“Black holes” and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli (original) (raw)
Abstract
Plasmids, bacteriophages, and pathogenicity islands are genomic additions that contribute to the evolution of bacterial pathogens. For example, Shigella spp., the causative agents of bacillary dysentery, differ from the closely related commensal Escherichia coli in the presence of a plasmid in Shigella that encodes virulence functions. However, pathogenic bacteria also may lack properties that are characteristic of nonpathogens. Lysine decarboxylase (LDC) activity is present in ≈90% of E. coli strains but is uniformly absent in Shigella strains. When the gene for LDC, cadA, was introduced into Shigella flexneri 2a, virulence became attenuated, and enterotoxin activity was inhibited greatly. The enterotoxin inhibitor was identified as cadaverine, a product of the reaction catalyzed by LDC. Comparison of the S. flexneri 2a and laboratory E. coli K-12 genomes in the region of cadA revealed a large deletion in Shigella. Representative strains of Shigella spp. and enteroinvasive E. coli displayed similar deletions of cadA. Our results suggest that, as Shigella spp. evolved from E. coli to become pathogens, they not only acquired virulence genes on a plasmid but also shed genes via deletions. The formation of these “black holes,” deletions of genes that are detrimental to a pathogenic lifestyle, provides an evolutionary pathway that enables a pathogen to enhance virulence. Furthermore, the demonstration that cadaverine can inhibit enterotoxin activity may lead to more general models about toxin activity or entry into cells and suggests an avenue for antitoxin therapy. Thus, understanding the role of black holes in pathogen evolution may yield clues to new treatments of infectious diseases.
Virulence genes of bacterial pathogens may be encoded on plasmids, bacteriophages, or the chromosome. Virulence is often multifactorial and coordinately regulated, and virulence genes tend to be clustered in the genome. Recently, the acquisition of pathogenicity islands was proposed as a major mechanism in pathogen evolution (1). Pathogenicity islands are regions on the genomes of certain pathogenic bacteria that are absent in nonpathogenic strains of the same or closely related species and that contain large contiguous blocks of virulence genes. The addition of pathogenicity islands is being recognized as an important element in the evolution of bacterial pathogens through horizontal spread of virulence genes similar to the horizontal transfer mediated by plasmids and bacteriophages. In this study, we report the existence of a complementary but inverse pathway that may enable commensal bacteria to evolve toward a pathogenic lifestyle: the formation of “black holes,” i.e., deletions of genes that are detrimental to a pathogenic lifestyle.
The four species of Shigella are so closely related to Escherichia coli that all of these bacteria could be considered members of a single species. They share greater than 90% homology by DNA–DNA reassociation analysis (2) and display colinearity of their chromosomes such that gene transfer by conjugation and transduction and formation of recombinants between Shigella and E. coli occur with high efficiency (3). Nevertheless, Shigella spp. are frank pathogens that cause bacillary dysentery, whereas E. coli (with the exception of certain pathogenic clones) are commensals of the human intestine. Of interest, one class of pathogenic E. coli, the enteroinvasive E. coli (EIEC), resembles a genetic hybrid between E. coli and Shigella. EIEC carry plasmids with extensive homology to the virulence plasmid of Shigella and cause a diarrheal disease that is clinically similar to dysentery caused by Shigella (4). One of the striking biochemical features shared by EIEC and Shigella is a lack of lysine decarboxylase (LDC) activity. Whereas almost 90% of E. coli strains are LDC+ (5), all strains of EIEC and Shigella spp. are LDC− (6). This observation suggested the possibility that absence of LDC activity may be important for Shigella and EIEC virulence.
In this paper, we demonstrate that expression of LDC activity by Shigella has no adverse effects on the invasive capability of this organism. However, cadaverine, produced by the decarboxylation of lysine, acts as an inhibitor of Shigella enterotoxin activity. We further show that Shigella spp. and EIEC have irreversibly lost LDC activity by genome deletion. These observations suggest that the creation of black holes (genome deletions) is a pathway that complements gene acquisition in the evolution of bacterial pathogens.
MATERIALS AND METHODS
Bacterial Strains and Media.
The strains used in this study are listed in Table 1. Strains were grown at 37°C in Luria–Bertani medium (LB) with aeration, on LB agar, or on M9 minimal salts with glucose (12). Media were supplemented with thiamine (50 μg/ml), spectinomycin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (15 μg/ml) as required. To optimize enterotoxin production, bacteria were grown in LB with ethylenediamine-N,_N_′-diacetic acid to chelate iron. Supernatants from overnight cultures of bacteria were harvested by centrifugation, filter-sterilized, and placed on ice until used.
Table 1.
