Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis (original) (raw)
Abstract
The nuclear bile acid receptor FXR has been proposed to play a central role in the feedback repression of the gene encoding cholesterol 7α-hydroxylase (CYP7A1), the first and rate-limiting step in the biosynthesis of bile acids. We demonstrate that FXR directly regulates expression of fibroblast growth factor-19 (FGF-19), a secreted growth factor that signals through the FGFR4 cell-surface receptor tyrosine kinase. In turn, FGF-19 strongly suppresses expression of CYP7A1 in primary cultures of human hepatocytes and mouse liver through a c-Jun N-terminal kinase (JNK)-dependent pathway. This signaling cascade defines a novel mechanism for feedback repression of bile acid biosynthesis and underscores the vital role of FXR in the regulation of multiple pathways of cholesterol catabolism in the liver.
Keywords: CYP7A1, FXR, bile acid, fibroblast growth factor, JNK
The catabolism of cholesterol to bile acids represents a major pathway for the elimination of this potentially pathogenic sterol fromthe body. Bile acids subserve a number of important physiological functions, including the solubilization of cholesterol, fat soluble vitamins, and other lipids in the intestine (Vlahcevic et al.1994,1996). In addition, bile acids contribute to the generation of bile flow and promote the secretion of lipids, notably phosphatidylcholine and cholesterol, fromthe canalicular membrane into the bile canaliculus. However, because of their intrinsic toxicity, intracellular levels of bile acids must be tightly regulated, which is largely accomplished by transcriptional regulation of genes encoding proteins involved in bile acid biosynthesis, transport, and metabolism.
The conversion of cholesterol to the primary bile acids, cholic acid and chenodeoxycholic acid (CDCA), involves at least 14 distinct enzymes and is accomplished via 2 pathways (Bjorkhem 1985; Russell and Setchell 1992). The first and rate-limiting step in the neutral (classic) pathway of bile acid biosynthesis is catalyzed by cholesterol 7 α-hydroxylase (CYP7A1; Bjorkhem 1985; Russell and Setchell 1992; Chiang 1998). Expression of the gene encoding CYP7A1 is known to be suppressed by a number of factors including insulin, protein kinase C activators, cytokines such as tumor necrosis factor α (TNF-α), steroid hormones, and, importantly, bile acids (for review, seeChiang 1998). The bile acid-dependent feedback repression of CYP7A1 is important in preventing a potentially harmful expansion of the bile acid pool. A number of studies have focused on characterizing the molecular mechanisms by which bile acids suppress CYP7A1 expression, and it is now apparent that multiple, redundant pathways exist (Stravitz et al. 1995;Antes et al. 2000;Goodwin et al. 2000;Lu et al. 2000;Miyake et al. 2000;De Fabiani et al. 2001;Kerr et al. 2002;Wang et al. 2002). Notably, these signaling cascades converge on a common bile acid responsive element (BARE) in the CYP7A1 promoter (Chiang and Stroup 1994;Stroup et al. 1997). This element is highly conserved across species and is a well-documented binding site for members of the nuclear receptor superfamily of ligand activated transcription factors, including liver receptor homolog-1 (LRH-1, NR5A2) and hepatocyte nuclear factor 4α (HNF-4α, NR2A1;Crestani et al. 1998;Nitta et al. 1999;Lu et al. 2000;Stroup and Chiang 2000;De Fabiani et al. 2001;Chiang 2002).
The farnesoid X receptor (FXR; NR1H4) is a bile acid-activated transcription factor that also belongs to the nuclear receptor family (Makishima et al. 1999;Parks et al. 1999;Wang et al. 1999). FXR binds DNA as an obligate heterodimer with the retinoid X receptors (RXRs;Forman et al. 1995;Seol et al. 1995). The FXR/RXR heterodimer typically binds to an inverted repeat of the hexanucleotide motif AGG/TTCA separated by a single nucleotide, a so-called IR-1 (Forman et al. 1995;Seol et al. 1995). FXR is known to be expressed in tissues that are exposed to bile acids, including liver, intestine, gallbladder (C. Housset, pers. comm.), kidney, and adrenal gland (Forman et al. 1995;Seol et al. 1995). In liver, the biological consequences of FXR activation have recently become increasingly clear. Upon activation, FXR initiates transcription of a cohort of genes that function to decrease the concentration of bile acids within the hepatocyte. Specifically, FXR induces the expression of ATP-binding cassette (ABC) transporters bile salt export pump (BSEP; ABCB11;Sinal et al. 2000;Ananthanarayanan et al. 2001;Plass et al. 2002), multidrug resistance protein 3 (MDR3, ABCB4; B. Goodwin and S.A. Jones, unpubl.), and multidrug resistance-associated protein 2 (MRP2; ABCC2;Kast et al. 2002). These transporters function to transport bile acids and bile constituents fromthe hepatocytes into the bile. In addition, activation of FXR by both naturally occurring (CDCA) and synthetic ligands leads to the repression of two important genes in the bile acid biosynthetic pathway, namely CYP7A1 and CYP8B1, which encodes oxysterol 12α hydroxylase (Goodwin et al. 2000;Lu et al. 2000;Sinal et al. 2000;del Castillo-Olivares and Gil 2001; Zhang and Chiang 2001). The FXR-dependent suppression of CYP7A1 is mediated by the transcriptional repressor short heterodimer partner-1 (SHP; NR0B2), an atypical nuclear receptor that lacks a DNA-binding domain (Goodwin et al. 2000;Lu et al. 2000). Thus, activation of FXR results in increased expression of the SHP gene. In turn, SHP interacts with LRH-1, a known positive regulator of CYP7A1 (discussed above) and represses its transcriptional activity. Elegant studies performed in mice harboring a disrupted SHP gene confirmthe importance of the FXR–SHP–LRH-1 cascade in suppression of_CYP7A1_, however, they also demonstrate the existence of additional SHP-independent pathways, possibly involving the c-Jun N-terminal kinase (JNK) mitogen-activated protein kinase (Kerr et al. 2002; Wang et al. 2002).
In this study, we describe the discovery of a novel FXR-dependent signaling cascade for the suppression of CYP7A1. We show that FXR directly regulates expression of FGF-19, a member of the fibroblast growth factor (FGF) family of signaling molecules (Nishimura et al. 1999; Xie et al. 1999). The FGFs bind the extracellular domain of their cognate cell surface receptor (FGFR) and induce receptor dimerization and tyrosine kinase phosphorylation, which, in turn, leads to the activation of a number of intracellular pathways (Goldfarb 2001; Ornitz and Itoh 2001). For many years, it has been understood that the FGFs regulate cell growth, differentiation, and morphogenesis, however, it is now apparent that some of these proteins are also important components of specific homeostatic pathways (Yu et al. 2000; Shimada et al. 2001; Tomlinson et al. 2002). We demonstrate that FGF-19, acting as an FXR-induced signaling molecule, represses expression of the CYP7A1 gene. Our findings define a novel regulatory pathway for the suppression of bile acid biosynthesis.
