Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration (original) (raw)
Abstract
The biological role of the protein tyrosine kinase, Pyk2, was explored by targeting the Pyk2 gene by homologous recombination. Pyk2–/– mice are viable and fertile, without overt impairment in development or behavior. However, the morphology and behavior of Pyk2–/– macrophages were impaired. Macrophages isolated from mutant mice failed to become polarized, to undergo membrane ruffling, and to migrate in response to chemokine stimulation. Moreover, the contractile activity in the lamellipodia of Pyk2–/– macrophages was impaired, as revealed by measuring the rearward movement toward the nucleus of fibronectin-coated beads on the lamellipodia in opposition to an immobilizing force generated by optical tweezers. Consistently, the infiltration of macrophages into a carageenan-induced inflammatory region was strongly inhibited in Pyk2–/– mice. In addition, chemokine stimulation of inositol (1, 4, 5) triphosphate production and Ca2+ release, as well as integrin-induced activation of Rho and phosphatidyl inositol 3 kinase, were compromised in Pyk2–/– macrophages. These experiments reveal a role for Pyk2 in cell signaling in macrophages essential for cell migration and function.
Focal adhesion kinases (FAK) and proline-rich tyrosine kinases (Pyk2, also designated RAFTK, CAK, and CADTK), are two members of a family of nonreceptor protein tyrosine kinases that are activated by a variety of extracellular stimuli (1–4). FAK and Pyk2 exhibit ≈45% amino acid sequence identity and similar domain structure; an N terminus with similarity to band 4.1 homology domain, a centrally located protein tyrosine kinase domain (5), and two proline-rich regions at the C terminus (1).
FAK is ubiquitously expressed and has been shown to be tyrosine phosphorylated in response to integrin-mediated cell adhesion to the extracellular matrix (6). Targeting the FAK locus by homologous recombination resulted in embryonic lethality, characterized by impaired fibroblast migration (7–9). Pyk2, by contrast, exhibits a more restricted tissue expression pattern primarily in neuronal and hematopoetic tissues (1). Pyk2 was shown to be stimulated in response to a broad range of stimuli, including extracellular signals that elevate intracellular Ca2+ concentration, agonists of G protein-coupled receptors, engagements of the antigen receptors on T cells (1, 10), B cells, and mast cells (11, 12). Pyk2 is also activated in response to inflammatory cytokines (13–15), by stress signals (13), and by integrin-mediated cell adhesion (11). Both Pyk2 and FAK bind to proteins that interact with the cytoskeleton, such as paxillin (16), p130cas, the Rho-GAP protein Graf (17), and a LIM domain-containing protein (18, 19), suggesting a role in regulation of the cytoskeleton and cellular morphology in response to extracellular stimuli. Pyk2 also interacts with a family of phosphatidylinositol (PI) transfer proteins, designated Nirs (20), and with a protein with an Arf GAP activity, designated Pap (21), both in vitro and in vivo. The role of these proteins in vesicle transport raises the possibility that Pyk2 may also play a role in the regulation of membrane flow within cells. Finally, it has been proposed that Pyk2 acts in concert with Src to link Gi- or Gq-coupled receptors with the mitogen-activated protein (MAP) kinase signaling pathway (22). Autophosphorylation of Y402 on Pyk2 generates a binding site for the Src homology 2 domain of the docking protein Grb2, and subsequent recruitment of the Grb2/Sos complex leads to activation of the Ras/MAP kinase signal transduction cascade (22).
To explore the biological functions of Pyk2 in vivo, we have created Pyk2-deficient mice by introducing a mutation into the murine Pyk2 gene. In contrast to the embryonic lethality in FAK-deficient mice, Pyk2–/– mice are viable and fertile, with no gross anatomical alterations. In this report, we demonstrate that chemokine-induced migration of Pyk2–/– macrophages is impaired because of altered cell polarization and diminished cell contractility. In addition, Pyk2-deficient macrophages exhibit multiple defects in cell signaling in response to integrin-mediated cell adhesion or chemokine stimulation, demonstrating that Pyk2 plays an important role in regulating a variety of macrophage responses to extracellular stimuli.
Materials and Methods
Reagents. Rabbit polyclonal anti-Pyk2 antibodies that were raised against the peptide corresponding to 17 C-terminal residues (23) were affinity-purified and used for immunostaining. Immunoblotting experiments were performed with anti-Pyk2 antibodies purchased from Transduction Laboratories (Lexington, KY). Anti-Rho and anti-p85 antibodies were purchased from Santa Cruz Biotechnology. Rhodamin conjugated Phalloidin was purchased from Sigma. SDF1α and MIP1α were purchased from R & D Systems.
Construction of Targeting Vector. A mouse genomic DNA clone corresponding to the N-terminal domain and the kinase domain of Pyk2 was isolated from a 129-strain P1 phage library (Genome Systems). A 4.5-kb _Bam_HI–_Acc_I genomic DNA fragment encoding the 5′ terminus of the kinase domain was inserted to a _Xho_I site flanking a neo expression cassette in the pPNT vector (24), and a 2-kb _Acc_I–_Sac_I genomic DNA fragment was inserted to a _Bam_HI site flanking the herpes simplex virus thymidine kinase expression cassette in the pPNT vector.
Electroporation of Embryonic Stem (ES) Cells and Generation of Pyk2–/– Mice. R1 ES cells (7 × 106) were electroporated in 800 μl of PBS with 32 μg of _Not_I-linealized targeting vector DNA at 240 V, 500 mF by using Gene Pulser (Bio-Rad) and plated on gelatin coated plastic dishes. After 48 h, the cells were transferred to growth medium supplemented with G418 (150 mg/ml) (GIBCO), and gancyclovir (2 mg/ml) (Syntex). G418- and gancyclovir-resistant colonies were picked up 10–12 days after electroporation. Homologous recombination was screened by Southern blot hybridization. Four independently mutated ES cell clones were used in embryo aggregation experiments for generation of mice. Chimeric mice were crossed to 129Sv/Ev females, and germ-line transmission in heterozyous mice was identified in two independent ES-cell-clone-derived F1 mice by Southern blot analysis. Heterozygote mice were intercrossed to produce homozygote mice.