Bacterial strains
| Strain | Description | Source or reference |
|---|---|---|
| 2457T | S. flexneri 2a wild type | 7 |
| BS103 | Plasmid-cured derivative of 2457T | 8 |
| BS529 | 2457T transformed with pCADA (_cadA_†) | This study |
| BS573 | BS103 _zii-215_∷Tn_10_dCamRCP2 _zjh-225_∷Tn_10_dSpcRCP2 | This study |
| MC4100 | E. coli K-12 prototype | 9 |
| MG1655 | E. coli K-12 prototype | B. Bachmann* |
| CAG18427 | MG1655 zje-2241_∷Tn_10 | 10 |
| χM2115 | MG1655 _zii-215_∷Tn_10_dCamRCP2 | 11 |
| χM2125 | MG1655 _zjh-225_∷Tn_10_dSpcRCP2 | 11 |
| χM2500 | MG1655 _zii-215_∷Tn_10_dCamRCP2 _zjh-225_∷Tn_10_dSpcRCP2 | This study |
pCADA is a plasmid that contains the wild-type cadA gene from E. coli K-12 under the transcriptional control of the lac promoter (13). The cadA gene in pCADA is expressed constitutively in Shigella flexneri because of the absence of lac repressor in the organism and the plasmid vector. Cadaverine (>98% pure) was obtained from Sigma. Cultures of E. coli for measurement of LDC activity under inducing conditions were grown in medium buffered with 100 mM 4-morpholineethanesulfonic acid to pH 5.5 (13).
Bacterial Genetics Techniques and Biochemical Assays.
Generalized transduction with P1 was as described (12). E. coli MG1655 mutants containing Tn_10_dSpcRCP2 or Tn_10_dCamRCP2 insertions were generated by electroporation with plasmids pGI300 or pGI310 as described (14). MG1655 double insertion mutants and single and double insertion mutants of S. flexneri 2a strain BS103 were generated by transducing recipient strains with P1Δ_dam rev6_ lysates of MG1655 insertion mutants (15).
Assays for LDC activity were those of Falkow (16) and Phan et al. (17). The former assay provides a qualitative measure of LDC activity based on a shift in pH from acid to alkaline due to the production of cadaverine from the decarboxylation of lysine. The latter assay measures cadaverine produced from lysine based on the differential solubility of the reaction products of 2,4,6-trinitrobenzenesulfonic acid with cadaverine and lysine. Medium blanks were used to control for trace amounts of amines and amino acids in the culture supernatant.
Virulence Assays.
The HeLa cell invasion and plaque assays have been described (18). The rabbit ileal loop assay was performed as described (19). Adult New Zealand white rabbits were starved for 24 h, but were allowed water ad libitum, and then were anesthetized with ketamine (50 mg/kg body weight) and acepromazine (1 mg/kg), followed by xylazine (7 mg/kg) i.m. Uninoculated LB and sterile culture supernatants (1 ml) were injected into the lumen of the intestine proximal to a tie placed near the mesoappendix. A second tie isolated the site of inoculation. Proceeding proximally along the ileum, five loops (7–8 cm long and separated by double ties) were isolated and inoculated. The sequence of inoculation of loops with the different preparations was randomized so that it varied from rabbit to rabbit. After 18 h, the animals were killed, and fluid volume and the length of the loops were measured. Ussing chamber experiments were performed as described (20). In brief, adult New Zealand white rabbits were killed by cervical dislocation, and a 20-cm segment of distal ileum was excised quickly and cut open along the mesenteric border. The ileum was rinsed free of luminal contents, stripped of the muscular and serosal layers, and mounted in Lucite Ussing chambers of aperture 1.12 cm2 (World Precision Instruments, Sarasota, FL). The tissue was bathed in Ringer’s solution at 37°C and gassed with 95% O2/5% CO2. Once the tissue reached a steady-state condition, 300 μl of sterile culture supernatant was added to the mucosal side of the tissue. Sterile culture supernatant (300 μl) also was added to the serosal side to preserve the osmotic balance. Supernatants of S. flexneri strain 2457T also were tested in the presence of either 300 μl of supernatant from S. flexneri strain BS529 or increasing concentrations of cadaverine. In a subset of experiments, the intestinal epithelium was pretreated for 30 min with cadaverine (300 μM), washed twice with fresh Ringer’s, and then exposed to 300 μl of 2457T supernatant.
Once the tissues were exposed to the above treatments, the potential difference (PD; the difference in voltage between the mucosal and serosal sides of the tissue) was measured under open-circuited conditions. The increase in voltage resulting from the passage of 100 μA current was used to calculate the short circuit current (Isc; the amount of current needed to nullify the PD) and the tissue resistance from Ohm’s law (Isc = PD/tissue resistance) (20).
Genomic DNA Biophysical Techniques.