Results
Induction of FGF-19 by FXR agonists
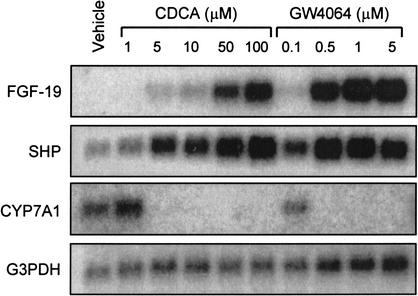
In experiments aimed at identifying changes in gene expression following FXR activation in liver cells, primary human hepatocytes were treated with the potent, selective, synthetic FXR agonist GW4064 (Maloney et al. 2000). Samples were then examined for differential gene expression using the CuraGen GeneCalling technology (Shimkets et al. 1999). The most highly induced gene following GW4064 treatment was a member of the FGF family of secreted signaling molecules, FGF-19. To further examine the FXR-dependent regulation of FGF-19, primary human hepatocytes were cultured in the presence of various concentrations of GW4064 or the naturally occurring FXR agonist chenodeoxycholic acid (CDCA). FGF-19 mRNA was undetectable in control cultures; however, treatment with either GW4064 or CDCA caused a robust, dose-dependent, increase in FGF-19 mRNA levels (Fig. 1). Such increases were not seen for other members of the FGF family, including FGF-1, FGF-2, FGF-9, FGF-21, or FGF-22. In parallel, expression of the FXR target gene SHP (Goodwin et al. 2000;Lu et al. 2000;Sinal et al. 2000) was also induced. As expected, activation of FXR by either GW4064 or CDCA resulted in a profound suppression of CYP7A1 expression (Fig. 1). Pretreatment of primary human hepatocytes with the protein synthesis inhibitor cycloheximide did not block the induction of FGF-19 by GW4064, indicating that the induction is likely to be a direct effect (data not shown).
Figure 1.
FXR-dependent induction of FGF-19. Primary cultures of human hepatocytes were treated for 48 h with the indicated concentrations of CDCA (1–100 μM) or GW4064 (0.1–5 μM). Control cultures received vehicle (0.1% DMSO) alone. Total RNA was isolated and Northern analysis performed using cDNA probes for FGF-19, SHP, CYP7A1, and G3PDH.
Identification of an FXRE in the FGF-19 gene
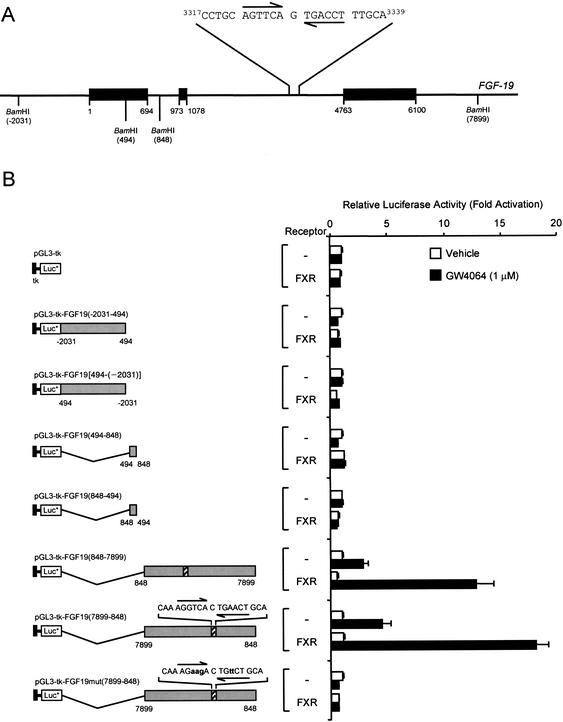
The ability of GW4064 and CDCA to strongly induce expression of_FGF-19_ suggested that this gene is directly regulated by FXR. To delineate potential FXR-responsive elements (FXRE) in the FGF-19 promoter, multiple fragments, up to 6.3 kb in size, of the FGF-19 5′-flanking region were isolated. Chimeric _FGF-19_-luciferase reporter gene constructs were prepared, and the ability of FXR to transactivate luciferase expression was examined by transient transfection into a human liver-derived cell line, HuH7. Despite extensive analysis, we were unable to identify an FXRE in this region of the FGF-19 gene (data not shown). To investigate the possibility that FXR-responsive regions lay downstreamof the transcription initiation site, within the FGF-19 structural gene, three Bam_HI fragments encompassing the entire_FGF-19 gene were isolated (Fig. 2A). These fragments, corresponding to bases –2031 to 494, 494 to 848, and 848 to 7904 (relative to the FGF-19 transcription initiation site, Fig. 2A) were inserted into the downstreamenhancer region of the pGL3-tk-LUC vector, which contains a minimal thymidine kinase (tk) promoter linked to a luciferase reporter gene. Upon transient transfection into HuH7 cells, only constructs harboring the 7051-bp Bam_HI fragment [pGL3-tk-FGF19(848–7899) and pGL3-tk-FGF19(7899–848)] exhibited FXR-responsiveness (Fig. 2B). Regardless of orientation, >10-fold induction was observed when these constructs were cotransfected into HuH7 cells with an FXR expression vector and exposed to GW4064, indicating that this region contained an FXR-responsive element (Fig. 2B). This 7051-bp_Bam_HI fragment contains a portion of intron 1, all of exons 2 and 3 including intron 2, and ∼2.8 kb of the 3′-flanking region of the_FGF-19 gene (Fig. 2A). Typically, FXR-response elements are comprised of inverted repeats of the AG(G/T)TCA hexad with a single nucleotide spacer (IR-1 element; Forman et al. 1995;Seol et al. 1995). Inspection of the 7051-bp _Bam_HI region revealed the presence of a perfect IR-1 motif located in intron 2 of the FGF-19 gene (bases 3322–3334 relative to the transcription initiation site,Fig. 2A). To investigate whether this site mediated the FXR response, mutations were introduced into the IR-1 of the 7051-bp fragment. Mutation of the putative FXRE completely abolished FXR-responsiveness of this construct, demonstrating that this site mediated the FXR response. Thus, the FGF-19 gene contains a functional FXR responsive element within the second intron.
Figure 2.
Identification of an FXR response element in the FGF-19 gene. (A) Structure of the FGF-19 gene showing the position of the intron–exon boundaries, restriction endonuclease sites used in the generation of reporter gene constructs, and the sequence of the FXR response element located in intron 2. Exons are shown as solid bars and all coordinates are relative to the FGF-19 transcription initiation site. (B) Delineation of an FXR response element. Three _Bam_HI fragments encompassing the entire FGF-19 structural gene were inserted downstreamof the reporter gene in the pGL3-tk-Luc, which contains a minimal thymidine kinase (tk) promoter (bases –105 to +51, solid box) linked to luciferase reporter gene. Site-directed mutagenesis of the putative FXRE was performed as described in Materials and Methods to produce pGL3-tk-FGF19mut(7899–848). All constructs were transfected in HuH7 cells and exposed to GW4064 (1 μM) for 24 h prior to harvest and determination of luciferase and SPAP activities. Luciferase values are normalized to SPAP and expressed as fold activation compared with control cells transfected with the same reporter, but not receptor, and which received vehicle (0.1% DMSO) alone. Data represent the mean ± S.E.M. from eight individual transfections.