Isolation of Peritoneal Macrophage and Ectopic Expression of Pyk2. A total of 2.0 ml of 3% Brewer thioglycollate in medium was injected into peritoneal cavity 4 days before cell harvest. The inflammatory cells, comprised of a mixture of macrophages and neutrophils, were harvested from killed animals as lavage by PBS. Macrophages were isolated as an adherent cells after plating harvested cells onto tissue-cultured dishes.
Bone marrow cells were suspended in single cells, plated onto plastic dish, and cultured with 100 nM macrophage colony-stimulating factor for 3 days. The cultured cells were infected with the supernatant of EcoPack packaging cells (CLONTECH) transfected pBABE retrovirus vector encoding for Pyk2-GFP. GFP-positive cells were isolated by fluorescence-activated cell sorter and subsequently subjected to further analyses.
Supporting Information. For details of immunofluorescence microscopy, cell migration assay, experimental inflammation in the air pouch model, cell signaling analyses, and laser-trapping experiments, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
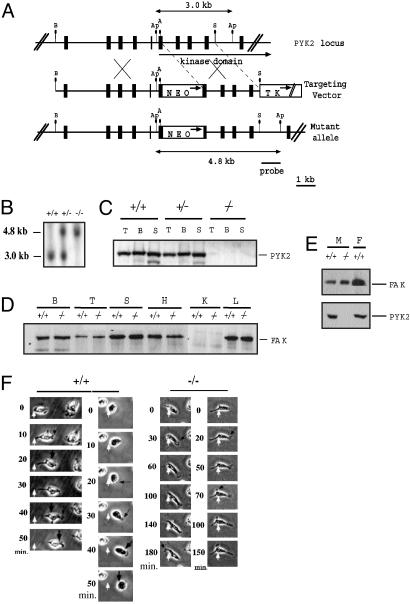
A targeting vector for Pyk2 mice was generated by inserting a neo-expressing cassette into the 5′ portion of Pyk2 sequence encoding for the protein tyrosine kinase domain. The targeting vector also contained a herpes virus thymidine kinase gene to allow for negative selection of nonhomologous recombinants (Fig. 1_A_). ES cells were transfected and clones were selected with G418 and gancyclovir. Drug-resistant clones were analyzed by Southern blot analysis using a probe specific for the 3′ region of the targeting vector (Fig. 1 A and B). Of the 300 drug-resistant ES cell clones that were isolated, six clones were identified as homologous recombinants. Germ-line transmission was obtained with two clones. The chimeric mice were crossed with 129Sv/Ev mice to obtain F1 progeny. Genotyping of mouse tail DNA indicated that the litters of all Pyk2+/– crosses displayed a 1:2:1 ratio of Pyk2+/+, Pyk2+/–, and Pyk2–/– offspring, indicating that the targeted allele does not compromise embryonic development and viability. Immunoblot analysis with anti-Pyk2 antibodies that recognize specifically an epitope in the C terminus of the protein did not show any Pyk2 immunoreactivity in the brain, thymus, and spleen tissue of Pyk2–/– mice (Fig. 1_C_). Similarly, Northern blot hybridization analysis of total RNA isolated from Pyk2–/– brain, thymus, and spleen tissues with a probe encoding for the N terminus of Pyk2 (residues 3–227), did not reveal any Pyk2 expression (data not shown). In addition, immunoblotting with anti-FAK antibodies demonstrated that FAK is similarly expressed in various tissues from wild-type or Pyk2–/– mice (Fig. 1_D_).
Fig. 1.
Generation of Pyk2 knockout mice. (A) Schematic diagrams and partial restriction maps of Pyk2 locus, the Pyk2 targeting vector, and the predicted structure of the Pyk2 locus after homologous recombination with targeting vector. The filled box represents Pyk2 coding sequence, the bar is the 0.8-kb _Sac_I–_Apa_I flanking probe used to identify the targeted Pyk2 locus, the open box indicates the PGK–Neo expression cassette and PGK–TK expression cassette. The arrows in open box indicate the direction of transcription. A, Ap, B, and S represent _Acc_I, _Apa_I, _Bam_HI, and _Sac_I, respectively. (B) Autoradiogram of Southern blot analysis of tail DNA from F2 progeny. Genomic DNA was digested with _Apa_I and hybridized with the radiolabeled probe shown in A. +/+, +/–, and –/– denote DNA from wild-type, heterozygous, and homozygous F2 littermates, respectively. (C) Immunoblot analysis of tissue lysates. Lysates from thymus, brain, and spleen were subjected to immunoprecipitation with Pyk2 antibodies followed by immunoblotting with Pyk2 antibodies. T, B, and S represent thymus, brain, and spleen, respectively. (D) Immunoblot analysis of FAK expression in tissues from wild-type or Pyk2–/– mice. B, brain; T, thymus; S, spleen; H, heart; K, kidney; L, lung. (E) Immunoblot analysis of FAK or Pyk2 expression in macrophages (M) or fibroblasts (F) isolated from wild-type or Pyk2–/– mice. (F) Migration of wild-type or Pyk2–/– macrophages in response to SDF1α stimulation. White arrows mark the initial location of the cell, large black arrows mark cell contraction, and small black arrows mark lamellipodia.