Genomic DNA was purified from 5.0-ml overnight cultures of _E. coli_∷Tn_10_dRCP2 and _S. flexneri_∷Tn_10_dRCP2 mutants in a manner suitable for yielding macrorestriction fragments (0.05–1.0 Mb) as described (21). After digestion of agarose-embedded DNA with I-_Sce_I (Boehringer Mannheim) for 1 hr or with I-_Ceu_I (Panvera, Madison, WI) overnight, according to the manufacturers’ directions, reaction buffer was decanted, and dots were melted (70°C) and gently pipetted into sample wells in 1.3% agarose (Fastlane, FMC) gels for electrophoresis in a Bio-Rad DR-III pulse field gel apparatus. Pulse times were ramped from 10 to 13 s over 10 h and 60 to 65 s over 12 h at a field strength of 6 V/cm. After electrophoresis of samples, gels were analyzed as described (22).
Genomic DNA Preparation and Southern Hybridization.
Genomic DNA was prepared by standard methods (23). Southern hybridization of dot blots was done by spotting 10 μg of genomic DNA onto filters (GeneScreen, Dallas, and Dupont/NEN) and hybridizing with oligonucleotide probes end labeled with digoxigenin (DIG/Genius labeling kit, Boehringer Mannheim). Blots were processed with CSPD (Boehringer Mannheim) for chemiluminescence according to the manufacturer’s instructions.
RESULTS
Effect of cadA Expression on Shigella Virulence.
We initially attempted to construct an LDC+ derivative of S. flexneri by transducing cadA [E. coli_ K-12 map position 93.83 min (24)], the gene for LDC, into 2457T. After infection with a P1 lysate grown on E. coli K-12 strain CAG18427 (Tn_10 inserted at 94.5 min.), tetracycline-resistant transductants were selected and screened for LDC activity. Transduction between E. coli K-12 and S. flexneri is normally fairly efficient, and cadA should cotransduce with tetracycline resistance at ≈23%. However, we were never able to isolate more than 10 tetracycline-resistant transductants per experiment, and none coinherited LDC activity. By contrast, 150–250 tetracycline-resistant transductants routinely were obtained when a donor strain with Tn_10_ inserted at 57.5 min was used. Failure to transduce cadA into 2457T suggested that the S. flexneri chromosome may have a large deletion in the cadA region relative to the E. coli K-12 genome and that the rare transductants recovered were the result of illegitimate recombination events (see below).
As an alternative approach, a cloned copy of cadA from E. coli K-12 was transformed into S. flexneri 2a strain 2457T. The resulting transformant, BS529, expressed LDC activity, and when tested for expression of virulence phenotypes, it invaded HeLa cells and produced plaques as efficiently as the wild-type parent strain. Thus, expression of cadA had no discernible effect on Shigella virulence in tissue culture invasion assays.
Wild-type S. flexneri 2a produce enterotoxins whose activity can be measured by the ability to cause fluid secretion in ligated rabbit intestinal loops. Previous studies (25) demonstrated that E. coli–Shigella hybrids from matings between a S. flexneri 2a donor and an E. coli K-12 recipient failed to induce fluid secretion in ligated rabbit ileal loops if the recombinants retained the LDC+ phenotype of the E. coli recipient. By contrast, hybrids that inherited the S. flexneri region around 90 min and became LDC− induced fluid secretion as efficiently as the wild-type S. flexneri parent (25). Because of the large, undefined size of the DNA transferred in these experiments, it could not be determined whether the fluid secretion ability of the LDC− transconjugants was caused by the absence of cadA or inheritance of an unlinked toxin gene. Therefore, we sought to determine whether BS529 expressing cadA could still induce fluid secretion by injecting supernatants of this strain into ligated rabbit ileal loops. Whereas the wild-type S. flexneri 2a parent 2457T caused an average of 0.6-ml fluid accumulation/cm, the LDC+ strain BS529 induced no fluid accumulation in the ligated loop. These results indicated that expression of cadA alone was sufficient to block the ability of Shigella to induce fluid secretion in this assay.
At least two enterotoxins produced by S. flexneri 2a are thought to be responsible for fluid accumulation in the ligated ileal loop assay. ShET1 is encoded chromosomally and present in all strains of S. flexneri, and ShET2 is encoded on the large virulence plasmid found in all strains of Shigella and EIEC (26–28). Because the activity of these and other enterotoxins can be measured more precisely in Ussing chambers (29), supernatants of BS529 were tested in this assay. Table 2 shows that the presence of the cadA+ gene in BS529 significantly inhibited enterotoxin activity relative to the parent strain. The ΔIsc values were even lower than the plasmid-cured strain (BS103) and suggested that the presence of the cadA+ gene reduced both plasmid- and chromosome-encoded enterotoxin activities. Supernatants of BS529, when mixed with supernatants of wild-type S. f lexneri 2457T, also were able to reduce dramatically the ΔIsc in a time-dependent fashion. This latter result suggested that the effect of cadA+ was not at the level of toxin gene expression in BS529 but rather that it acted in trans on toxin that was present in the cell free supernatants.