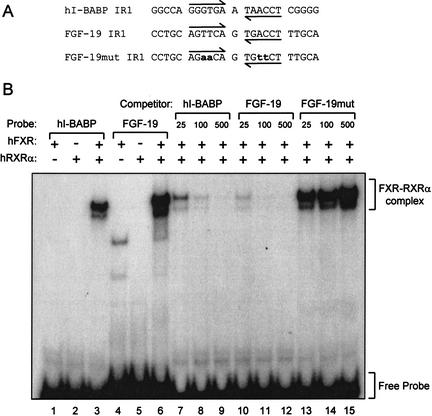
To determine whether FXR–RXRα heterodimers could directly bind the FXRE identified in the second intron of FGF-19, electrophoretic mobility-shift assays were performed. Radiolabeled probes corresponding to the putative FXRE fromthe FGF-19 gene (FGF-19 IR-1) and a previously characterized FXRE fromthe human I-BABP gene (hI-BABP IR-1;Grober et al. 1999) were prepared and examined for their ability to bind FXR or RXRα alone or in combination (Fig. 3A). Neither FXR nor RXRα were capable of binding to these elements when added to the binding reaction alone (Fig. 3B). However, when both proteins were present, a robust complex formed on the control h_I-BABP_ IR-1 and the IR-1 from the_FGF-19_ gene (Fig. 3B). Competition binding analyses showed that these interactions were specific; no competition was seen when a mutated derivative of the IR-1 motif derived from the FGF-19 gene was added to the binding reaction (Fig. 3B). Thus, FXR–RXRα heterodimers bind and activate an FXRE within intron 2 of the FGF-19 gene.
Figure 3.
The FXR response element in Intron 2 binds FXR–RXRα. The ability of FXR–RXRα-heterodimers to bind the IR-1 motif in intron 2 of the FGF-19 gene was examined by EMSA as outlined in Materials and Methods. (A) Sequence of the oligonucleotide probes (sense strand only) corresponding to a previously characterized FXR-binding site in the human I-BABP promoter (hI-BABP IR1), the putative FXR response element from intron 2 of FGF-19 (FGF-19 IR1), and a mutated derivative of this site (FGF-19mut IR1, mutated bases are shown in lowercase). (B) EMSA was performed with recombinant human FXR and/or RXRα as indicated and radiolabeled h-IBABP IR1 or FGF-19 IR1 (lanes_1–3,4–6_, respectively). Competition EMSA (lanes_7–15_) was performed using in vitro synthesized FXR and RXRα, radiolabeled hI-BABP IR1, and the indicated molar excess of unlabeled hI-BABP IR1 (lanes 7–9), FGF-19 IR1 (lanes_10–12_), or FGF-19mut IR1 (lanes 13–15). The position of the shifted FXR–RXRα complex and free probes are indicated.
Repression of CYP7A1 by FGF-19
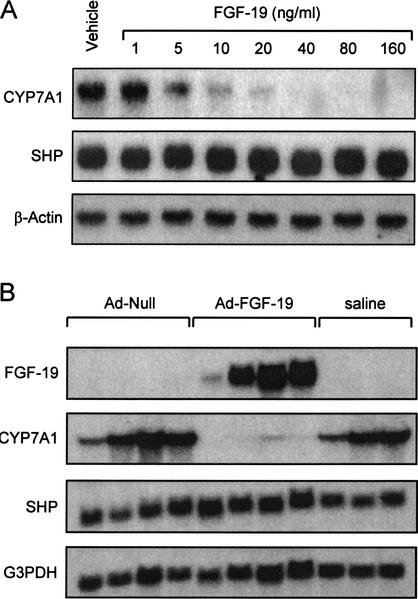
FGFs signal by activating transmembrane tyrosine kinase receptors. Four FGF receptors (FGFR1–FGFR4) have been identified (Goldfarb 2001;Ornitz and Itoh 2001). FGF-19 is a high-affinity ligand for FGFR4, and unlike other FGF family members, exhibits exclusive binding to FGFR4 (Xie et al. 1999). Interestingly, mice harboring a disrupted_FGFR4_ gene exhibit elevated expression of CYP7A1 and an expanded bile acid pool (Yu et al. 2000). These data suggested that FGFR4 plays a key role in the regulation of CYP7A1, and as a result, bile acid biosynthesis (Yu et al. 2000). Because FGFR4 is the FGF-19 receptor, we hypothesized that FGF-19 might play a role in the bile acid-mediated repression of CYP7A1. Accordingly, primary human hepatocytes were treated with purified recombinant human FGF-19 and analyzed for CYP7A1 expression. In preliminary studies,CYP7A1 expression was strongly suppressed after 3–6 h of treatment with FGF-19 (data not shown). Thus, all subsequent experiments were performed within this time frame. Treatment of human hepatocytes with recombinant FGF-19 resulted in a profound dose-dependent suppression of_CYP7A1_ expression (Fig. 4A). In parallel, expression of the gene encoding SHP was not altered by FGF-19 treatment (Fig. 4A). This observation suggests that FGF-19 represses expression of_CYP7A1_ through a novel mechanismthat does not involve induction of_SHP_ expression. Thus, SHP induction is not a prerequisite for CYP7A1 repression and FXR can mediate bile-acid feedback repression of CYP7A1 via two distinct mechanisms.
Figure 4.
FGF-19 suppresses expression of CYP7A1 in primary cultures of human hepatocytes and in vivo. (A) Primary cultures of human hepatocytes were treated with the indicated concentration of recombinant FGF-19 for 6 h prior to harvest and isolation of total RNA. Northern blot analysis of CYP7A1, SHP, and β-actin expression was performed as described in Materials and Methods. Results froma single hepatocyte donor are shown, but are typical of three separate preparations. (B) Total RNA was isolated from the livers of adult male FVB mice injected intravenously with saline (200 μL), or 2 × 1011 units of either null adenovirus or FGF-19-expressing adenovirus as outlined in Materials and Methods. Northern analysis was performed sequentially with cDNA probes corresponding to FGF-19, mouse CYP7A1, mouse SHP, and G3PDH.
To determine whether bile acid biosynthesis can be regulated by FGF-19 in vivo, it was necessary to express FGF-19 without induction of SHP. Accordingly, mice were infected with an adenovirus expressing human FGF-19 and the levels of CYP7A1 expression determined. Three days after infection, livers were collected for mRNA analysis (Fig. 4B). Northern blot analysis showed the expected appearance of FGF-19 message in the livers of the exposed mice. In agreement with experiments performed in primary hepatocytes, overexpression of FGF-19 resulted in a marked suppression of CYP7A1 mRNA levels compared with mice infected with null virus. Importantly, expression of_SHP_ was unchanged in these animals. These data demonstrate that FGF-19 is capable of down-regulating the expression of the CYP7A1 gene in vivo.
