is underscored by the exacerbated degeneration observed in 127Q-expressing flies crossed with the line P3728, in which one copy of the nejire gene is disrupted by a P-element insertion (Fig. 1D). These results suggest that the level of dCBP expression is an important factor in determining the severity of polyglutamine toxicity.
Figure 1.
Polyglutamine-induced eye degeneration is rescued by up-regulation of dCBP. (A) Control fly expressing the transcription factor GAL4 under control of the eye-specific promoter GMR. (B) Expression of 127Q driven by GAL4 leads to progressive eye degeneration. (C) Up-regulation of dCBP expression in concert with 127Q prevents polyglutamine toxicity. (D) Down-regulation of dCBP expression in concert with 127Q exacerbates polyglutamine toxicity. LM, light microscopy; SEM, scanning electron microscopy. (E) Rescue of phototactic behavior by augmentation of dCBP expression. In the multiple-trial countercurrent distribution experiment, the ordinate represents flies that responded positively, and the number of positive responses from 1 to 6 is indicated by the abscissa. GMR-GAL4;UAS-127Q flies showed poor performance. GMR-GAL4;UAS-127Q;EP1179 dCBP flies perform as well as control GMR-GAL4 flies. n = 100 flies, 5d of age, for each graph. (F) 127Q mRNA expression is not significantly altered by up-regulation of dCBP as demonstrated by real-time PCR quantitation. (G) Polyglutamine-induced degeneration is accompanied by reduced dCBP expression. Rescue by up-regulation of endogenous dCBP is accomplished by normalization of dCBP expression.
Prior studies have identified modifiers of polyglutamine toxicity in Drosophila based on morphological criteria, but the strength of the suppressors has been mixed, and none has been shown to rescue eye function. To test whether dCBP overexpression is able to rescue the degenerative eyes functionally, we used the countercurrent phototaxis test in which populations of flies are characterized according to the number of positive responses in movement towards light (Min and Benzer 1999). Flies that have normal vision move repeatedly to light in each trial, whereas blind flies do not detect light and distribute randomly. As shown in Figure 1E, GMR-GAL4/UAS-127Q flies did not respond to light, exhibiting random distribution in the countercurrent device, indicating that they are essentially blind. In contrast, polyglutamine-expressing flies rescued by up-regulation of dCBP showed normal phototaxis, demonstrating that up-regulation of dCBP also prevents the functional defect caused by polyglutamine toxicity.
To examine the levels of 127Q and dCBP transcripts, we used quantitative real-time PCR analysis. Figure 1F shows that 127Q transcript levels are not significantly altered by up-regulation of dCBP, demonstrating that the rescue of polyglutamine-mediated degeneration observed here is not a consequence of reduced 127Q expression. Polyglutamine-induced degeneration is accompanied by a reduction in levels of dCBP transcript, and this is restored to normal levels in the rescued flies heterozygous for (EP1179) dCBP (Fig. 1G). Proper levels of dCBP appear to be critical for normal eye development because overexpression of dCBP in the absence of 127Q (Fig. 1G) leads to a small eye with a disorganized ommatidial array (data not shown). Thus, overexpression of both 127Q and dCBP in the eye results in normalization of dCBP levels and mutual suppression of the degenerative and developmental phenotypes.