Table 2.
Enterotoxin activity of S. flexneri strains as measured in Ussing chamber assay*
| Strain | Enterotoxin produced | ΔIsc | P vs. 2457T† |
|---|---|---|---|
| 2457T (Δ_cadA_) | ShET1; ShET2 | 103.0 ± 19.0 | |
| BS103 (Δ_cadA_) | ShET1 | 78.1 ± 3.03 | not significant |
| BS529 (pCADA+) | ShET1; ShET2 | 33.8 ± 13.1 | 0.04 |
| 2457T + BS529‡ | ShET1; ShET2 | 56.0 ± 12.3 | 0.02 |
| 2457T + BS529§ | ShET1; ShET2 | 19.5 ± 4.3 | 0.0013 |
Cadaverine Is the Enterotoxin Inhibitor Secreted in Supernatants of cadA+ Strains.
The above results indicated that the inhibiting factor associated with expression of cadA in S. flexneri strain BS529 was present in culture supernatants. We hypothesized that this factor could either be LDC or a product of the reaction it catalyzes. LDC is a cytoplasmic protein and is not likely to be exported from the bacteria. In contrast, cadaverine, a product of the decarboxylation of lysine, is secreted from LDC+ cells (13). We measured supernatants of BS529 grown in LB for the presence of cadaverine by using a quantitative spectrophotometric assay (17). Whereas cultures of wild-type S. flexneri 2a strain 2457T contained no measurable cadaverine, BS529 supernatants contained 225–300 μM cadaverine. These levels are comparable to the amount of cadaverine produced by E. coli K-12 MC4100 under inducing conditions (data not shown and ref. 13). To determine whether cadaverine showed the same inhibitory effects on Shigella enterotoxins in Ussing chamber assays, supernatants of 2457T were mixed with cadaverine, and toxin activity was measured. Table 3 shows that inhibition of enterotoxin activity of 2457T supernatants increased with increasing concentrations of cadaverine. When 300 μM cadaverine alone was added to the Ussing chamber, no difference in ΔIsc was observed, as compared with uninoculated LB. Thus, despite the charged nature of cadaverine, it has no electrical signaling effect in the Ussing chamber when used at a concentration that showed complete inhibition of the ΔIsc induced by 2457T supernatants.
Table 3.
Effect of increasing concentrations of cadaverine on S. flexneri 2a enterotoxin activity in Ussing chamber assay
| Concentration*, μM | ΔIsc | P vs. 2457T alone† |
|---|---|---|
| No cadaverine | 68.6 ± 6.5 | |
| 50 | 68.7 ± 21.5 | not significant |
| 100 | 52.1 ± 12.6 | not significant |
| 200 | 42.9 ± 5.5 | 0.039 |
| 300 | 2.7 ± 8.9 | 0.009 |
| 500 | −18.7 ± 14.1 | 0.007 |
| Uninoculated LB | −3.0 ± 14.9 | 0.005 |
| 300 cadaverine alone‡ | 7.9 ± 4.3 | 0.001 |
To determine whether cadaverine directly inhibited the toxin(s) or acted to protect the host cells, tissues in the Ussing chamber were pretreated with 300 μM cadaverine for 30 min before being washed twice with Ringer’s solution and then subjected to enterotoxin-containing supernatants from S. flexneri 2457T. Pretreatment with cadaverine reduced the ΔIsc to 40% of the value observed in tissues that had not been pretreated (61.5 ± 19.8 vs. 153.6 ± 25.5). This result suggested that cadaverine acted to protect the host cells before exposure to the toxins.
cadA Is Deleted in Shigella spp. and EIEC as Part of a Large Deletion in the Genome.
PCR primers flanking the coding sequence of cadA (GenBank database accession no. M76411 and ref. 13) were used to amplify this gene from genomic DNA of representative isolates of Shigella spp. (four strains) and EIEC (one strain). The PCRs failed to yield any product, whereas the positive control, E. coli K-12 strain MC4100, gave the expected 1.9-kb PCR product. Further evidence for the absence of the entire cadA gene was the failure to detect a hybridizing band in a Southern blot of genomic digests from the same Shigella and EIEC strains by using a 2.6-kb fragment containing cadA from the plasmid pCADA as a probe. The only exception was a strain of S. sonnei that yielded a positive hybridization signal when the cadA probe was used. Nevertheless, this strain was consistently negative in assays for LDC activity. These results and the inability to transduce markers from this region of E. coli K-12 into S. flexneri 2a (see above) suggested that Shigella spp. and EIEC have deleted a large region of the chromosome around the cadA locus.