Suppression of CYP7A1 by FGF-19 is mediated by the JNK-signaling cascade
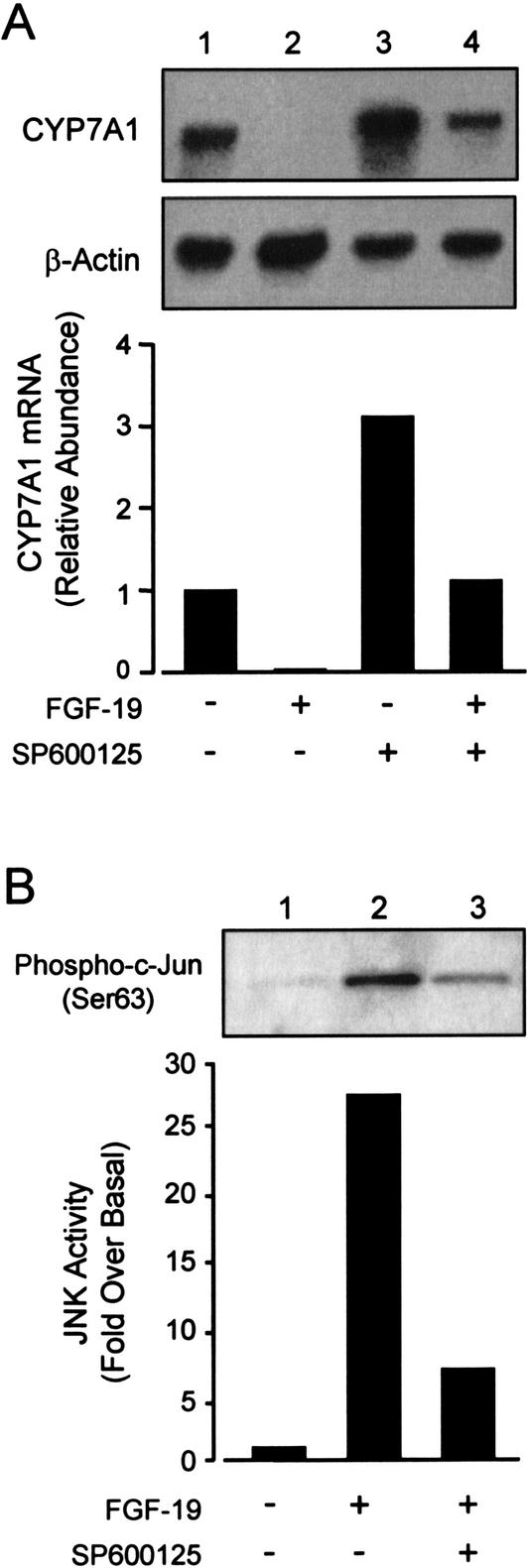
Upon ligand binding, FGF receptors activate a number of pathways, including the extracellular signal-regulated kinases (ERK) and JNK phosphoprotein-signaling cascades (Goldfarb 2001;Sheikh et al. 2001). In addition, several reports have implicated the JNK pathways in the bile acid-dependent suppression of CYP7A1 (De Fabiani et al. 2001;Gupta et al. 2001;Wang et al. 2002). Therefore, we examined the ability of a specific JNK inhibitor to block the effects of FGF-19 treatment on CYP7A1 expression. Hepatocytes were cotreated with either recombinant FGF-19 alone or in combination with the JNK inhibitor SP600125 (Bennett et al. 2001). As expected, FGF-19 treatment completely repressed CYP7A1 expression. Pretreatment of hepatocytes with the JNK-specific inhibitor SP600125 resulted in inhibition of the FGF-19-dependent suppression of CYP7A1. Consistent with the negative effects of JNK activation on CYP7A1 expression, cells treated with SP600125 exhibited an approximately threefold induction in CYP7A1 mRNA levels (Fig. 5A). A similar observation has been reported recently by Moore and coworkers (Wang et al. 2002). SP600125-treatment did not completely prevent the suppression of_CYP7A1_ by FGF-19, suggesting the existence of additional JNK-independent CYP7A1 regulatory pathways.
Figure 5.
Activation of the JNK signaling cascade by FGF-19 causes suppression of_CYP7A1_. (A) Primary cultures of human hepatocytes were treated with FGF-19 (80 ng/mL) alone or in combination with the JNK inhibitor SP600125 (10 μM). SP600125 was added to the culture medium as a 1000× stock in DMSO 30 min before the addition of recombinant FGF-19. Control cultures received vehicle (0.1% DMSO) alone. Cells were cultured for a further 6 h prior to harvest, and Northern blot analysis of CYP7A1 and β-actin expression. (B) FGF-19 activates the JNK-signaling cascade. Total cell lysates were prepared from hepatocytes treated for 3 h as described above. The level of activated c-Jun was analyzed by immunoblotting using antibodies specific for Ser 63-phosphorylated c-Jun. Data are representative of a least three individual hepatocyte preparations.
To determine whether JNK activity is elevated by exposure of primary human hepatocytes to FGF-19, a JNK activity assay was performed. Cell extracts were prepared fromprimary hepatocytes and incubated with recombinant c-Jun. An antibody that specifically detects Ser 63-phosphorylated c-Jun was used to visualize the activated protein prior to quantitation. Exposure of hepatocytes to FGF-19 resulted in a 25–30-fold increase in the level of phosphorylated c-Jun, indicating activation of the JNK pathway (Fig. 5B). Increased JNK activity was also seen following GW4064 treatment (data not shown). Cotreatment with the JNK inhibitor, SP600125, strongly inhibited the FGF-19 induced c-Jun phosphorylation. Thus, FGF-19 treatment of primary human hepatocytes is able to activate the JNK pathway. This indicates that the JNK pathway is involved in the suppression of CYP7A1 by FGF-19 and is consistent with previous observations that activation of the JNK pathway by bile acids and cellular stress can repress CYP7A1 expression.
Discussion
Bile acids are physiologically important amphipathic molecules that subserve a number of functions. To prevent their accumulation to potentially harmful levels, bile acids coordinately regulate genes involved in their biosynthesis, transport, and metabolism. Notably, bile acids suppress expression of the CYP7A1 gene, which encodes the first and rate-limiting step in the neutral (classic) pathway of bile acid biosynthesis. Studies performed in a number of laboratories have demonstrated the existence of multiple, redundant pathways for the bile acid-dependent repression of this gene. Previously, we and others described a pathway in which induction of the repressor protein SHP by bile acid-activated FXR leads to suppression of_CYP7A1_ promoter activity through a direct interaction with LRH-1 (Goodwin et al. 2000;Lu et al. 2000). This pathway relies on a network of nuclear receptor interactions that presumably all occur within the nucleus of an individual hepatocyte. Thus, within a single cell, when bile acid concentrations are high enough to activate FXR, that individual cell can suppress bile acid biosynthesis through the FXR–SHP–LRH-1 mechanism. The pathway described in this report is distinct in that it provides a mechanism for cell-to-cell signaling and coordinate down-regulation of CYP7A1 among neighboring hepatocytes and for a potential paracrine pathway to reinforce the activity of the FXR–SHP–LRH-1 mechanism. Here, we report that bile acid activation of FXR results in the induction of_FGF-19_ gene expression and that the FGF-19-signaling pathway represses CYP7A1 gene expression without elevating SHP expression, via a JNK-dependent pathway.
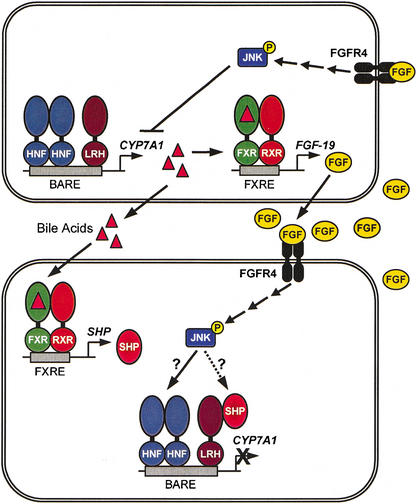
FGF-19 is a member of the FGF family of secreted signaling molecules. In the adult, FGF-19 mRNA has been reported to be localized in the brain as well as adult liver, gallbladder, kidney, spleen, heart, and leukocytes (Nishimura et al. 1999;Xie et al. 1999). Unlike other members of the FGF family, FGF-19 displays little mitogenic activity toward fibroblasts and binds exclusively to FGFR4, one of the four known FGFRs (Xie et al. 1999). Thus, bile acid activation of FXR would lead to secretion of FGF-19, which could then bind to FGFR4 and initiate a signaling pathway that results in suppression of_CYP7A1_ in neighboring cells (Fig. 6). Importantly, FGFR4 is known to be expressed on the cell surface of hepatocytes and bile duct epithelium (Partanen et al. 1991;Hughes 1997). Moreover, McKeehan and coworkers demonstrated that mice harboring a disrupted_FGFR4_ gene exhibit elevated levels of CYP7A1 compared with their wild-type littermates, resulting in an enlarged bile acid pool (Yu et al. 2000). However, cholic acid feeding resulted in suppression of CYP7A1 in both the wild-type and FGFR4-deficient mice (Yu et al. 2000). Taken together, these observations suggest that FGFR4 plays a role in the suppression of CYP7A1 expression, but other FGF-independent pathways are also active in this process.