Eradication of polyglutamine aggregates
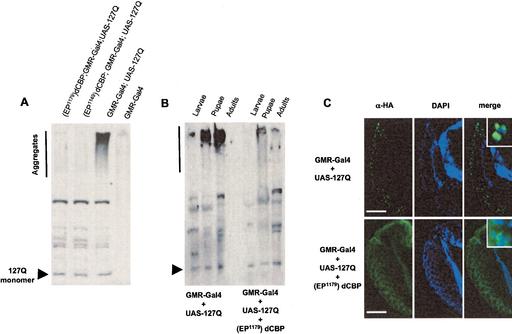
To investigate whether the morphological and functional rescue of polyglutamine toxicity by dCBP overexpression is accompanied by inhibition of polyglutamine aggregation or effects downstream from the aggregate formation, we used both Western blot analysis and immunohistochemistry to detect HA-tagged 127Q. In striking contrast to previously known modifiers of polyglutamine toxicity, such as dHSP70, dHDJ1, dTPR2, and dMLF (Warrick et al. 1999; Kazemi-Esfarjani and Benzer 2000, 2002), dCBP overexpression was associated with no accumulation of polyglutamine aggregates. As seen in Figure 2A, 127Q protein was present in high-molecular-weight, insoluble protein complexes that create a smear in the stacking gel. These high-molecular-weight complexes likely reflect aggregates of 127Q, and they were found to accumulate gradually throughout the life span of degenerating flies (Fig. 2B). A small fraction of 127Q protein migrated at the expected molecular weight for monomeric species with polyglutamine gel electrophoresis. Strikingly, 127Q aggregates did not accumulate to a significant extent in rescued flies, while the amount of monomeric 127Q was unchanged (Fig. 2A,B). To verify these results in the eyes of rescued flies, we further investigated retinal sections of the flies overexpressing both dCBP and 127Q, and control flies containing 127Q alone. Consistent with the Western blot results, immunofluorescence demonstrated that 127Q accumulates in nuclear inclusions in the eye of degenerating flies, but not in flies rescued by up-regulation of dCBP (Fig. 2C). These results suggest that dCBP rescues toxicity at a point prior to polyglutamine aggregation. The mechanism by which dCBP rescues toxicity may be direct (as with the azo-dye Congo red; Sánchez et al. 2003) or indirect by influencing the expression of genes that regulate polyglutamine aggregation. Reduced aggregation would result in a greater proportion of monomeric 127Q, which is expected to be an unstable peptide that is rapidly degraded as is its nonexpanded counterpart 20Q (Kazemi-Esfarjani and Benzer 2000). Alternatively, dCBP may influence the expression of genes that contribute to the degradation of 127Q.
Figure 2.
127Q aggregation and inclusion formation are prevented by increased dCBP. (A) Western blot of adult head tissue demonstrates that up-regulation of dCBP prevents the formation of high-molecular-weight, insoluble 127Q protein complexes. (B) Western blot of whole animal tissue demonstrates that in degenerating flies, polyglutamine aggregate accumulates gradually throughout development, while the steady-state level of polyglutamine monomer remains constant. Flies rescued by up-regulation of dCBP do not show this accumulation of aggregate. (C) Immunofluorescence demonstrates that up-regulation of dCBP prevents the formation of intranuclear polyglutamine inclusions. (Insets) Diffuse polyglutamine staining remains visible in the cytoplasm and nucleus. Bar, 20 μm.
Restoration of the histone acetylation level
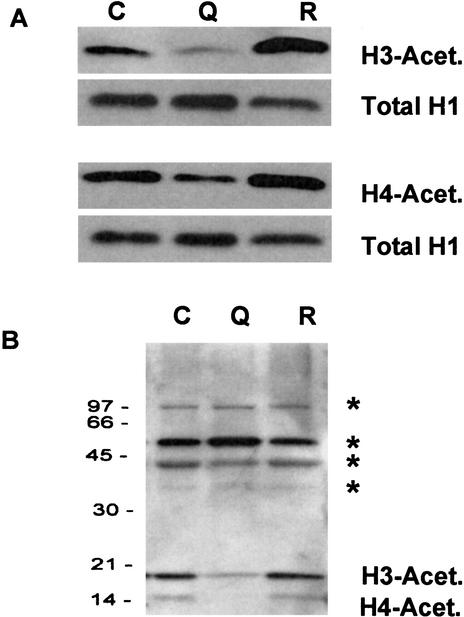
Polyglutamine-induced cell death in cultured cells is associated with defective histone acetylation (McCampbell et al. 2001; Steffan et al. 2001). To investigate whether a similar defect occurs in animals with polyglutamine expansion, we examined the acetylation level of histones H3 and H4 extracted from the nuclear fraction of flies expressing 127Q. We found that the level of histone acetylation was significantly reduced; moreover, this defect was reversed in flies rescued by up-regulation of dCBP (Fig. 3A). The acetylation defect appeared to be specific to histones, as evidenced by the immunoblot pattern of fly heads with an antibody against proteins containing acetyl lysine. This antibody also detected the altered H3 and H4 acetylation pattern, but other prominent acetylation targets showed no change with degeneration or rescue (Fig. 3B).
Figure 3.
Histone acetylation in 127Q and rescued flies. (A) The level of acetylated H3 and H4 is decreased in flies having 127Q, but restored in flies overexpressing CBP. (B) No changes in the acetylation level of other nuclear proteins (as indicated by *) were observed. C, GMR-Gal4; Q, GMR-Gal4;127Q; R, (EP1179)CBP;GMRGal4;127Q.