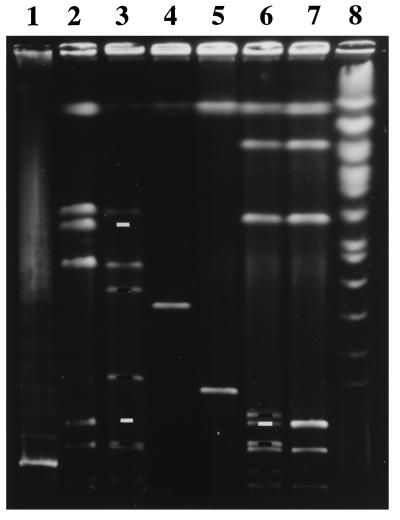
To obtain further evidence of the Shigella deletion detected above, E. coli insertion alleles carrying rare restriction sites were used to measure chromosomal distances between corresponding loci in E. coli K-12 and S. flexneri 2a. Two mini Tn_10_dRCP2 insertions flanking cadA in E. coli, carrying both the I-Sce I and I-Ceu I restriction sites and different antibiotic resistance genes (11), were mobilized individually by P1 transduction from E. coli K-12 strains χM2115 and χM2125 into MG1655 (E. coli K-12) and BS103 (S. flexneri 2a). Any change in chromosomal distance between these two insertions, from the E. coli to S. flexneri backgrounds, then could be measured by a difference in the length of genomic DNA separating the new restriction sites introduced by them. Genomic DNAs from E. coli MG1655 and S. flexneri BS103 double-insertion mutants (χM2500 and BS573) were digested with I-Sce I, whose recognition site of 18 bp is extremely rare and occurs only at the insertion sites (30). As shown in Fig. 1, these digests yielded I-Sce I restriction fragments, the sizes of which indicated the distances between the two insertions in each background. These sizes of ≈398 and 205 kb, respectively (lanes 4 and 5), were consistent with an S. flexneri 2a deletion as large as 190 kb relative to E. coli K-12. Identical I-Sce I fragments of 205 kb obtained from six separate BS103 transductants (data not shown) and similarities in the chromosomal organization of strains MG1655 and BS103 by I-Ceu I restriction (Fig. 1) were consistent with P1 transduction fidelity and genetic map conservation, respectively, between strains.
Figure 1.
Pulse-field gel electrophoresis separation of genomic segments delimited by a pair of Tn_10_dRCP2 insertions. Lanes: 1 and 8, yeast-chromosome and λ-concatemer standards; 2, 3, 6, and 7, I-Ceu I digests of MG1655 (parent E. coli K-12), χM2500 (K-12 double-insertion mutant), BS573 (S. flexneri 2a double-insertion mutant), and BS103 (parent S. flexneri 2a), respectively; 4 and 5, χM2500 and BS573 digested with I-Sce I. The MG1655 native I-Ceu I fragments of 130 and 670 kb were cleaved into pairs of subfragments of 30 and 100 kb and 235 and 435 kb, respectively, in the double insertion mutant χM2500 [as predicted from the _Not_I and _Bln_ I map coordinates of its insertions (11) and the native I-Ceu I map (31)]. The I-Ceu I pattern of genomic DNA from strain BS573 (containing the identical two insertions in the BS103 background) showed a new subfragment of 135 kb (consistent with cleavage and lack of change in migration of the largest native I-Ceu I fragment) and a pair of subfragments of 40 and 100 kb from cleavage of one of two 140-kb native I-Ceu I fragments, consistent with P1 transduction fidelity between strains and with genetic map conservation judged by rrl gene architecture (31). Digestion of χM2500 and BS573 with I-Sce I resulted in isolated bands allowing side-by-side comparison of a pair of I-Sce I restriction fragments (398 kbMG1655 and 205 kbBS103) with corresponding end points from the MG1655 and BS103 backgrounds. White bars indicate the wild-type I-Ceu I fragments missing because of Tn_10_dRCP2 insertion, and black bars indicate the corresponding I-Ceu I subfragments generated.