Pyk2–/– homozygous mice were fertile and did not show gross anatomical abnormalities compared with wild-type littermates, including tissues that normally express high levels of Pyk2 such as brain, thymus, and spleen. Further analysis of lymphoid subpopulations by flow cytometry did not reveal any obvious difference in the distribution of T cells, B cells, macrophage monocytes, or natural killer cells in any lymphoid tissue including spleen, lymph node, thymus, bone marrow, and peritoneal cavity. Serum levels of IgG1, IgG2a, IgG3, IgM, and IgA were also similar in Pyk2–/– and wild-type littermates.
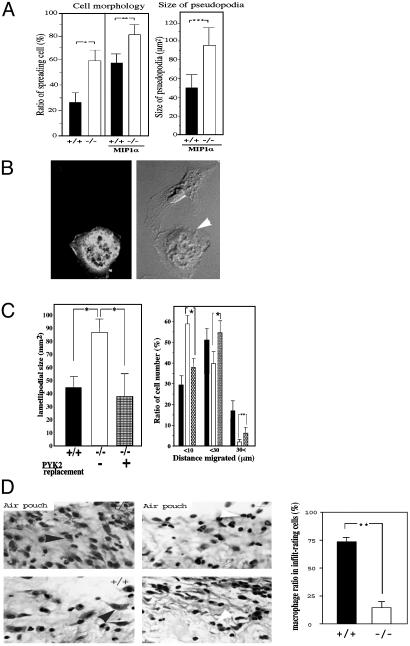
Impaired Migration in Vitro and in Vivo of Pyk2–/– Macrophages. Previous studies have implicated both FAK and Pyk2 in the control of cell migration (7–9). Because Pyk2 is expressed in cells from the immune system, we have analyzed the effect of Pyk2 deficiency on the morphology and migration of macrophages. An immunoblotting experiment with anti-FAK antibodies (Fig. 1_E_) showed similar expression of FAK in wild-type or Pyk2–/– macrophages, demonstrating that Pyk2 deficiency is not compensated for by overexpression of FAK in these cells. The experiment presented in Fig. 1_F_ depicts comparison of migration of wild-type or Pyk2–/– macrophages in response to chemokine stimulation by using video microscopy. In this experiment, wild-type or Pyk2 macrophages were monitored by video microscopy after stimulation with the chemokine SDF1α. This experiment showed that, within 10 min, wild-type macrophages became polarized, developing lamellipodia at one side of the cell. At later time points (>20 min), the cell body moves in the direction established by the leading edge, detaching from the substrate at the trailing edge. In contrast, formation of the new lamellipodia by Pyk2–/– macrophages in response to chemokine stimulation was delayed as compared with that of wild-type macrophages. Furthermore, the cell body of Pyk2–/– macrophages showed reduced ability to follow the leading edge and failed to detach from the substratum. Over time, Pyk2–/– macrophages extended multiple lamellipodia in several directions with similar failure to detach from the substrate. Eventually, most Pyk2–/– macrophages developed several pseudopodia-like processes with minimal net migration. Overall, Pyk2–/– macrophages are able to form a leading edge in response to a chemotactic stimulus, albeit with slower kinetics. However, these cells are unable to move the cell body into the leading edge efficiently and fail to detach the lagging edge from the substratum. Quantification of the number of spreading macrophages (Fig. 2_A_ Left) shows a strong increase among unstimulated and chemokine stimulated Pyk2–/– versus the number of spreading wild-type macrophages. In addition, the average size of lamellipodia of wild-type macrophages was almost half the size of the lamellipodia of Pyk2–/– macrophages (Fig. 2_A_ Right). The morphological changes and migration of Pyk2–/– macrophages were rescued upon infection of bone marrow-derived Pyk2–/– macrophages with a retrovirus that directs the expression of a Pyk2–GFP fusion protein (Fig. 2_B_). Quantitation of the number and size of the lamellipodias of wild-type or mutant macrophages (Fig. 2_C_), showed that the size of the lamellipodia of Pyk2–/– macrophages expressing Pyk2-GFP is similar to the size of lamellipodia of wild-type macrophages (Fig. 2_C_ Left). Similarly, the impaired migration of Pyk2–/– macrophages was restored in mutant macrophages expressing Pyk2-GFP (Fig. 2_D_ Right).
Fig. 2.
Quantitation of impaired cell migration and altered cell morphology of Pyk2–/– macrophages. (A) Quantitative analysis of cell morphology (Left) and size of lamellipodia (Right). Wild-type or Pyk2–/– macrophages were plated on tissue culture dishes, and 200 cells in each genotype were categorized by their morphology and ability to spread and produce cell processes. Macrophage images were captured by using a charge-coupled device camera equipped on an inverted microscope. The size of lamellipodia was measured by NIH image software. Mean values and standard deviation of three experiments are presented. (B) Morphology of Pyk2–/– macrophage ectopically expressing Pyk2-GFP (Left). The morphology of macrophage expressing Pyk2-GFP (Right, white arrow) is similar to the morphology of wild-type macrophages. (C) Rescue of lamellipodia size and migration upon ectopic expression of Pyk2-GFP in Pyk2–/– macrophages. Quantitative analysis of the size of lamellipodia in wild-type (filled bar), Pyk2–/– (open bar), or Pyk2–/– macrophages expressing Pyk2-GFP. Quantitative analysis of cell migration (distance migrated in μM) of wild-type (filled bar), Pyk2–/– (open bar), or Pyk2–/– macrophages expressing GFP-Pyk2 (Right). *, P < 0.05. (D) Carageenen was applied into an air pouch generated under the skin of three wild-type or Pyk2–/– mice. The inflammatory region surrounding the air pouch was fixed and stained by hematoxylin and eosin. Black arrowheads mark representative macrophages and white arrowheads mark representative polymorphic neutrophils. Exude in the air pouch of wild-type or Pyk2–/– mice was recovered, and ratios of number of macrophages over a total of 900 infiltrating cells in the inflammatory region surrounding the air pouch per each wild-type or Pyk2–/– mouse were scored. (Right) Mean values and standard deviations from three wild-type and three Pyk2–/– mice are presented. Statistical analyses were done by unpaired t test. **, Analyzed groups with P < 0.01. Experiments were repeated twice with similar results.