Transcriptional dysregulation in flies with 127Q is restored by dCBP overexpression
CBP has intrinsic acetyltransferase activity and works in conjunction with sequence-specific transcription factors to regulate the expression of target genes by acetylating histones (Ogryzko et al. 1996). Covalent modification of histones has emerged as an important mechanism controlling the structure and function of chromatin (Cheung et al. 2000). Acetylation of histones appears to alter chromatin structure in a way that promotes increased accessibility of the chromatin to transcription factors. Thus, hyperacetylation of histones marks transcriptionally active regions of chromatin, and hypoacetylation of histones is associated with transcriptionally silent chromatin domains and is a mechanism of transcriptional repression. Two opposing classes of proteins determine histone acetylation status: the histone acetyltransferases and the histone deacetylases. Histone acetyltransferases, such as CBP, are recruited to target promoters by interaction with sequence-specific transcription factors (Marmorstein 2001). Histone deacetylases function as components of multiprotein repressosomes, which include DNA-binding proteins and corepressors (Ng and Bird 2000). The fine balance between histone acetylation and deacetylation plays a major role in determining the patterns of expressed and silent genes that characterize cell physiology.
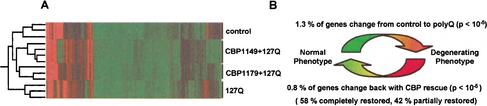
We reasoned that because polyglutamine-induced degeneration in Drosophila is associated with defective histone acetylation, this should be reflected by an altered pattern of gene expression. To investigate this, we examined global expression patterns with whole genome oligonucleotide microarrays. Total RNA was isolated from the heads of GMR-Gal4;UAS-127Q (degenerating) flies during the first 1–2 d of adult eclosion while degeneration was still progressing. Total RNA was also isolated from the heads of GMR-Gal4 (control), EP1179;GMRGal4;UAS-127Q (rescue 1), and EP1149;GMR-Gal4;UAS-127Q (rescue 2) flies of the same age. At least three independent microarray experiments were run for each genotype (four for rescue 2), and the resulting expression profiles were compared by pairwise hierarchical analysis and expressed as a heat map with an accompanying dendrogram to show relatedness (Fig. 4A). Replicates of each genotype showed a high degree of concordance and clustered together. The greatest dissimilarity in expression patterns was seen between the GMR-Gal4 (control) genotype and the GMR-Gal4;UAS-127Q (degenerating) genotype, demonstrating that degeneration is accompanied by an altered expression profile (Fig. 4A). Both rescued genotypes showed high concordance with the control genotype, indicating that the rescued expression profiles had partially normalized (Fig. 4A). The number of genes that showed significantly different expression between the control genotype and the degenerating genotype was relatively small [116 genes (1.3% of 8548 present calls) using Student's t-test with the Bonforoni correction setting, _p_ < 10-5; Fig. 4B]. This relatively small degree of altered expression with mutant polyglutamine is similar to that observed in expression profiles of Saccharomyces cerevisiae mutants with defective acetylation machinery and in cultured cells treated with histone deacetylase inhibitors (Bernstein et al. 2000). Rescue of polyglutamine-induced degeneration by up-regulation of dCBP was accompanied by a similarly small change in the gene expression pattern [68 genes (0.8% of 8548 present calls) using Student's t-test with the Bonforoni correction setting, p < 10-5]. Of note, nearly all of the genes that show altered expression between degenerating → rescued genotypes represented reversals in the direction of change for genes that were altered between control → degenerating genotypes (Fig. 4B). Approximately 58% of these reversals represent complete restoration, and 42% represent partial restoration. Thus, the expression profiles analysis demonstrates that in addition to morphological and functional rescue, up-regulation of dCBP largely restores the normal pattern of gene expression. In essence, analysis of the expression profile patterns permits high-resolution phenotypic analysis documenting strong suppression of the degenerative phenotype induced by polyglutamine. Cluster analysis showing the patterns of altered expression, and lists of those that show reversal, are provided as Supplemental Material. It will be of great interest to implicate specific genes as participating directly in polyglutamine pathogenesis by comparing these alterations with those resulting from degeneration induced by other means.
Figure 4.
Expression profiles of degenerating and rescued flies. (A) Each row shows patterns of gene expression in GMR-Gal4 (1,2,3), (EP1149)CBP;GMR-Gal4;127Q (4,5,6,7), (EP1179)CBP;GMR-Gal4;127Q (8,9,10), and GMR-Gal4;127Q (11,12,13) flies. The expression ratios of two rescued fly lines (1179 and 1149) are closely related to control flies, but are distant from degenerated flies. (B) Transcriptional dysregulation of degenerating flies is restored in the rescued flies.