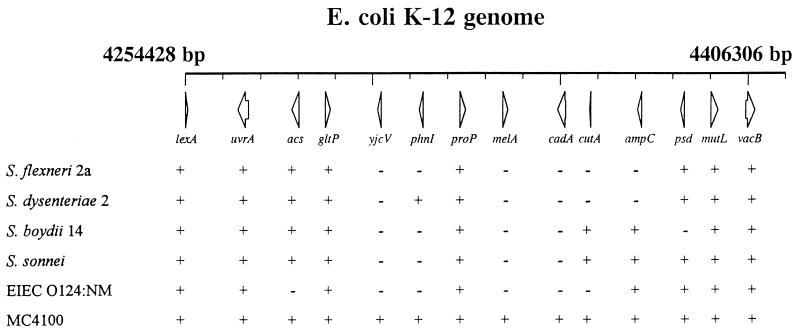
We attempted to define roughly the limits of this large deletion in Shigella spp. and EIEC by hybridizing genomic DNA with oligonucleotide probes from 14 different genes 1–2 min clockwise and counter-clockwise of cadA. The results are shown in Fig. 2 and, as expected, indicated a large deletion (up to ≈90 kb) with variable end points in these representative strains. The hybridization pattern of the 14 probes in S. flexneri accounted only in part for the change detected by comparative macrorestriction mapping and suggested additional event(s) beyond the deletion of a contiguous segment in the region covered by the probes. Also, the retention in all isolates of hybridization to proP, which is surrounded on the K-12 map by deleted regions, suggested either a proP “island” flanked by deleted segments or the dislocation of proP by transposition or inversion to elsewhere in the Shigella spp. genome. In either case, retention of the proP gene would argue that the gene’s function (low affinity transport for glycine betaine and proline) is beneficial to the bacteria. Experiments are underway to resolve the nature of the positive hybridization signal with the proP probe and to define precisely the end points of the deletion in S. flexneri 2a. Nevertheless, the hybridization results are consistent with the results in Fig. 1, and the large size of the deletion also explains the inability to transduce markers from this region of E. coli K-12 into S. flexneri.
Figure 2.
Schematic representation of black holes in Shigella spp. and EIEC. Oligonucleotide probes from 14 different genes of E. coli K-12 between basepair 4254428 and basepair 4406306 were hybridized to genomic DNA of representative strains of Shigella spp. and EIEC. +, positive hybridization; −, no hybridization. All strains (with the exception of 2457T) were from Nancy Strockbine, Centers for Disease Control and Prevention.
A constitutive ldc gene was recently identified in E. coli K-12 (32). However, little activity is detectable from the single chromosomal copy, and the biological function of this protein is not known. Although we cannot exclude the possibility that a homolog of ldc exists in Shigella, it is unlikely that the extremely low LDC activity attributable to this second locus would inhibit enterotoxin activity.
DISCUSSION
In contrast to the almost universal expression of LDC activity by strains of E. coli, the absence of LDC activity in the closely related but pathogenic Shigella spp. and EIEC strains suggested that expression of LDC from cadA might be incompatible with virulence. In support of this hypothesis is the report of Sansonetti et al. (25), which showed that conjugal transfer of the Shigella chromosomal region near 90 min and inheritance of the LDC− marker are necessary for E. coli–Shigella hybrids to induce fluid secretion in ligated rabbit ileal loops (25). By using BS529, a wild-type strain of S. flexneri 2a transformed with a cloned copy of cadA (the structural gene for LDC), we showed that expression of LDC attenuated the ability of the organism to induce fluid secretion in rabbit ileal loops and reduced its ability to cause a change in short circuit current in Ussing chambers. Further examination of the inhibitory effect of LDC expression showed that supernatants from BS529 contained a factor that inhibited the enterotoxin activity of supernatants from LDC− strains. LDC decarboxylates lysine to yield cadaverine and CO2, and the LDC+ transformant of S. flexneri secreted large quantities of cadaverine into the supernatant. Equivalent concentrations of cadaverine, when added to supernatant from wild-type S. flexneri, displayed the same ability to inhibit enterotoxin activity in the Ussing chamber assay.
Strains of S. flexneri 2a produce two iron-regulated enterotoxins. The chromosomally encoded ShET1 rarely is found in other serotypes of Shigella, whereas the virulence plasmid-encoded ShET2 is present in >80% of Shigella tested (28). Both toxins irreversibly alter electrolyte and water transport in rabbit intestine in vitro and in vivo (26, 28, 29). Our results suggest two possible models for the action of cadaverine: First, cadaverine inactivates the toxins synthesized by Shigella, or, second, cadaverine acts directly on the target cell to protect it. The first model would require that cadaverine be able to inhibit both ShET1 and ShET2 (and other undefined enterotoxins) produced by S. flexneri. Although the molecular mechanism of action of these two toxins has yet to be determined, they appear to act via different pathways (A.F., unpublished data). By contrast, the second model is supported by our results, which show that pretreatment of rabbit mucosa in the Ussing chamber with cadaverine protected the mucosa from the effect(s) of enterotoxins added after the cadaverine was washed from the tissue (see Results). Polyamines such as cadaverine are absorbed from the lumen by rabbit small intestine cells (33), and it has been proposed that intracellular polyamines might act as second messengers in the eukaryotic cell by modulation of extracellular signals transduced through G protein-coupled receptors (34). In this light, cadaverine could protect the cell by closing ion channels induced by bacterial toxins, altering intracellular signaling, or displacing toxin from cellular receptors. These possibilities await experimental testing.