The acute inflammatory response to s.c. carrageenan (23) was next tested to assess the in vivo consequence of impaired migration of Pyk2–/– macrophages. Ten hours after carrageenan injection into wild-type or Pyk2–/– mice, tissue sections of the injected lesion were examined microscopically for the presence of infiltrating macrophages and neutrophils (Fig. 2_D_ Left). The average number of infiltrating cells in wild-type mice was 4.8 × 106 per injected area, whereas the number of infiltrating cells in Pyk2–/– mice was reduced to 2.8 × 106 cells per injected area. Morphological examination of the infiltrating cells indicated that macrophages comprised ≈70% of the infiltrate in wild-type mice but only 20% of the infiltrate in the Pyk2–/– mice, the remaining cells were primarily neutrophils (2_D Right_). The failure of Pyk2–/– macrophages to migrate effectively in vitro is thus correlated with a deficit in inflammatory infiltration in vivo.
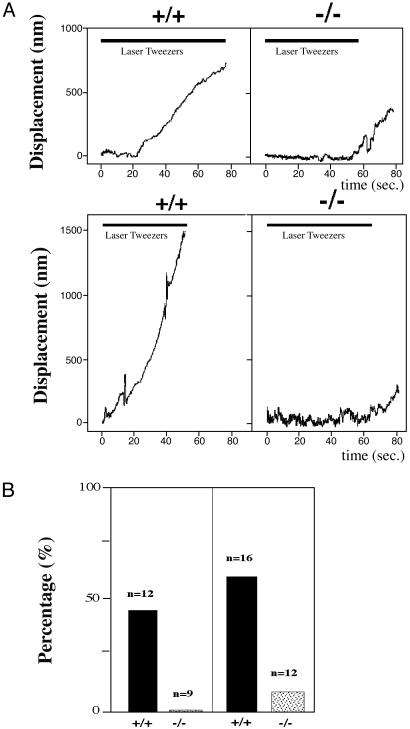
Impairment in Contractile force in Pyk2–/– Macrophages Revealed by Optical Tweezers Analysis. Microscopic observation of migrating macrophages revealed that Pyk2–/– cells could extend lamellipodia but the cell body failed to flow into the newly formed leading edge. This observation suggested that the contractile activity of the cytoskeleton in the lamellipodia was impaired in Pyk2–/– macrophages. The contractile force was determined by measuring the rearward movement toward the nucleus of beads coated with recombinant fragment of fibronectin (FN type III domains 7–10) on lamellipodia in opposition to an immobilizing force generated by optical tweezers (25–27). Immobilizing force by optical tweezers was applied to the beads on the lamellipodia of the cells and the movement of the bound beads was monitored. Representative plots of the distance of bead displacement versus time in wild-type or Pyk2–/– macrophages is presented in Fig. 3_A_. The beads on the lamellipodia from wild-type macrophages exhibited rearward movement and escaped from the force field of the laser trap. After chemokine stimulation, the velocity of bead movement on the lamellipodia of wild-type macrophages was increased and the beads escaped more quickly. With the immobilizing force exerted by the optical tweezers, >50% of the beads that were attached to the lamellipodia of either stimulated or unstimulated wild-type macrophages were able to escape from the force field of the optical trap. In contrast, beads bound to the lamellipodia of Pyk2–/– macrophages did not exhibit rearward movement in presence or absence of chemokine stimulation (Fig. 3_A_). No beads that were attached to the lamellipodia of unstimulated Pyk2–/– macrophages were able to escape from the optical trap, and only 10% of the beads that were attached to the lamellipodia of stimulated macrophage were able to escape from the optical trap (Fig. 3_B_). Overall, rearward movement, i.e., the contractile force generated by the cytoskeleton, is impaired in Pyk2–/– macrophages in comparison to the contractile force generated in wild-type macrophages.
Fig. 3.
Measurement of contractile capacity of lamellipodia in wild-type or Pyk2–/– macrophages by laser tweezers. (A) Plots of bead displacement from leading edge as a function of time. Fibronectin-coated beads were positioned with tweezers on the lamellipodia of Pyk2–/– or wild-type macrophages of unstimulated (Upper) or MIP1α-stimulated (Lower) cells near the leading edge at time 0. The trap remained on for ≈60 sec, as indicated by shaded area. (Upper) Bead displacement on nonstimulated macrophages. (Lower) Displacement on MIP1α-stimulated macrophages. (B) Histogram of ratio (%) of beads displaying escape from trapped field by laser tweezers. All of the beads on the lamellipodia of Pyk2–/– or wild-type macrophages before and after MIP1α stimulation were subjected to a restraining force after initial bead– cell contact for 60 sec. Ratios of beads that escaped and moved rearward were scored. (Left) Score of nonstimulated macrophages. (Right) Score of MIP1α-stimulated macrophages.