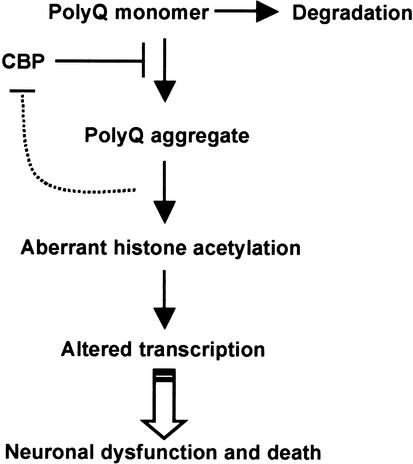
The results presented here demonstrate that CBP is a potent modifier of polyglutamine-induced neurodegeneration in vivo and support the hypothesis that histone acetylation may be a target of polyglutamine toxicity. Alterations in CBP activity by sequestration (McCampell et al. 2000; Steffan et al. 2000; Nucifora et al. 2001), increased degradation (Jiang et al. 2003), or decreased expression as seen here may contribute to altered acetylation. The loss of polyglutamine aggregation seen with rescue by CBP suggests that CBP itself, as a polyglutamine protein, may directly block accumulation of polyglutamine monomers into aggregates, similar to a blocking peptide (Kazantsev et al. 2002), making it more susceptible to degradation. Alternatively, genes regulated by CBP may influence aggregation or accelerate degradation of polyglutamine (Fig. 5). The findings presented here add to the accumulating evidence that pharmacologics capable of influencing histone acetylation may be of benefit in polyglutamine disease (Taylor and Fischbeck 2002).
Figure 5.
Aggregation of polyglutamine is associated with neuronal dysfunction and death. It remains unclear whether the toxic species is an unfolded intermediate, oligomer, or large, organized aggregate. The data presented here suggest that CBP rescues toxicity at an upstream point by preventing aggregation. CBP may inhibit aggregation directly (like a blocking peptide) or indirectly (by influencing the expression of genes that regulate aggregation). Reduced aggregation results in a higher proportion of monomeric 127Q, an unstable peptide that is rapidly degraded. Alternatively, CBP may work indirectly by influencing the expression of genes involved in 127Q degradation.
Materials and methods
Drosophila stocks
Flies were maintained on a standard mixture of corn meal, yeast, and agar at room temperature. The UAS-127Q and GMR-GAL4 flies were described previously (Kazemi-Esfarjani and Benzer 2000). Fly lines P(EP)1179, P(EP)1149, and P3728 were obtained from Exelixis Inc. and the Bloomington fly stock center.
Microscopy
For scanning electron microscopy, 5-day-old adult flies were fixed in 1% gluteraldehyde, dehydrated through a graded ethanol series, immersed in hexamethyldisilazane for 5min, and air-dried. The flies were then mounted and sputter-coated with gold. Polaroid images were obtained on a Hitachi S4500 field emission scanning electron microscope.
Behavioral tests
Phototaxis was performed as described previously (Min and Benzer 1999). Three independent experiments were performed for each strain of flies (∼100 flies total per strain). Standard errors were calculated from these averages.
Real-time quantitative PCR
Total RNA was isolated from ∼100 flies with TRIzol reagent (Life Sciences). The primers and probes for Drosophila GAPDH2 and CBP (product numbers CT25538 and CT35306, respectively) were purchased from Applied Biosystems. Custom primers and the probe for 127Q (F-CAATC TGCAGGCCG, R-GACGGTGGGCCTCCCA, P-CCTGATAAGTGAA TTCGCCGCCACC) were also purchased from Applied Biosystems. All probes were labeled with the fluorescent reporter dye FAM (6-carboxyfluorscein) and the quencher TAMRA (6-carboxy-tetramethyl-rhodamine). A Taqman assay was performed using 50-ng aliquots of total RNA and 0.5μL of Superscript One-Step RT–PCR with Platinum Taq (Invitrogen Life Sciences) in a total volume of 25μL using a separate tube, comparative CT protocol on an ABI 7700 sequence detector. Two independent experiments were carried out with 3–6 replicates, and the data are presented as mean ± standard deviation.
Western blotting
Western blotting was performed with α-HA antibody (Santa Cruz Biotechnology) after SDS-PAGE separation of 50 μg of isolated head lysate (Fig. 2A) or 150 mg of whole animal lysate (Fig. 2B). Immunofluorescence and Western blotting of histones were performed as described previously (Kazemi-Esfarjani and Benzer 2000; Kang et al. 2002).
