Helicobacter pylori genetic diversity and risk of human disease (original) (raw)
Microbes residing on a mucosal surface may communicate in complex ways with their neighboring host tissue. Signals derived from a bacterium may include metabolites or specific host-directed substances such as secreted toxins, or they may result from direct contact with host cells. Conversely, certain host activities or responses may be considered as signals to the microbe, including production of molecules that can be used by the microbe as substrates, release of defense molecules (e.g., antibodies, defensins, mucus), or organ-specific physiologic traits, including peristalsis and acid secretion in the stomach. In the case of Helicobacter pylori, a common resident of the human stomach, these bidirectional signals appear to have coevolved over long periods such that, in most cases, carriage of H. pylori is, if not mutually beneficial, at least well tolerated by the human host. Here, we examine the relationship of H. pylori and humans in the context of these diverse signals.
Linkage of microbial and host signals
Signals to and from the host can be considered either unlinked or linked. In the “unlinked signal” model, both parties, microbe and host, respond to signals from the other in an independent fashion. Such a relationship can be invoked for an organism that is “accidentally” or recently acquired by humans, or that is present in an atypical niche. The interactions tend to become “arms races” in which the host either ultimately eliminates the microbe or suffers severe injury or death. Examples of this phenomenon include human infection with Ebola virus or with non-typhi Salmonella, agents that can be carried long-term by their natural hosts (rodents and poultry, respectively) but that usually cause acute short-term infections in humans.
Alternatively, the signaling between the microbe and host may be linked. This model generally reflects an evolutionary history of interaction, with selection for traits in the microbe and host that allow for their long-term coexistence and optimize the outcome of interactions between them. Microbes are selected to be able to evade the host’s immune responses, to use the host’s resources efficiently, and to limit the toxicity of interactions. The host, conversely, may acquire mechanisms to minimize tissue injury or to exclude more virulent organisms specifically.
One example of coevolution with humans is provided by the Bacteroides species that are among the most numerous colonizers of the colon. These organisms benefit humans by producing vitamin K and by enhancing resistance to high-grade enteric pathogens. That the LPS of these Gram-negative bacteria has a markedly reduced ability to activate inflammatory cells — a property that may facilitate long-term residence in the colon — may represent an adaptation selected over millennia. Nevertheless, despite the overwhelmingly benign interaction between Bacteroides species and humans, lethal infections sometimes occur. The contrast between HIV infections in their natural host, monkeys, in whom infection is essentially benign, and in humans, in whom lethal AIDS occurs, further illustrates the importance of host-specific adaptation. HIV, which has been present in humans for less than 100 years, has already selected for specific changes in particular host determinants, such as the Δ32 mutation in the CCR5 chemokine receptor gene, which confers partial or absolute resistance to HIV infection of targeted cells. Thus, HIV may become a powerful selector of particular human genotypes.
H. pylori and illness in humans
More than half of all humans are colonized in their stomachs by H. pylori (1). That carriage is nearly universal among adults in developing countries suggests that in earlier times most humans carried these organisms. However, it is not known whether H. pylori has been with all humans for hundreds of years, or for millions of years (see refs. 2, 3 for different views). In either case, with the changing conditions associated with improved socioeconomic status, H. pylori is disappearing in industrialized countries (4) and declining in prevalence in some developing countries as well.
The ability to identify persons who do or do not harbor H. pylori permits us to examine the consequences of its carriage. Over the past decade, it has become clear that gastric H. pylori carriage is associated with increased risk for the development of peptic ulcer disease and noncardia gastric adenocarcinoma, and that removal of H. pylori favorably influences the natural history of peptic ulceration (5). More recently, it also has become evident that individuals without H. pylori are at greater risk for gastroesophageal reflux disease and its sequelae, Barrett’s esophagus and adenocarcinoma of the esophagus (5). Thus, from clinical and epidemiological studies, the pathogenetic significance of H. pylori is mixed.