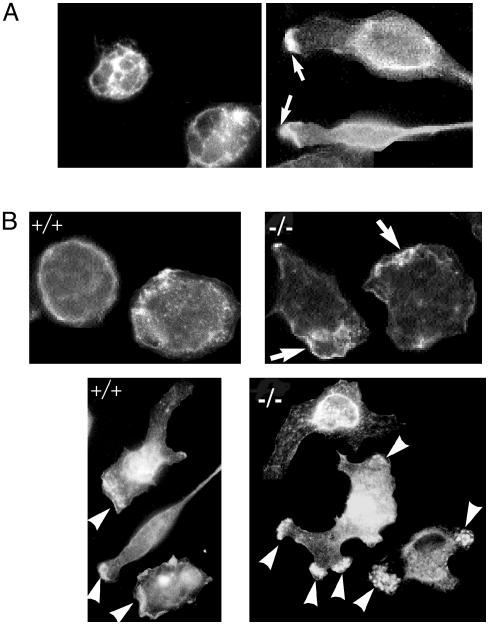
Impaired F-Actin Localization in Pyk2–/– Macrophages. We next analyzed the subcellular localization of Pyk2 in unstimulated or chemokine-stimulated macrophages by using indirect immunofluorescence microscopy of permeabilized macrophages labeled with anti-Pyk2 antibodies. This experiment shows that in unstimulated cells, Pyk2 is predominantly localized at the cell membrane and cytoplasm (Fig. 4_A_ Left), whereas after chemokine stimulation, Pyk2 is localized in cell membrane and cytoplasm as well as in the leading edge of the macrophages at the region exposed to the highest chemokine concentration (Fig. 4_A_ Right). We next used fluorescently labeled phalloidin to compare actin localization in unstimulated or chemokine stimulated wild-type or Pyk2–/– macrophages. Fig. 4_B_ demonstrates that unstimulated Pyk2–/– macrophages show more pronounced F-actin staining in ruffles as compared with F-actin staining in unstimulated wild-type macrophages. In response to localized chemokine stimulation, F-actin was found to be concentrated at the edge of the cells at the region exposed to the highest chemokine concentration. In contrast, in Pyk2–/– macrophages that were also subjected to localized chemokine stimulation, F-actin was found to be accumulated at multiple sites along the cell periphery (Fig. 4_B_ Lower). The accumulation of F-actin at multiple sites in Pyk2–/– macrophages is consistent with the extension of multiple lamellipodia toward different directions that was observed in the Pyk2-deficient macrophages. These experiments demonstrate that the Pyk2–/– macrophages fail to become properly oriented toward a chemotactic gradient.
Fig. 4.
Pyk2 and F-actin localization in response to chemokine stimulation. (A) Unstimulated or MIP1α stimulated cells were fixed, permeabolized, labeled with anti-Pyk2 antibodies, followed by labeling with fluorescently conjugated anti-rabbit antibodies. In addition to membrane and cytoplasmic localization, Pyk2 is localized in the leading edge of stimulated cells pointing toward the chemokine gradient. Arrows mark the leading edge. (B) Altered F-actin localization in Pyk2–/– macrophages. Wild-type or Pyk2–/– macrophage were plated on cover glasses, fixed by 4% paraformaldehyde, and stained with fluorescently labeled phalloidin. (Upper) F-actin distribution in unstimulated wild-type or Pyk2–/– macrophages. (Lower) F-actin distribution after stimulation under MIP1α gradient concentration. The bottom left part of the field was exposed to the highest concentration of MIP1α. White arrows mark regions with strong F-actin staining.
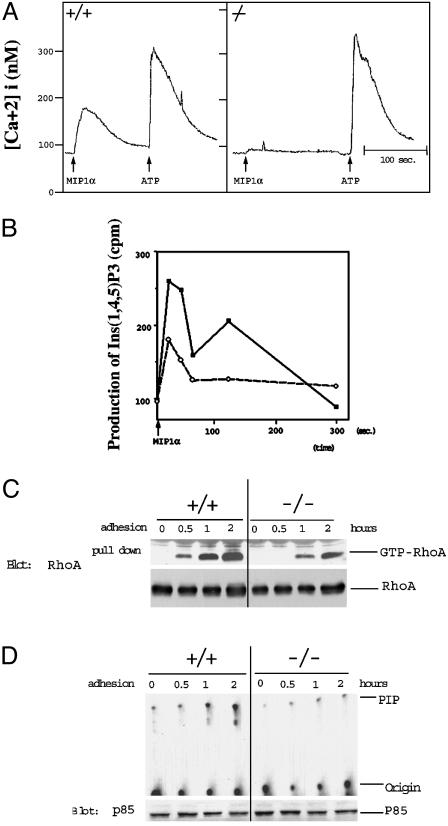
Reduced Intracellular Calcium Release and Inositol (1, 4, 5) Triphosphate [Ins(1,4,5)P3] Production in Pyk2–/– Macrophages after Chemokine Stimulation. Calcium plays an important role in the control of a variety of intracellular events as well as in the control of cell shape and movement (28). Therefore, we measured cytoplasmic calcium release in single cells in response to chemokine stimulation by using quantitative fluorescence microscopy of Fura-2-loaded cells. Treatment of wild-type macrophages attached to cover slips showed maximum increase in cytoplasmic [Ca2+] concentration at ≈300 nM chemokine. By contrast, Pyk2–/– macrophages did not show an obvious increase in [Ca2+] concentration (Fig. 5_A_). However, both wild-type and Pyk2–/– macrophages showed a similar rise in intracellular [Ca2+] concentration in response to ATP stimulation (Fig. 5_A_). This experiment shows that Pyk2 plays an important role in the control of chemokine-induced Ca2+ release in adherent macrophages. Defects in calcium release may contribute toward the failure of the cells to detach at the rear end leading to impairment in cell migration because proteins that regulate the degradation of focal contact components and disassembly of F-actin require calcium for their action (29, 30).
Fig. 5.