Figure. 6.
Role of FXR in the suppression of CYP7A1. Activation of FXR by bile acids leads to induction of the FGF-19 gene. FGF-19 is secreted fromthe hepatocyte and can signal to other hepatocytes through its cell-surface receptor FGFR4. This results in suppression of CYP7A1 expression through a JNK-dependent signaling cascade. FXR can also activate expression of the SHP gene, which can directly suppress the CYP7A1 gene through an interaction with LRH-1 (Goodwin et al. 2000;Lu et al. 2000). SHP itself may be a component of the JNK-signaling pathway (Wang et al. 2002).
The importance of kinase-signaling pathways in the regulation of_CYP7A1_ gene expression has been recognized for several years. Phorbol ester activation of protein kinase C (PKC), specifically PKCδ, results in repression of CYP7A1 expression (Stravitz et al. 1996;De Fabiani et al. 2001). Additionally, TNF-α has been shown to repress CYP7A1 gene expression by activating the JNK pathway (Feingold et al. 1996;Miyake et al. 2000;De Fabiani et al. 2001;Gupta et al. 2001). More recently, two groups have demonstrated that bile acid activation of the JNK pathway is able to mediate repression of CYP7A1 gene expression (De Fabiani et al. 2001;Gupta et al. 2001). Although it is possible that FGF-19 regulates CYP7A1 mRNA levels through post-transcriptional mechanisms, JNK signaling is thought to exert its effects at the level of the CYP7A1 promoter, and we suggest that a similar mechanism is responsible for mediating the effects of FGF-19 on_CYP7A1_ expression. DeFabiani et al. (2001) reported that the HNF-4α-binding site in the BARE of the CYP7A1 promoter was necessary for TNF-α and JNK-mediated repression of CYP7A1. Further, they demonstrated that inactivation of this signaling pathway by a dominant negative JNK kinase prevented the repression of CYP7A1 by bile acids (De Fabiani et al. 2001). The precise mechanism of HNF-4α inactivation by the JNK pathway is not yet clear, but it could be analogous to the negative regulation of RXR via MKK4 and JNK phosphorylation of RXR that inhibits retinoid signaling (Lee et al. 2000). Importantly, studies by Wang et al. (2002) in mice lacking a functional SHP gene suggest that SHP itself may be a component of the JNK signaling cascade. It is also noteworthy that whereas the bile acid-dependent suppression of CYP7A1 expression is largely intact in the SHP-knockout mice, repression of this gene by the synthetic FXR agonist GW4064 is completely lost (Kerr et al. 2002; Wang et al. 2002). In addition, it is intriguing that DeFabiani et al. (2001) only observed the JNK-dependent repression of CYP7A1 in HepG2 cells, which express high constitutive levels of SHP (B. Goodwin, unpubl.). Here, we demonstrate that GW4064 is a strong inducer of FGF-19 expression and that FGF-19 is capable of acting directly on the hepatocyte to suppress_CYP7A1_ expression. Taken together, these observations suggest that the FXR–FGF-19–JNK signal cascade may converge on the nuclear receptor SHP (Fig. 6). However, despite extensive analysis, we have been unable to demonstrate modulatory activity of FGF-19 on the CYP7A1 promoter or on proteins known to regulate CYP7A1 expression, including SHP, LRH1, and HNF-4α. Nuclear extracts prepared from primary cultures of human hepatocytes treated with vehicle or FGF-19 show no difference in the binding of LRH-1 or HNF-4α to the BARE in the CYP7A1 promoter (data not shown). Additionally, protein–protein interaction studies failed to show an increased affinity of SHP for LRH-1 or HNF-4α in nuclear extracts from FGF-19-treated human hepatocytes, and chromatin immunoprecipitation experiments did not detect any changes in the occupancy of the CYP7A1 promoter by these nuclear receptors (data not shown). Thus, the mechanism by which FGF-19 suppresses CYP7A1 expression remains obscure and is the focus of ongoing studies.
Recently, transgenic mice expressing FGF-19 under the control of the myosin light-chain promoter have been reported to have increased metabolic rate, decreased adiposity, and increased insulin sensitivity compared with control mice (Tomlinson et al. 2002). These mice also develop hepatocellular carcinomas as they age (Nicholes et al. 2002). On the basis of their observations with the FGF-19 transgenic mouse, Tomlinson et al. propose the liver as the primary site of action of FGF-19. Our studies further support this hypothesis by showing that FGF-19 is under the transcriptional control of FXR, a liver-enriched nuclear receptor, and by showing that it is a key regulator of cholesterol and bile acid homeostasis in primary human hepatocytes. Tomlinson et al. (2002) suggest increased brown adipose tissue mass and decreased liver expression of acetyl-CoA carboxylase 2 (ACC2) as a mechanism whereby FGF-19 modulates metabolismand insulin sensitivity in the transgenic mouse. ACC2 catalyzes the conversion of acetyl-CoA to malonyl-CoA. Decreased expression of ACC2 relieves malonyl CoA repression of carnitine palmitoyl transferase 1 (CPT1) initiation of fatty acid oxidation in the mitochondria. It will be interesting to see whether physiological concentrations of FGF-19, such as would be expected following activation of FXR, elicit similar changes in metabolic rate and insulin sensitivity. It may not be straightforward to ascertain this in rodent models, as a rodent homolog of FGF-19 has not been identified.
In summary, the observations reported here connect, for the first time, the activity of the bile acid receptor, FXR, with the JNK pathway and the negative regulation of CYP7A1 gene expression. Bile acid activation of FXR initiates two distinct pathways that lead to repression of bile acid biosynthesis. One is strictly intracellular and consists of the regulatory cascade of the nuclear receptors SHP and LRH-1. The second pathway provides for extracellular signaling and regulation of bile acid homeostasis across colocalized cells. This work illustrates an alternate pathway for bile acid feedback regulation of CYP7A1.
Materials and methods
Materials
Primary cultures of human hepatocytes were obtained from Dr. Steve Strom(University of Pittsburgh, PA), BioWhittaker, or In Vitro Technologies. CDCA, dexamethasone, and charcoal-stripped, delipidated calf serumwere acquired fromthe Sigma Chemical Co. Charcoal/dextran-treated FBS was from Hyclone Laboratories Inc. Trizol, insulin-transferrin-selenium(ITS-G) and all other tissue culture reagents were from Invitrogen. Hybond N+, [α32P]dCTP, poly(dI-dC) · poly(dI-dC), and the Mega-prime DNA Labeling Systemwere purchased from Amersham Pharmacia Biotech., Inc. Recombinant human FGF-19 was provided by R&D Systems. Restriction endonucleases were provided by Roche Molecular Biochemicals. The BAC clone RP11-300I6 (GenBank accession no. AP001888) was obtained from BACPAC Resource Center (Children's Hospital Oakland Research Institute). GW4064 was prepared at GlaxoSmithKline as described elsewhere (Maloney et al. 2000).