Both illness and protection from illness occur in only a small fraction of those carrying H. pylori, whereas for most carriers, morbidity and mortality are unaffected by their H. pylori status. In that sense, H. pylori can be considered a type of commensal, or more properly, has an “amphibiotic” relationship with humans (as with Bacteroides species), whose pathogenic significance is determined by context (6). Differences in the consequences of colonization could depend on variation among colonizing H. pylori strains, on environmental cofactors, or on factors determined by the individual host. In particular, DNA-level analyses have indicated that H. pylori is one of the most genetically diverse of bacterial species (3, 7, 8). Although much of the diversity may be neutral, some will affect the ability of these bacteria to colonize, persist, and/or affect disease in particular hosts. We aim to assess how H. pylori DNA sequence diversity (polymorphism) may affect disease risk in the broader context of signaling between microbes and hosts.
A general model _of H. pylori_–human interactions
H. pylori colonization of the gastric mucosa typically occurs in early childhood and may persist for decades or for life, unless eradicated by antimicrobial treatment (5). Even in high-risk environments, colonization occurs with an estimated probability of far less than 1% per day, but most children become colonized within their first few years of life because of the cumulative effect of a low daily risk of colonization. H. pylori seems to proliferate almost exclusively on human gastric epithelial cell surfaces and in the overlying mucus. It does not invade epithelial tissue aggressively, as do bona fide intracellular pathogens such as Shigella and Listeria (although there are reports of occasionally finding some H. pylori within host cells), nor does it seem to colonize any other tissue type or to proliferate in the environment. Nevertheless, many H. pylori strains can adhere to gastric epithelial cells (9), into which they can inject at least one bacterial protein, the CagA protein discussed below (see also ref. 10).
The evidence that H. pylori and humans have coevolved includes consideration of the H. pylori LPS, an important cell-surface structure (11, 12). For example, there is considerable diversity among H. pylori strains in the types of (Lewis) fucosylated oligosaccharide antigen structures expressed on their LPS, and a corresponding diversity among humans in the types of Lewis antigens on gastric epithelial cells (11). Similarly, the LPS of Helicobacter mustelae, which obligately colonizes the ferret stomach, expresses Lewisa, the predominant antigen on ferret gastric epithelial cells. Possible advantages to microbes of expressing the same surface antigen as host cells (13) include camouflage from host detection and defense, and enhanced adherence to cells, using host lectins that normally mediate Lewis antigen–based cell-cell recognition or adherence. H. pylori also has evolved a form of lipid A to which human phagocytic cells barely respond (12).
We infer that H. pylori may be nourished by exudate from host tissue, rather than directly by what we eat and drink (14, 15). If this view is correct, then H. pylori may need to induce host tissue injury to survive. However, hypervirulence (excessive tissue damage) could result in more severe gastritis, atrophy, and hypochlorhydria, which permits overgrowth of more pH-sensitive competing bacterial species, and loss of habitat by replacement of gastric-type by intestinal-type epithelia (intestinal metaplasia), a tissue type that H. pylori cannot colonize. Hence, H. pylori may walk a tightrope — needing to induce sufficient response to permit nutrient release and regulation of habitat, but not so much as to result in loss of its niche (14, 15).
Such a coevolution model predicts that H. pylori benefits its human hosts, and indeed, these bacteria produce antibacterial substances (16) and may help suppress more virulent species. For example, gastric colonization induces an augmented enteral and systemic response to Vibrio cholerae (17), and recent epidemiologic data indicate that colonization is associated with protection from childhood diarrhea (18). Protection against diarrheal pathogens (parallel to that observed for the colonic flora) would be strongly selected whenever enteric diseases are a major contributor to childhood mortality. The increased susceptibility to pathogens and to gastric neoplasia occurring later in life, in _H. pylori_–induced atrophic gastritis and hypochlorhydria, may be seen as a deferred cost to the host, after reproductive age, for the early benefit of the interaction.