Impaired cellular signaling in Pyk2–/– macrophages. (A) Single cell measurements of Ca2+ release of Fura-2 loaded wild-type or Pyk2–/– macrophages in Ringer's solution containing 2 mM calcium. Changes in fluorescence intensity as a function of time were monitored in response to MIP1α or ATP stimulation. Similar traces were obtained in >30 recordings of different cells. (B) Production of Ins(1,4,5)P3 in wild-type or Pyk2–/– macrophages. Wild-type or Pyk2–/– macrophages were labeled with myo-[3H]inositol for 24 h. After MIP1α stimulation, the lipid fraction was extracted and analyzed by HPLC. Filled squares indicate production of Ins(1,4,5)P3 in wild-type macrophages, and open circles indicate production of Ins(1,4,5)P3 in Pyk2–/– macrophages. The experiment shows mean values of [3H]Ins(1, 4, 5)P3 produced in duplicate samples. This experiment was repeated three times with similar results. (C) Reduced integrin-induced Rho activation in Pyk2-null macrophages. Wild-type or Pyk2–/– macrophages were plated on fibronectin-coated dishes for 30 min, 1 h, or 2 h incubation. Lysates from unstimulated or stimulated cells were treated with GST-RBD (Rho-binding domain) bound to glutathione beads as described in Materials and Methods. The amount of RhoA:GTP complex was determined by immunoblotting with anti-RhoA antibodies. (D) Integrin-stimulation of PI-3 kinase activity is compromised in Pyk2-null macrophages. Cell lysates were immunoprecipitated with anti-p85 antibodies followed by measurement of activity of PI-3 kinase as described (42).
A significant proportion of intracellular Ca2+ released in response to extracellular signals is mediated by Ins(1,4,5)P3 production (31). We therefore analyzed the production of Ins(1,4,5)P3 in these cells. In this experiment, wild-type or Pyk2–/– macrophages were labeled with [3H]myo-inositol and then stimulated with MIP1α. At various times after chemokine stimulation, the production of Ins(1,4,5)P3 was determined by HPLC analysis (32). Wild-type macrophages showed a biphasic production of Ins(1,4,5)P3 with peaks at 20 sec and 2 min after chemokine stimulation. The experiment presented in Fig. 5_B_ shows that Ins(1,4,5)P3 production was compromised in Pyk2–/– macrophages; the peak at 20 sec after stimulation was reduced by ≈50% as compared with Ins(1,4,5)P3 production in wild-type macrophages, and no Ins(1,4,5)P3 was generated after 2 min of chemokine stimulation (Fig. 5_B_).
Impaired Rho and PI-3 Kinase Activation in Response to Integrin-Mediated Cell Adhesion of Pyk2–/– Macrophages. The small G protein Rho has been implicated in the control of actin polymerization, the contractile activity of the acto-myosin system (33), and the control of integrin binding to the extracellular matrix (34). Therefore, we have examined the status of Rho in response to integrin-mediated adhesion of wild-type or Pyk2–/– macrophages. In this experiment, either wild-type or Pyk2–/– macrophages were plated on substratum coated with fibronectin. Unstimulated or stimulated cells were solubilized, and cell lysates were subjected to a “pull-down” assay using a GST fusion protein containing the CRIB domain of Rhotekin as a measure for GTP-bound state of Rho. The experiment presented in Fig. 5_C_ shows that Rho is activated after integrin-mediated attachment to the fibronectin-coated substratum of wild-type macrophages. By contrast, Rho activation was compromised after integrin-mediated attachment of Pyk2–/– macrophages.
The activation of PI-3 kinase in wild-type or Pyk2–/– macrophages in response to integrin-mediated cell adhesion was also compared. The experiment presented in Fig. 5_D_ shows cell adhesion-dependent activation of PI-3 kinase activity in wild-type macrophages. By contrast, PI-3 kinase activity was strongly impaired in macrophages isolated from Pyk2–/– mice, demonstrating that Pyk2 is required for cell adhesion-dependent PI-3 kinase stimulation in macrophages.
Discussion
Pyk2 and FAK are two members of a family of protein tyrosine kinases that are activated by a variety of extracellular stimuli. Pyk2 was shown to be activated by peptide hormones that bind to G protein-coupled receptors that mediate their intracellular responses via Gi and Gq type G proteins (1). In certain cells, Pyk2 is activated by integrin-mediated cell adhesion (11), and in other cells Pyk2 is strongly activated by a variety of extracellular stimuli that result in elevation of intracellular [Ca2+] concentration (1).
Pyk2 deficiency results in compromised chemokine-induced macrophage migration. The migration of cells in culture in response to extracellular stimuli can be divided into a five-step cycle: (i) extension of the leading edge toward stimulus; (ii) adhesion of the leading edge to the substrate; (iii) movement of the cytoplasm toward the leading edge; (iv) release from contact sites at the lagging edge; and (v) recycling of membrane receptors from the lagging edge to the leading edge of the cell (27). Each step of the cycle requires orderly changes in cytoskeletal structures and focal contacts, a process that is regulated by a variety of intracellular enzymes, including protein tyrosine kinases and protein tyrosine phosphatases (35). Several steps of the migration cycle are impaired by Pyk2 deficiency: in Pyk2–/– macrophages, extension of the leading edge is delayed and multiple extensions are generated (steps i and ii). Furthermore, Pyk2–/– macrophages inefficiently detach the lagging edge from the matrix to allow net movement (steps iv and v). It appears that the migration of the cytoplasm is also impaired in the Pyk2-deficient macrophages. Alterations in the migration cycle are particularly evident after the initial extension of lamellipodia and altered cell polarization, which could be observed as multi directional lamellipodia and redistribution of F-actin in multiple sites.