Primary culture of human hepatocytes
Primary human hepatocytes were cultured on Matrigel-coated 6-well plates at a density of 1.5 × 106 cells per well. Culture medium consisted of serum-free Williams' E medium supplemented with 100 nM dexamethasone, 100 U/mL penicillin G, 100 μg/mL streptomycin and ITS-G. Twenty-four hours after isolation, cells were treated with the indicated concentration of either GW4064 or CDCA, which were added to the culture mediumas 1000× stocks in DMSO. Control cultures received vehicle (0.1% DMSO) alone. Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Differentially regulated genes were identified using CuraGen GeneCalling Technology (Shimkets et al. 1999). Recombinant human FGF-19 was reconstituted in PBS containing 0.1% BSA, and added directly to the cell culture medium to produce the indicated final concentration. The phosphorylation inhibitor SP600125, a selective JNK inhibitor (BioMol), was added to the medium 30 min prior to treatment with recombinant FGF-19 as a 1000× stock in DMSO (final concentration 10 μM). RNA was isolated as described above and resolved on a 1% agarose/2.2 M formaldehyde denaturing gel, then transferred to a Hybond N+ membrane, and a Northern Blot analysis was performed. Blots were hybridized with32P-labeled cDNA probes corresponding to human CYP7A1, SHP (Goodwin et al. 2000), FGF-19 (bases 464–1114, GenBank accession no. NM_005117), glyceraldehyde-3-phosphate (G3PDH), or β-actin (BD Biosciences Clon-tech).
Plasmid constructs
Chimeric FGF-19-reporter gene constructs were prepared by subcloning fragments of genomic DNA from the BAC clone RP11-300I6 into pGL3-tk-LUC, which contains the minimal thymidine kinase promoter (bases –105 to +51) linked to a luciferase reporter gene. Three _Bam_HI fragments of genomic DNA corresponding to bases –2031 to 499, 494 to 853, and 848 to 7904 (relative to the FGF-19 transcription initiation site) were subcloned in both orientations into the _Bam_HI site of pGL3-tk-Luc, which lies downstreamof the luciferase reporter gene. These constructs, designated pGL3-tk-FGF19(–2031–499), pGL3-tk-FGF19[499–(–2031)], pGL3-tk-FGF19(494–853), pGL3-tk-FGF19(853–494), pGL3-tk-FGF19(848–7904), and pGL3-tk-FGF19(7904–848), encompass the entire FGF-19 structural gene. Mutations were introduced in the FXRE in the pGL3-tk-FGF19(7904–848) using the Quick Change XL Site-Directed Mutagenesis Kit (Stratagene) and the oligonucleotide 5′-CAC CCACCTGCAGaaCAGTcttCTTTGCAGACTTAG-3′. Mutant constructs were sequenced and verified to be free of nonspecific base changes.
Transient transfection assay
A human liver-derived cell line, HuH7, was used to delineate FXR-responsive regions of the FGF-19 gene. Cells were maintained in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin. Plasmid DNA was transfected into HuH7 cells using the Lipofectamine Plus Reagent (Invitrogen) according to the manufacturer's instructions. Thus, 96-well plates were inoculated with 20,000 cells per well in phenol red-free DMEM/F-12 nutrient mixture containing 15 mM HEPES, 2 mM glutamine, and 10% charcoal/dextran-treated FBS 24 h prior to transfection. Cells were transfected for 4 h in OptiMEM with 2 ng of human FXR expression vector (pSG5-hFXR), 8 ng of the control plasmid pβ-Actin-SPAP, 8 ng of luciferase reporter gene construct. All transfection mixes were complemented with the pBluescript plasmid to an identical total amount of DNA (65 ng). Following transfection, cells were incubated for 24 h in DMEM/F-12 medium containing 10% heat-inactivated, charcoal-stripped, delipidated bovine calf serum. Cells were incubated for a further 24 h in the same medium supplemented with either GW4064 (1 μM) or vehicle alone (0.1% DMSO). An aliquot of medium was assayed for SPAP activity and the cells were lysed prior to determination of luciferase expression. Luciferase values were normalized to SPAP.
Electrophorectic mobility-shift assay
Human FXR and RXRα proteins were synthesized in vitro using the TNT rabbit reticulocyte lysate coupled in vitro transcription/translation system (Promega) according to the manufacturer's instructions. Double-stranded oligonucleotides were end labeled with [α-32P]dCTP using Klenow enzyme (Promega). Reactions contained 10 mM HEPES (pH 7.8), 60 mM KCl, 0.2% Nonidet P-40, 6% glycerol, 2 mM dithiothreitol, 1 μg poly(dI-dC) · poly(dI-dC), and recombinant FXR and/or RXRα proteins (2.5 μL each) in a final volume of 20 μL. Double-stranded, unlabeled, competitor oligonucleotides, corresponding to the wild-type FXRE (FGF-19 IR-1; 5′-GATCCCTGCAGTTCAGTGACCTTTGCA-3′), mutated wild-type element (Mut IR-1; 5′-GATCCCTGCAGaaCAGTGttCTTTGCA-3′), and a previously characterized FXRE from the human I-BABP promoter (I-BABP IR-1; 5′-GATCGGCCAGGGTGAATAACCTCGGGG-3′) were included at 25-, 100-, or 500-fold excess. After a 10-min incubation on ice, 25 nM of probe was added, and the incubation continued for an additional 20 min on ice, followed by 3 min at room temperature. DNA–protein complexes were resolved on a 4% polyacrylamide gel in 0.5× TBE. Gels were dried and subjected to autoradiography at –70°C.
FGF-19 adenovirus
Recombinant adenovirus expressing FGF-19 was made using standard protocols. Briefly, FGF-19 cDNA was subcloned into pAd.CMVlink and recombinant adenovirus was generated by cotransfection of 293 cells as described previously (Kozarsky et al. 1997). Plaque purification, expansion, and purification of the recombinant adenovirus yielded sufficient virus for in vivo studies. Transgene expression was confirmed by Western blotting of medium from Ad-FGF-19-infected HeLa cells. Six-week-old male FVB mice were injected with 200 μL saline, null virus (2 × 1011 particles in saline) or Ad-FGF-19 expressing virus (2 × 1011 particles in saline) via tail vein. Mice were housed in the AAALAC-accredited GlaxoSmithKline Research Triangle Park facility under a 12-h light/dark cycle at 72°F ± 2°F, 50% humidity, and allowed food and water ad libitum. Experimental protocols were approved by the GlaxoSmithKline Institutional Animal Care and Use Committee. Mice were sacrificed 72 h after injection, and livers were removed for analysis of gene expression. RNA isolation and Northern Blot analysis are described above. Blots were hybridized with 32P-labeled cDNA probes corresponding to mouse CYP7A1 (bases 772–2256 of GenBank accession no. L23754), mouse SHP (Goodwin et al. 2000), FGF-19 (bases 464–1114, GenBank accession no. NM_005117), and glyceraldehyde-3-phosphate (G3PDH).