The potent inhibitory effect of cadaverine on Shigella enterotoxin activity poses a potential obstacle to full expression of the virulent phenotype in Shigella spp. and thus might lead to evolutionary pressure to mutate to _cadA_−. Deletion of the cadA gene could be favored mechanistically by occurring at a higher rate than point mutations and also would be favored if other genes in the same region inhibited virulence and needed to be inactivated. Unlike point mutations, deletions have the added advantage of being nonrevertible. We determined that all four species of Shigella and EIEC have undergone large deletions covering the region around cadA. When compared with the closely related nonpathogenic commensal E. coli, this deletion represents a black hole in the genome that was selected to enhance pathogenicity of Shigella. As such, we propose that the deletion of commonly inherited genes that inhibit virulence is a complement to the acquisition of genes, such as pathogenicity islands, that augment virulence. Our preliminary definition of black holes is drawn from the criteria used to define pathogenicity islands: (i) The deleted sequence is absent from the genome of pathogenic strains but is present in nonpathogenic variants of the same or closely related species, and (ii) the deleted region is large. Additional work in progress will determine whether association with tRNA genes at the boundaries holds true for the black holes as it does for pathogenicity islands (1).
The strategy of black hole formation may not be as pivotal an event in the creation of pathogenic bacteria as the acquisition of pathogenicity islands. Rather, this mechanism may represent a subtle fine-tuning of the genome repertoire of a newly created pathogen to delete genes that are detrimental to the new pathogenic lifestyle. Black holes could precede additions of pathogenicity islands, making the recipient background more favorable to expression of virulence genes. However, it is likely that the complementary strategy of black hole formation follows additions. Another example of the mechanism by which loss of function may lead to enhanced virulence can be found in Shigella. Deletion of a 21-kb segment from Shigella, representing a cryptic prophage (DLP12) present in E. coli K-12 at 12 min, eliminated a protease activity that degrades an outer membrane protein (VirG) needed for cell-to-cell spread (35). The insertion site for the prophage and a boundary of this black hole is the tRNA gene argU. One major difference between this example and the black hole around cadA is that the latter represents a considerably larger deletion. Another difference is that the sequence of the E. coli K-12 genome that corresponds to the deleted black hole around cadA in S. flexneri does not contain any sequences known to be bacteriophage attachment sites.
The enhancement of virulence through formation of black holes invites comparison to the avirulence genes of plant pathogens. These genes allow certain plant hosts to resist bacterial infection by evoking a hypersensitive response in the plant (36). Mutation of these genes can attenuate virulence but sometimes results in broadening of the host range of the bacteria, thus improving its fitness as a pathogen. Further study is needed to determine any potential role of black hole formation in the evolution of bacterial plant pathogens.
In conclusion, this study presents two major findings: (i) As Shigella spp. evolved from E. coli to become pathogenic, it not only acquired genes on a virulence plasmid but also shed genes on deletions at cadA; and (ii) cadaverine, generated as a product of the decarboxylation of lysine, is a potent inhibitor of Shigella enterotoxin activity. These findings provide a perspective on the development of antibacterial therapies. The observation that a simple, natural polyamine can inhibit multiple enterotoxins may have future applications in treatment of diarrheal diseases and in the construction of vaccine strains that are better tolerated. The formation of black holes to enhance expression of virulence, as reported here for Shigella spp. and EIEC, is a mechanism that deserves consideration in the study of bacterial pathogen evolution. The comparison of genomes of nonpathogens with closely related pathogens, particularly those with pathogenicity islands, may reveal as yet undetected black holes. Our findings suggest the exciting possibility that such studies may identify other natural inhibitors of virulence, which may be exploited in the treatment of infectious diseases caused by these pathogens.
Acknowledgments
We thank George Bennett for the plasmid pCADA and Nancy Strockbine for the Shigella and EIEC strains. Thanks to Lee Metcalf and Robin Sandlin for their thoughtful reading of the manuscript. This work was supported by National Institutes of Health Grants AI24656 (A.T.M.), AI31419 (C.A.B.), and AI35740 (A.F.) and by Uniformed Services University of the Health Sciences Grant R07385 (A.T.M.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: EIEC, enteroinvasive E. coli; LDC, lysine decarboxylase; LB, Luria–Bertani medium; PD, potential difference; Isc, short circuit current.