The experiments presented in this report show that several intracellular signals that are stimulated by chemokines are impaired in Pyk2–/– macrophages, including chemokine-induced production of PI hydrolysis and Ca2+ release. We have also demonstrated that chemokine stimulation of tyrosine phosphorylation of phospholipase Cγ is compromised in Pyk2-null macrophages (data not shown), suggesting a potential mechanism for the diminished PI hydrolysis and Ca2+ release in mutant macrophages. The diminished Ca2+ release in Pyk2–/– macrophages is of particular interest because there is good evidence that, in certain cell types, intracellular release of Ca2+ leads to Pyk2 activation (1), suggesting that Pyk2 can function both as a Ca2+ sensor as well as an element crucial for the control of Ca2+ release in response to extracellular cues.
The strength of the traction force in the lamellipodia of wild-type or Pyk2–/– macrophages was analyzed by applying the laser tweezer method (25). The ability of plasma membrane-bound beads to move rearward on the cell surface in opposition to the restraining force imposed by the force field of the laser provides a measurement for the ability of cells to migrate over a fixed point on the matrix. Measurements of bead movements on wild-type or Pyk2–/– lamellipodia revealed the diminished capacity of integrin/cytoskeletal complex in Pyk2–/– macrophages to supply the contractile force necessary for cell migration. This can be caused by decreased tightness of the linkage between integrin and the cytoskeleton or decreased contractile force of the cytoskeleton or decreases in both processes. However, no difference was detected in the expression level of β1, β2, and αM (CD11β) integrins as revealed by fluorescence-activated cell sorter analysis (data not shown). Therefore, it appears that diminished traction force in Pyk2–/– macrophages will stabilize lamellipodia that would normally be retracted, resulting in the disruption of cellular polarization and migration.
It is thought that cells subjected to a chemotactic gradient are “sampling” the environment by extending lamellipodia in several directions. Stable attachments to the extracellular matrix are formed in the direction of the highest concentration of the stimulus, whereas other lamellipodia eventually retract into the cell. The maintenance of an appropriate cellular polarization in a gradient thus requires regulation of contact stability and contractile capacity of the lamellipodia. In Pyk2–/– macrophages, both processes are impaired. Cell morphological change, such as contraction of lamellipodia or formation of contact, require continuous changes in the assembly of the cytoskeleton. Members of the Rho family of GTPases play an important role in modulating the cytoskeleton in response to extracellular stimuli. Furthermore, Rho GTPases were implicated in the control of cytoskeletal organization, actomyosin contraction, vesicle transport, phospholipid production and in the control of integrin clustering (36–39). We have demonstrated that tyrosine phosphorylation of Vav in response to cell adhesion is reduced in Pyk2-null macrophages (data not shown), suggesting a potential mechanism for reduced Rho-activation in mutant macrophages. The reduced activation of Rho and PI-3 kinase activity induced by integrin-mediated adhesion of Pyk2–/– macrophages may lead to impairment of a variety of cellular responses. Moreover, because activation of Rho has been shown to be critical for contraction of lamellipodia (39), the decreased contractility in Pyk2–/– macrophages could be due in part to the decreased activity of Rho. Although it is not yet clear how Pyk2 is linked to the multiple signaling pathways that depend, as shown in this report, on the integrity of this tyrosine kinase, it is clear, however, that Pyk2 plays a role in the control of macrophage migration and function.
Supplementary Material
Supporting Text
Acknowledgments
We thank M. Ittmann, A. Weiss, D. Qian, A. Zychlinsky, M. Schwartz, and F. Maxfield for advice and help.
Abbreviations: FAK, focal adhesion kinase; PI, phosphatidylinositol; ES, embryonic stem; Ins(1,4,5)P3, inositol (1, 4, 5) triphosphate.
References
- 1.Lev, S., Moreno, H., Martinez, R., Canoll, P., Peles, E., Musacchio, J. M., Plowman, G. D., Rudy, B. & Schlessinger, J. (1995) Nature 376**,** 737–745. [DOI] [PubMed] [Google Scholar]
- 2.Avraham, S., London, R., Fu, Y., Ota, S., Hiregowdara, D., Li, J., Jiang, S., Pasztor, L. M., White, R. A., Groopman, J. E. & Avraham, H. (1995) J. Biol. Chem. 270**,** 27742–27751. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki, H., Nagura, K., Ishino, M., Tobioka, H., Kotani, K. & Sasaki, T. (1995) J. Biol. Chem. 270**,** 21206–21219. [DOI] [PubMed] [Google Scholar]
- 4.Yu, H., Li, X., Marchetto, G. S., Dy, R., Hunter, D., Calvo, B., Dawson, T. L., Wilm, M., Anderegg, R. J., Graves, L. M. & Earp, H. S. (1996) J. Biol. Chem. 271**,** 29993–29998. [DOI] [PubMed] [Google Scholar]
- 5.Girault, J. A., Costa, A., Derkinderen, P., Studler, J. M. & Toutant, M. (1999) Trends Neurosci. 22**,** 257–263. [DOI] [PubMed] [Google Scholar]