One possible criticism of such coevolutionary models stems from the view that competition among microbes will select for variants that can best exploit the available resources, regardless of consequences to the host. At least in its simplest form, however, this model does not seem to apply to H. pylori, given the tightrope that H. pylori must walk and the extraordinary chronicity of successful H. pylori colonization. In natural or experimental mixed infections, different genotypes apparently predominate at different sites (19). We propose that the stomach consists of multiple microniches, and that the bacteria found in individual niches may often descend from single founding cells. If this is correct, a possible long-term consequence of emergence of a highly aggressive variant would be to render the entire stomach unfit for H. pylori, and to bring about its clearance from the colonized person and lack of transmission to new hosts. Aggressive variants that remain in their microniche only would destroy local habitat (producing local patches of intestinal metaplasia), and thus would decrease in number without directly affecting the less virulent strains that occupy other gastric sites. In this model, there would be continual selection against hypervirulent organisms, unless changing conditions were to allow for a sudden increase in the bacterial population, which might facilitate their transmission to a new host.
Types of genetic diversity among H. pylori strains
H. pylori populations are very diverse genetically, due to point mutations and larger substitutions, insertions, or deletions that may involve one or more genes or multigene segments (including restriction/modification genes and at least one pathogenicity island). Chromosomal rearrangements (mostly inversions) are also seen, as are a variety of auxiliary DNAs, including various insertion sequence elements and plasmids (7, 8, 20–23). Although we do not have good estimates of the rate of mutation of H. pylori during acute or chronic colonization, this great diversity suggests that the species is ancient and that its effective population size is large. We propose that this diversity also reflects a striking lack of direct competition between most strains, even when resident in different persons in the same community. Diversity would be accentuated by the large geographical separation of many of the world’s peoples (e.g., those of East Asia from those of Western Europe), and thus a corresponding separation of the H. pylori strains they carry (3). These features effectively diminish the kind of population-wide “selective sweeps” that routinely reduce the diversity of populations of several other microbes that have been studied in this way (24).
Divergence among H. pylori strains would be further increased by differences among people in traits important to individual strains (25). Such traits might include types of Lewis antigens available for H. pylori adherence, the specificity and intensity of immune and inflammatory responses, and the complex regulation of gastric acid secretion. As one consequence of human diversity, any strain that has become well-adapted to its current host (reflecting decades of selection) will often be less well-suited to others who might ingest it. Its establishment in its new host would result in selection for genotypes better suited to its new environment, and thus further divergence from ancestral genotypes.
One special group of polymorphisms involves repeated sequences (mononucleotide or dinucleotide repeats) near the 5′ ends of genes dubbed “contingency genes” (refs. 20, 21, 26; see also Bayliss et al., this Perspective series, ref. 27). Slipped strand mispairing in such repeats during DNA synthesis is frequent and generates frameshift mutations that turn translation of the affected protein ON or OFF. Genes containing such repeats tend to be ones for which such metastable ON/OFF switches are highly advantageous (11, 26). Depending on the number (n) of these contingency genes, an H. pylori population colonizing a host’s stomach may consist of as many as 2_n_ competing bacterial lineages. This phenomenon, a type of “quasispecies” development analogous to that seen with HIV, allows for optimal use of the host’s resources, and the possibility of a pool of variants that can effectively compete in the next host (28). In this model, each member of the population competes with each other member, allowing conditions in the host to select for optimal fitness. Of course, if specific microniches exist, the criteria for fitness could be local. Criteria also might change over time, for example, due to cofactor shifts or the onset of specific immune responses by the host.
Given the divergence among H. pylori strains, the population diversity and overall fitness of the species can be further enhanced by recombination among different bacterial lineages (which may often occur by DNA transformation and/or conjugation), because it generates new potentially advantageous genotypes far more efficiently than is possible by recurrent mutation alone. Analysis of DNA sequences from representative strains implicates such recombination as a major factor in H. pylori population genetic structure, far more important for H. pylori than for most other bacterial species (8). Evidence of gene exchange and selection for particular recombinant genotypes also can be detected by analyses of bacteria recovered from mixed natural infections (19).