Microarray analysis
The heads of ∼600 flies were isolated, and total RNA for microarray analysis was extracted using Trizol. Then 30 μg of total RNA from each fly genotype was labeled using the Affymetrix protocol. The intensity values of individual genes from the Affymetrix arrays were quantified using MAS 5.0 (Affymetrix Inc.). Genes were called present (P) if the p-value of the PM/MM probe pairs derived from the MAS 5.0 output was <0.05. The raw intensity values for each array were imported into Excel (Microsoft). Genes that were not called present (P) were set to 0.01. The remaining positive genes from each array were scaled to a percent value using the highest positive signal on that array. To correct for variation between arrays, global normalization to the 75th percentile was performed as described previously (Hegde et al. 2000). Genes that were never present were eliminated from further analysis. The Student's t-test was used to do pairwise comparisons of the remaining 8548 genes between the 127Q mutant strain and the control and rescue strains. P-values were corrected for multiple comparisons using the Bonferroni correction and were considered significant at a p-value <1.17 × 105. Genes were grouped according to the patterns they exhibited (up, down, or neutral changes) between these pairwise comparisons of the mutant strain with the control and rescue strains. Genes were considered rescued if a significant difference was observed between the 127Q mutant and both the control and rescue strains. Genes were considered to be partially rescued if a trend in gene expression was noted in the rescue strain that appeared in the same direction as that seen in the control.
Acknowledgments
We thank N. Bonini for helpful comments and Y. Shi for technical assistance. Support was provided by NIH intramural funds to K.H.F. and K.-T.M., the Ellison Medical Foundation to K.-T.M., and an NIH grant to P.K.-E. J.P.T. was supported by a physician postdoctoral fellowship from the Howard Hughes Medical Institute and by K22 NS44125from NINDS.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1087503.
Footnotes
Supplemental material is available at http://www.genesdev.org.
References
- Bernstein B.E., Tong, J.K., and Schreiber, S.T. 2000. Genome wide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. 97**:** 13708–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Shao, J., Miller, V.M., Williams, A., and Paulson, H.L. 2002. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc. Natl. Acad. Sci. 99**:** 9310–9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Allis, C.D., and Sassone-Corsi, P. 2000. Signaling to chromatin through histone modifications. Cell 103**:** 263–271. [DOI] [PubMed] [Google Scholar]
- Chrivia J.C., Kwok, R.P., Lamb, N., Hagiwara, M., Montminy, M.R., and Goodman, R.H. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365**:** 855–859. [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P., Nino-Rosales, M.L., de Gouyon, B., She, W.C., Luchak, J.M., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P.J., et al. 2000. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408**:** 101–106. [DOI] [PubMed] [Google Scholar]
- Hegde P., Qi, R., Abernathy, K., Gay, C., Dharap, S., Gaspard, R., Hughes, J.E., Snesrud, E., Lee, N., and Quackenbush, J. 2000. A concise guide to cDNA microarray analysis. Biotechniques 29**:** 548–562. [DOI] [PubMed] [Google Scholar]
- Jackson G.R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P.W., MacDonald, M.E., and Zipursky, S.L. 1998. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21**:** 633–642. [DOI] [PubMed] [Google Scholar]
- Jiang H., Nucifora Jr., F.C., Ross, C.A., and DeFranco, D.B. 2003. Cell death triggered by polyglutamine-expanded huntingtin in a neuronal cell line is associated with degradation of CREB-binding protein. Hum. Mol. Genet. 12**:** 1–12. [DOI] [PubMed] [Google Scholar]
- Kang H.L., Seymour, B., and Min, K.-T. 2002. Life extension in Drosophila by feeding a drug. Proc. Natl. Acad. Sci. 99**:** 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A., Walker, H.A., Slepko, N., Bear, J.E., Preisinger, E., Steffan, J.S., Zhu, Y.Z., Gertler, F.B., Housman, D.E., Marsh, J.L., et al. 2002. A bivalent huntingtin binding peptide suppresses polyglutamine aggregation and pathogenesis in Drosophila. Nat. Genet. 30**:** 367–376. [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P. and Benzer, S. 2000. Genetic suppression of polyglutamine toxicity in Drosophila. Science 287**:** 1837–1840. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Suppression of polyglutamine toxicity by a Drosophila homolog of myeloid leukemia factor 1. Hum. Mol. Genet. 11**:** 2657–2672. [DOI] [PubMed] [Google Scholar]