- 6.Richardson, A. & Parsons, T. (1996) Nature 380**,** 538–540. [DOI] [PubMed] [Google Scholar]
- 7.Ilic, D., Furuta Y., Kanazawa, S., Taketa, N, Sobue, K., Nakatsuji, N., Ninomira, S., Fujimoto, J., Okada, M. & Yamamoto, T. (1995) Nature 377**,** 539–544. [DOI] [PubMed] [Google Scholar]
- 8.Sieg, D. J., Ilic, D., Jones, K. C., Damsky, H. C., Hunter, T. & Schlaepfer, D. D. (1998) EMBO J. 17**,** 5933–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieg, D. J., Hauck, C. R. & Schlaepfer, D. D. (1999) J. Cell Sci. 112**,** 2677–2691. [DOI] [PubMed] [Google Scholar]
- 10.Qian, D., Lev, S., van Oers, N. S., Dikic, I., Schlessinger, J. & Weiss, A. (1997) J. Exp. Med. 185**,** 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Astier, A., Avraham, H., Manie, S. N., Groopman, J., Canty, T., Avraham, S. & Freedman, A. S. (1997) J. Biol. Chem. 272**,** 228–232. [DOI] [PubMed] [Google Scholar]
- 12.Okazaki, H., Zhang, J., Hamawy, M. M. & Siraganian, R. P. (1997) J. Biol. Chem. 272, 32443–32447. [DOI] [PubMed] [Google Scholar]
- 13.Tokiwa, G., Dikic, I., Lev, S. & Schlessinger, J. (1996) Science 273**,** 792–794. [DOI] [PubMed] [Google Scholar]
- 14.Miyazaki, T., Takaoka, A., Nogueira, L., Dikic, I., Fujii, H., Tsujino, S., Mitani, Y., Maeda, M., Schlessinger, J. & Taniguchi, T. (1998) Genes Dev. 12**,** 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takaoka, A., Tanaka, N., Mitani, Y., Miyazaki, T., Fujii, H., Sato, M., Kovarik, P., Decker, T., Schlessinger, J. & Taniguchi, T. (1999) EMBO J. 18**,** 2480–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salgia, R., Avraham, S., Pisick, E., Li, J. L., Raja, S., Greenfield, E. A., Sattler, M., Avraham, H. & Griffin, J. D. (1996) J. Biol. Chem. 271, 31222–31226. [DOI] [PubMed] [Google Scholar]
- 17.Ohba, T., Ishino, M., Aoto, H. & Sasaki, T. (1998) Biochem. J. 330**,** 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuya, M., Sasaki, H., Aoto, H., Mitaka, T., Nagura, K., Ohba, T., Ishino, M., Takahashi, S., Suzuki, R. & Sasaki, T. (1998) J. Biol. Chem. 273**,** 1003–1014. [DOI] [PubMed] [Google Scholar]
- 19.Lipsky, B. P., Beals, C. R. & Staunton, D. E. (1998) J. Biol. Chem. 273**,** 11709–11713. [DOI] [PubMed] [Google Scholar]
- 20.Lev, S., Hernandez, J., Martinez, R., Chen, A., Plowman, G. & Schlessinger, J. (1999) Mol. Cell. Biol. 19**,** 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreev, J., Simon, J. P., Sabatini, D. D., Kam, J., Plowman, G., Randazzo, P. A. & Schlessinger, J. (1999) Mol. Cell. Biol. 19**,** 2338–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dikic, I., Tokiwa, G., Lev, S., Courtneidge, S. A. & Schlessinger, J. (1996) Nature 383**,** 547–550. [DOI] [PubMed] [Google Scholar]
- 23.Dikic I, Dikic, I. & Schlessinger, J. (1998) J. Biol. Chem. 273**,** 14301–14308. [DOI] [PubMed] [Google Scholar]
- 24.Soriano, P., Montgomery, C., Geske, R. & Bradley, A. (1991) Cell 64**,** 693–702. [DOI] [PubMed] [Google Scholar]
- 25.Choquet, D., Felsenfeld, D. P. & Sheetz, M. P. (1997) Cell 88**,** 39–48. [DOI] [PubMed] [Google Scholar]
- 26.Felsenfeld, D. P., Schwartzberg, P. L., Venegas, A., Tse, R. & Sheetz, M. P. (1999) Nat. Cell Biol. 1, 200–206. [DOI] [PubMed] [Google Scholar]
- 27.Sheetz, M. P., Felsenfeld, D., Galbraith, C. G. & Choquet, D. (1999) Biochem. Soc. Symp. 65**,** 233–243. [PubMed] [Google Scholar]
- 28.Lawson, M. A. & Maxfield, F. R. (1995) Nature 377**,** 75–79. [DOI] [PubMed] [Google Scholar]
- 29.Witke, W., Sharpe, A. H., Hartwig, J. H., Azuma, T., Stossel, T. P. & Kwiatkowski, D. J. (1995) Cell 81**,** 41–51. [DOI] [PubMed] [Google Scholar]
- 30.Kulkarni, S., Saido, T. C., Suzuki, K. & Fox, J. E. (1999) J. Biol. Chem. 274**,** 21265–21275. [DOI] [PubMed] [Google Scholar]
- 31.Furuichi, T., Yoshikawa, S., Miyawaki, A., Wada, K., Maeda, N. & Mikoshiba, K. (1999) Nature 342**,** 32–38. [DOI] [PubMed] [Google Scholar]
- 32.Falasca, M., Logan, S. K., Lehto, V. P., Baccante, G., Lemmon, M. A. & Schlessinger, J. (1998) EMBO J. 17**,** 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen, W. E., Jones, G. E., Pollard, J. W. & Ridley, A. J. (1997) J. Cell Sci. 110**,** 707–720. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz, M. A. & Shattil, S. J. (2000) Trends Biochem. Sci. 25**,** 388–391. [DOI] [PubMed] [Google Scholar]
- 35.Manes, S., Mira, E., Gomez-Mouton, C., Zhao, Z. J., Lacalle, R. A. & Martinez, A. C. (1999) Mol. Cell. Biol. 19**,** 3125–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Exton, J. H. (1997) Eur. J. Biochem. 243**,** 10–20. [DOI] [PubMed] [Google Scholar]
- 37.Hotchin, N. A. & Hall, A. (1995) J. Cell Biol. 131**,** 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renshaw, M. W., Toksoz, D. & Schwartz, M. A. (1996) J. Biol. Chem. 271**,** 21691–21694. [DOI] [PubMed] [Google Scholar]
- 39.Kimura, K., Ito, M., Amano, M., Chihara, K., Fukata, Y., Nakafuku, M., Yamamori, B., Feng, J., Nakano, T., Okawa, K., et al. (1996) Science 273**,** 245–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Text