JNK activity assay
JNK activity in primary human hepatocytes was determined using a commercially available assay system (Cell Signaling Technology). Briefly, mediumwas aspirated fromhuman hepatocytes cultured on Matrigel, and the cells were overlaid with 2 mL of ice-cold PBS supplemented with 5 mM EDTA. Following a 45-min incubation on ice to dissolve the Matrigel, the cells were pelleted by centrifugation at 2000 rpmfor 5 min. The supernatant was aspirated, and the cells were lysed in 20 mM Tris buffer (pH 7.4) supplemented with 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Nonidet P-40, 2.5 mM sodium pyrophosphate, 1 mM β-Glycerolphosphate, 1 mM sodium orthovanadate, 1 μg/mL leupeptin, and Complete, Mini, EDTA-free protease inhibitor cocktail tablets (Roche Molecular Biochemicals). Samples were sonicated and centrifuged to remove cellular debris. The supernatant was transferred to a fresh tube and the protein concentration determined. Samples were processed as directed by the manufacturer (Cell Signaling Technology) and electrophoresed on a 4%–20% Tris-Glycine gel (Invitrogen) and transferred to a nitrocellulose membrane filter (Invitrogen). The level of phosphorylated c-Jun was determined by standard techniques using a primary antibody that specifically recognizes Ser 63-phosphorylated c-Jun (Cell Signaling Technology). Blots were probed with an HRP-conjugated secondary antibody (Santa Cruz Biotechnology), and the bands visualized using a chemiluminescence reagent (Western Blotting Chemiluminescence Luminol, Santa Cruz Biotechnology). The signal was quantitated using a Molecular Dynamics Densitometer SI (Molecular Dynamics).
Acknowledgments
We thank Steven Haneline for assistance in obtaining FGF-19-containing BAC clones and John Moore for his input throughout this project. We also thank Kelvin Nurse, Michael McQueney, and Karen Kabnick for helpful discussions on FGF biology.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1083503.
Corresponding author.
References
- Ananthanarayanan M., Balasubramanian, N., Makishima, M., Mangelsdorf, D.J., and Suchy, F.J. 2001. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276**:** 28857–28865. [DOI] [PubMed] [Google Scholar]
- Antes T.J., Chen, J., Cooper, A.D., and Levy-Wilson, B. 2000. The nuclear matrix protein CDP represses hepatic transcription of the human cholesterol-7α hydroxylase gene. J. Biol. Chem. 275**:** 26649–26660. [DOI] [PubMed] [Google Scholar]
- Bennett B.L., Sasaki, D.T., Murray, B.W., O'Leary, E.C., Sakata, S.T., Xu, W., Leisten, J.C., Motiwala, A., Pierce, S., Satoh, Y., et al. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. 98**:** 13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I. 1985. Mechanism of bile acid biosynthesis in mammalian liver. In Sterols and bile acids. (eds. H. Danielson and J. Sjovall), pp. 231–278. Elsevier Science Publishers, Amsterdam, The Netherlands.
- Chiang J.Y. 2002. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr. Rev. 23**:** 443–463. [DOI] [PubMed] [Google Scholar]
- Chiang J.Y. and Stroup, D. 1994. Identification and characterization of a putative bile acid-responsive element in cholesterol 7 α-hydroxylase gene promoter. J. Biol. Chem. 269**:** 17502–17507. [PubMed] [Google Scholar]
- Chiang J.Y.L. 1998. Regulation of bile acid synthesis. Front. Biosci. 3**:** D176–D193. [DOI] [PubMed] [Google Scholar]
- Crestani M., Sadeghpour, A., Stroup, D., Galli, G., and Chiang, J.Y. 1998. Transcriptional activation of the cholesterol 7α-hydroxylase gene (CYP7A) by nuclear hormone receptors. J. Lipid Res. 39**:** 2192–2200. [PubMed] [Google Scholar]
- De Fabiani E., Mitro, N., Anzulovich, A.C., Pinelli, A., Galli, G., and Crestani, M. 2001. The negative effects of bile acids and tumor necrosis factor-α on the transcription of cholesterol 7α-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: A novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J. Biol. Chem. 276**:** 30708–30716. [DOI] [PubMed] [Google Scholar]
- del Castillo-Olivares A. and Gil, G. 2001. Suppression of sterol 12α-hydroxylase transcription by the short heterodimer partner: Insights into the repression mechanism. Nucleic Acids Res. 29**:** 4035–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold K.R., Spady, D.K., Pollock, A.S., Moser, A.H., and Grunfeld, C. 1996. Endotoxin, TNF, and IL-1 decrease cholesterol 7 α-hydroxylase mRNA levels and activity. J. Lipid Res. 37**:** 223–228. [PubMed] [Google Scholar]
- Forman B.M., Goode, E., Chen, J., Oro, A.E., Bradley, D.J., Perlmann, T., Noonan, D.J., Burka, L.T., McMorris, T., Lamph, W.W., et al. 1995. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81**:** 687–693. [DOI] [PubMed] [Google Scholar]
- Goldfarb M. 2001. Signaling by fibroblast growth factors: The inside story. Sci. STKE PE37. [DOI] [PMC free article] [PubMed]
- Goodwin B., Jones, S.A., Price, R.R., Watson, M.A., McKee, D.D., Moore, L.B., Galardi, C., Wilson, J.G., Lewis, M.C., Roth, M.E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6**:** 517–526. [DOI] [PubMed] [Google Scholar]
- Grober J., Zaghini, I., Fujii, H., Jones, S.A., Kliewer, S.A., Willson, T.M., Ono, T., and Besnard, P. 1999. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J. Biol. Chem. 274**:** 29749–29754. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stravitz, R.T., Dent, P., and Hylemon, P.B. 2001. Down-regulation of cholesterol 7α-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J. Biol. Chem. 276**:** 15816–15822. [DOI] [PubMed] [Google Scholar]
- Hughes S.E. 1997. Differential expression of the fibroblast growth factor receptor (FGFR) multigene family in normal human adult tissues. J. Histochem. Cytochem. 45**:** 1005–1019. [DOI] [PubMed] [Google Scholar]
- Kast H.R., Goodwin, B., Tarr, P.T., Jones, S.A., Anisfeld, A.M., Stoltz, C.M., Tontonoz, P., Kliewer, S., Willson, T.M., and Edwards, P.A. 2002. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J. Biol. Chem. 277**:** 2908–2915. [DOI] [PubMed] [Google Scholar]
- Kerr T.A., Saeki, S., Schneider, M., Schaefer, K., Berdy, S., Redder, T., Shan, B., Russell, D.W., and Schwarz, M. 2002. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell 2**:** 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarsky K.F., Donahee, M.H., Rigotti, A., Iqbal, S.N., Edelman, E.R., and Krieger, M. 1997. Overexpression of the HDL receptor SR-BI alters plasma HDL and bile cholesterol levels. Nature 387**:** 414–417. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Suh, Y.A., Robinson, M.J., Clifford, J.L., Hong, W.K., Woodgett, J.R., Cobb, M.H., Mangelsdorf, D.J., and Kurie, J.M. 2000. Stress pathway activation induces phosphorylation of retinoid X receptor. J. Biol. Chem. 275**:** 32193–32199. [DOI] [PubMed] [Google Scholar]