References
- 1.Hacker J, Blum–Oehler G, Mühldorfer I, Tschäpe H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 2.Brenner D J, Fanning G R, Johnson K E, Citarella R V, Falkow S. J Bacteriol. 1969;98:637–650. doi: 10.1128/jb.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Formal S B, Gemski P, Jr, Baron L S, LaBrec E H. Infect Immun. 1970;1:279–287. doi: 10.1128/iai.1.3.279-287.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansonetti P J, Hale T L, Oaks E V. In: Microbiology-1985. Schlesinger D, editor. Washington, DC: Am. Soc. Microbiol.; 1985. pp. 74–77. [Google Scholar]
- 5.Edwards P R, Ewing W H. Identification of Enterobacteriaceae. 3rd ed. Minneapolis: Burgess; 1972. [Google Scholar]
- 6.Silva R M, Toledo R F, Trabulsi L R. J Clin Microbiol. 1980;11:441–444. doi: 10.1128/jcm.11.5.441-444.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Formal S B, Dammin G J, LaBrec E H, Schneider H. J Bacteriol. 1958;75:604–610. doi: 10.1128/jb.75.5.604-610.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurelli A T, Blackmon B, Curtiss R., III Infect Immun. 1984;43:397–401. doi: 10.1128/iai.43.1.397-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban M J. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 10.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloch C A, Rode C K, Obreque V H, Mahillon J. Biochem Biophys Res Commun. 1996;223:104–111. doi: 10.1006/bbrc.1996.0853. [DOI] [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 13.Meng S-Y, Bennett G N. J Bacteriol. 1992;174:2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahillon J, Rode C K, Leonard C, Bloch C A. Gene. 1997;187:273–279. doi: 10.1016/s0378-1119(96)00766-4. [DOI] [PubMed] [Google Scholar]
- 15.Bloch C A, Rode C K, Obreque V H, Russell K Y. J Bacteriol. 1994;176:7121–7125. doi: 10.1128/jb.176.22.7121-7125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkow S. Am J Clin Pathol. 1958;29:598–600. doi: 10.1093/ajcp/29.6_ts.598. [DOI] [PubMed] [Google Scholar]
- 17.Phan A P H, Ngo T T, Lenhoff H M. Anal Biochem. 1982;120:193–197. doi: 10.1016/0003-2697(82)90336-0. [DOI] [PubMed] [Google Scholar]
- 18.Hromockyj A E, Maurelli A T. Infect Immun. 1989;57:2963–2970. doi: 10.1128/iai.57.10.2963-2970.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fasano A, Kay B A, Russel R G, Maneval D R, Jr, Levine M M. Infect Immun. 1990;58:3717–3723. doi: 10.1128/iai.58.11.3717-3723.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasano A, Baudry B, Pumplin D W, Wasserman S S, Tall B D, Ketley J M, Kaper J B. Proc Natl Acad Sci USA. 1991;88:5242–5246. doi: 10.1073/pnas.88.12.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rode C K, Obreque V H, Bloch C A. Gene. 1995;166:1–9. doi: 10.1016/0378-1119(95)00630-5. [DOI] [PubMed] [Google Scholar]
- 22.Heath J D, Perkins J D, Sharma B, Weinstock G M. J Bacteriol. 1992;174:558–567. doi: 10.1128/jb.174.2.558-567.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 24.Tabor H, Hafner E W, Tabor C W. J Bacteriol. 1980;144:952–956. doi: 10.1128/jb.144.3.952-956.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sansonetti P J, Hale T L, Dammin G J, Kapfer C, Collins H H, Jr, Formal S B. Infect Immun. 1983;39:1392–1402. doi: 10.1128/iai.39.3.1392-1402.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasano A, Noriega F R, Maneval D R, Jr, Chanasongcram S, Russel R, Guandalini S, Levine M M. J Clin Invest. 1995;95:2852–2861. doi: 10.1172/JCI117991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noriega F R, Liao F M, Formal S B, Fasano A, Levine M M. J Infect Dis. 1995;172:1408–1410. doi: 10.1093/infdis/172.5.1408. [DOI] [PubMed] [Google Scholar]
- 28.Nataro J P, Seriwatana J, Fasano A, Maneval D R, Guers L D, Noriega F, Dubovsky F, Levine M M, Morris J G., Jr Infect Immun. 1995;63:4721–4728. doi: 10.1128/iai.63.12.4721-4728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fasano A, Noriega F R, Liao F M, Wang W, Levine M M. Gut. 1997;40:505–511. doi: 10.1136/gut.40.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteilhet C, Perrin A, Thierry A, Colleau L, Dujon B. Nucleic Acids Res. 1990;18:1407–1413. doi: 10.1093/nar/18.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S–L, Hessel A, Sanderson K E. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikuchi Y, Kojima H, Tanaka T, Takatsuka Y, Kamio Y. J Bacteriol. 1997;179:4486–4492. doi: 10.1128/jb.179.14.4486-4492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brachet P, Debbabi H, Tome D. Am J Physiol. 1995;269:754–762. doi: 10.1152/ajpgi.1995.269.5.G754. [DOI] [PubMed] [Google Scholar]
- 34.Bueb J L, Da Silva A, Mousli M, Landry Y. Biochem J. 1992;282:545–550. doi: 10.1042/bj2820545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakata N, Tobe T, Fukuda I, Suzuki T, Komatsu K, Yoshikawa M, Sasakawa C. Mol Microbiol. 1993;9:459–468. doi: 10.1111/j.1365-2958.1993.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 36.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]