- Klement I.A., Skinner, P.J., Kaytor, M.D., Yi, H., Hersch, S.M., Clark, H.B., Zoghbi, H.Y., and Orr, H.T. 1998. Ataxin-1 nuclear localization and aggregation: Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95**:** 41–53. [DOI] [PubMed] [Google Scholar]
- Lin X., Antalffy, B., Kang, D., Orr, H.T., and Zoghbi, H.Y. 2000. Polyglutamine expansion down-regulates specific neuronal genes before pathologic changes in SCA1. Nat. Neurosci. 3**:** 157–163. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R., Strand, A., Peters, N.L., Solano, S.M., Hollingsworth, Z.R., Menon, A.S., Frey, A.S., Spektor, B.S., Penney, E.B., Schilling, G., et al. 2000. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet. 9**:** 1259–1271. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T., Lemberger, T., Bleckmann, S.C., Kern, H., Kretz, O., Martin Villalba, A., Tronche, F., Kellendonk, C., Gau, D., Kapfhammer, J., et al. 2002. Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 31**:** 47–54. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. 2001. Structure and function of histone acetyltransferases. Cell. Mol. Life Sci. 58**:** 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek K.W., Ng, N., Fetter, R., Smolik, S., Goodman, C.S., and Davis, G.W. 2000. A genetic analysis of synaptic development: Pre- and postsynaptic dCBP control transmitter release at the Drosophila NMJ. Neuron 25**:** 537–547. [DOI] [PubMed] [Google Scholar]
- McCampbell A., Taylor, J.P., Taye, A.A., Robitschek, J., Li, M., Walcott, J., Merry, D., Chai, Y., Paulson, H., Sobue, G., et al. 2000. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 9**:** 2197–2202. [DOI] [PubMed] [Google Scholar]
- McCampbell A., Taye, A.A., Whitty, L., Penney, E., Steffan, J.S., and Fischbeck, K.H. 2001. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc. Natl. Acad. Sci. 98**:** 15179–15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K.-T. and Benzer, S. 1999. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science 284**:** 1985–1988. [DOI] [PubMed] [Google Scholar]
- Ng H.H. and Bird, A. 2000. Histone deacetylases: Silencers for hire. Trends Biochem. Sci. 25**:** 121–126. [DOI] [PubMed] [Google Scholar]
- Nucifora Jr F.C., Sasaki, M., Peters, M.F., Huang, H., Cooper, J.K., Yamada, M., Takahashi, H., Tsuji, S., Troncoso, J., Dawson, et al. 2001. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 291**:** 2423–2428. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz, R.L., Russanova, V., Howard, B.H., and Nakatani, Y. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87**:** 953–959. [DOI] [PubMed] [Google Scholar]
- Sánchez I., Mahlke, C., and Yuan, J. 2003. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature 421**:** 373–379. [DOI] [PubMed] [Google Scholar]
- Saudou F., Finkbeiner, S., Devys, D., and Greenberg, M.E. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95**:** 55–66. [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Kazantsev, A., Spasic-Boskovic, O., Greenwald, M., Zhu, Y.Z., Gohler, H., Wanker, E.E., Bates, G.P., Housman, D.E., and Thompson, L.M. 2000. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. 97**:** 6763–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S., Bodai, L., Pallos, J., Poelman, M., McCampbell, A., Apostol, B.L., Kazantsev, A., Schmidt, E., Zhu, Y.Z., Greenwald, M., et al. 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413**:** 739–743. [DOI] [PubMed] [Google Scholar]
- Taylor J.P. and Fischbeck, K.H. 2002. Altered acetylation in polyglutamine disease: An opportunity for therapeutic intervention? Trends Mol. Med. 8**:** 195–197. [DOI] [PubMed] [Google Scholar]
- Taylor J.P., Lieberman, A.P., and Fischbeck, K.H. 2002. Repeat expansion and neurological disease. In Diseases of the nervous system, 3rd ed. (eds. A.K. Asbury et al.), pp. 32–54. Cambridge University Press, Cambridge, UK.
- Warrick J.M., Paulson, H.L., Gray-Board, G.L., Bui, Q.T., Fischbeck, K.H., Pittman, R.N., and Bonini, N.M. 1998. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93**:** 939–949. [DOI] [PubMed] [Google Scholar]
- Warrick J.M., Chan, H.Y., Gray-Board, G.L., Chai, Y., Paulson, H.L., and Bonini, N.M. 1999. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23**:** 425–428. [DOI] [PubMed] [Google Scholar]