- Lu T.T., Makishima, M., Repa, J.J., Schoonjans, K., Kerr, T.A., Auwerx, J., and Mangelsdorf, D.J. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6**:** 507–515. [DOI] [PubMed] [Google Scholar]
- Makishima M., Okamoto, A.Y., Repa, J.J., Tu, H., Learned, R.M., Luk, A., Hull, M.V., Lustig, K.D., Mangelsdorf, D.J., and Shan, B. 1999. Identification of a nuclear receptor for bile acids. Science 284**:** 1362–1365. [DOI] [PubMed] [Google Scholar]
- Maloney P.R., Parks, D.J., Haffner, C.D., Fivush, A.M., Chandra, G., Plunket, K.D., Creech, K.L., Moore, L.B., Wilson, J.G., Lewis, M.C., et al. 2000. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 43**:** 2971–2974. [DOI] [PubMed] [Google Scholar]
- Miyake J.H., Wang, S.L., and Davis, R.A. 2000. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7α-hydroxylase. J. Biol. Chem. 275**:** 21805–21808. [DOI] [PubMed] [Google Scholar]
- Nicholes K., Guillet, S., Tomlinson, E., Hillan, K., Wright, B., Frantz, G.D., Pham, T.A., Dillard-Telm, L., Tsai, S.P., Stephan, J.P., et al. 2002. A mouse model of hepatocellular carcinoma: Ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am. J. Pathol. 160**:** 2295–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Utsunomiya, Y., Hoshikawa, M., Ohuchi, H., and Itoh, N. 1999. Structure and expression of a novel human FGF, FGF-19, expressed in the fetal brain. Biochim. Biophys. Acta 1444**:** 148–151. [DOI] [PubMed] [Google Scholar]
- Nitta M., Ku, S., Brown, C., Okamoto, A.Y., and Shan, B. 1999. CPF: An orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc. Natl. Acad. Sci. 96**:** 6660–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D.M. and Itoh, N. 2001. Fibroblast growth factors. Genome Biol. 2**:** REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.J., Blanchard, S.G., Bledsoe, R.K., Chandra, G., Consler, T.G., Kliewer, S.A., Stimmel, J.B., Willson, T.M., Zavacki, A.M., Moore, D.D., et al. 1999. Bile acids: Natural ligands for an orphan nuclear receptor. Science 284**:** 1365–1368. [DOI] [PubMed] [Google Scholar]
- Partanen J., Makela, T.P., Eerola, E., Korhonen, J., Hirvonen, H., Claesson-Welsh, L., and Alitalo, K. 1991. FGFR-4, a novel acidic fibroblast growth factor receptor with a distinct expression pattern. EMBO J. 10**:** 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass J.R., Mol, O., Heegsma, J., Geuken, M., Faber, K.N., Jansen, P.L., and Muller, M. 2002. Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology 35**:** 589–596. [DOI] [PubMed] [Google Scholar]
- Russell D.W. and Setchell, K.D. 1992. Bile acid biosynthesis. Biochemistry 31**:** 4737–4749. [DOI] [PubMed] [Google Scholar]
- Seol W., Choi, H.S., and Moore, D.D. 1995. Isolation of proteins that interact specifically with the retinoid X receptor: Two novel orphan receptors. Mol. Endocrinol. 9**:** 72–85. [DOI] [PubMed] [Google Scholar]
- Sheikh F., Sontag, D.P., Fandrich, R.R., Kardami, E., and Cattini, P.A. 2001. Overexpression of FGF-2 increases cardiac myocyte viability after injury in isolated mouse hearts. Am. J. Physiol. Heart Circ. Physiol. 280**:** H1039–H1050. [DOI] [PubMed] [Google Scholar]
- Shimada T., Mizutani, S., Muto, T., Yoneya, T., Hino, R., Takeda, S., Takeuchi, Y., Fujita, T., Fukumoto, S., and Yamashita, T. 2001. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc. Natl. Acad. Sci. 98**:** 6500–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets R.A., Lowe, D.G., Tai, J.T., Sehl, P., Jin, H., Yang, R., Predki, P.F., Rothberg, B.E., Murtha, M.T., Roth, M.E., et al. 1999. Gene expression analysis by transcript profiling coupled to a gene database query. Nat. Biotechnol. 17**:** 798–803. [DOI] [PubMed] [Google Scholar]
- Sinal C.J., Tohkin, M., Miyata, M., Ward, J.M., Lambert, G., and Gonzalez, F.J. 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102**:** 731–744. [DOI] [PubMed] [Google Scholar]
- Stravitz R.T., Vlahcevic, Z.R., Gurley, E.C., and Hylemon, P.B. 1995. Repression of cholesterol 7 α-hydroxylase transcription by bile acids is mediated through protein kinase C in primary cultures of rat hepatocytes. J. Lipid Res. 36**:** 1359–1369. [PubMed] [Google Scholar]
- Stravitz R.T., Rao, Y.P., Vlahcevic, Z.R., Gurley, E.C., Jarvis, W.D., and Hylemon, P.B. 1996. Hepatocellular protein kinase C activation by bile acids: Implications for regulation of cholesterol 7 α-hydroxylase. Am. J. Physiol. 271**:** G293–G303. [DOI] [PubMed] [Google Scholar]
- Stroup D. and Chiang, J.Y. 2000. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7α-hydroxylase gene (CYP7A1). J. Lipid Res. 41**:** 1–11. [PubMed] [Google Scholar]
- Stroup D., Crestani, M., and Chiang, J.Y. 1997. Identification of a bile acid response element in the cholesterol 7 α-hydroxylase gene CYP7A. Am. J. Physiol. 273**:** G508–G517. [DOI] [PubMed] [Google Scholar]
- Tomlinson E., Fu, L., John, L., Hultgren, B., Huang, X., Renz, M., Stephan, J.P., Tsai, S.P., Powell-Braxton, L., French, D., et al. 2002. Transgenic mice expressing human fibroblast growth factor-19display increased metabolic rate and decreased adiposity. Endocrinology 143**:** 1741–1747. [DOI] [PubMed] [Google Scholar]
- Vlahcevic Z.R., Hylemon, P.B., and Chiang, J.Y. 1994. Hepatic cholesterol metabolism. In The liver: Biology and pathobiology (eds. I.M. Arias, J.L. Boyer, and N. Fausto), pp. 379–389. Raven Press, New York.
- Vlahcevic Z.R., Heuman, D.M., and Hylemon, P.B. 1996. Physiology and pathophysiology of enteric circulation of bile acids. In Hepatology: A textbook of liver disease (eds. D. Zakimand T.D. Boyer), pp. 376–417. W.B. Saunders Co., Philadelphia PA.
- Wang H., Chen, J., Hollister, K., Sowers, L.C., and Forman, B.M. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3**:** 543–553. [DOI] [PubMed] [Google Scholar]
- Wang L., Lee, Y.K., Bundman, D., Han, Y., Thevananther, S., Kim, C.S., Chua, S.S., Wei, P., Heyman, R.A., Karin, M., et al. 2002. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell 2**:** 721–731. [DOI] [PubMed] [Google Scholar]
- Xie M.H., Holcomb, I., Deuel, B., Dowd, P., Huang, A., Vagts, A., Foster, J., Liang, J., Brush, J., Gu, Q., et al. 1999. FGF-19, a novel fibroblast growth factor with unique specificity for FGFR4. Cytokine 11**:** 729–735. [DOI] [PubMed] [Google Scholar]
- Yu C., Wang, F., Kan, M., Jin, C., Jones, R.B., Weinstein, M., Deng, C.X., and McKeehan, W.L. 2000. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J. Biol. Chem. 275**:** 15482–15489. [DOI] [PubMed] [Google Scholar]
- Zhang M. and Chiang, J.Y. 2001. Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1): Roles of heaptocyte nuclear factor 4α in mediating bile acid repression. J. Biol. Chem. 276**:** 41690–41699. [DOI] [PubMed] [Google Scholar]