Especially interesting in the context of genetic/phenotypic diversity and interstrain recombination are the large number of restriction-modification (R-M) systems found in H. pylori strains (20, 21). Some seem to be complete, while others may be vestigial; the exact complement of functioning R-M systems varies among strains (20, 21, 29). Although their roles in genome function and stability are not known, several models merit consideration. In one model, R-M systems may be molecular parasites (selfish DNAs), whose presence in a lineage reflects evolutionary accident (30). A second view focuses on the restriction feature of R-M systems: DNA breakage, its molecular manifestation, interferes with the exchange of large (gene- or multigene-length) DNA segments, but may also stimulate exchange of short DNA segments, since DNA ends are excellent substrates for recombination (31). A third view focuses on how methylation (modification) of transcription promoters decreases promoter activity, thereby affecting gene expression. Diversity in expression of sets of coadapted genes might then be achieved through gain, loss, or changes in activity of particular methylase genes. The discovery of the iceA1 gene (32) raised yet a fourth possibility, the direct participation in _H. pylori_–host interaction, because iceA1 transcription seems to be specifically induced by H. pylori contact with gastric epithelial cells, and full-length iceA1 encodes a restriction endonuclease (NlaIII).
The significance of polymorphisms in an equilibrium model of microbial persistence
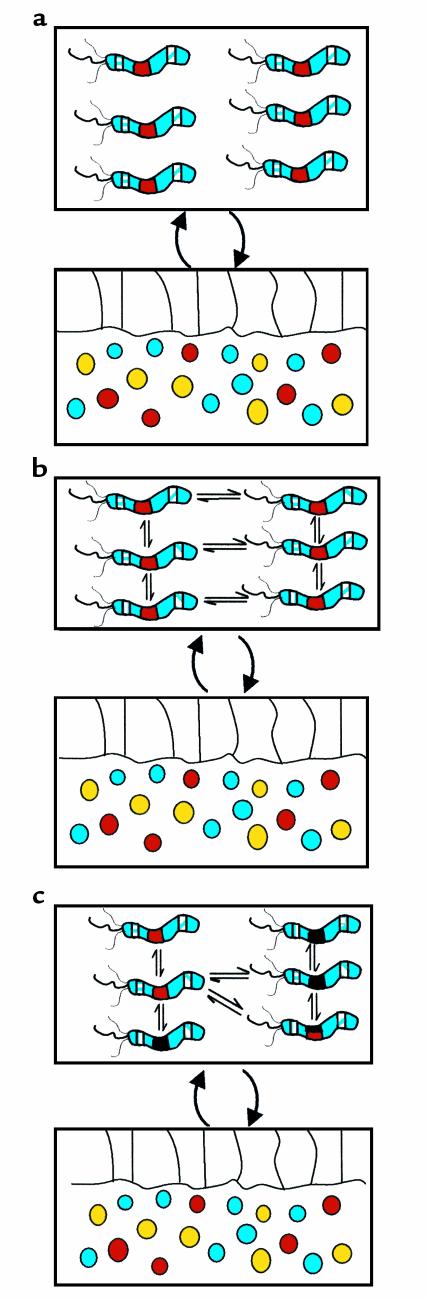
When all individuals in a microbial population are identical, the concept (Figure 1a) and the mathematical treatment (15) of the equilibrium established with the host are relatively simple. However, microbial variation and regional or temporal differences in the host gastric mucosa introduce much greater complexity and assure that the equilibrium will be dynamic. If a single H. pylori “strain” (quasispecies) colonizes a host, variants that arise may signal the host in different ways. The host’s local responses to these signals then will vary, creating local differences in milieu for the microbes, representing differential selective pressures on the resident H. pylori population (Figure 1b). These selective pressures will affect the relative proportions of the individuals in the population, or alleles of key genes, and thus move the system to new equilibria. Continued mutation ensures that the equilibrium remains dynamic.
Figure 1.
H. pylori polymorphisms affect the nature of the interaction with humans. In each panel, the H. pylori population is represented by the upper box, and the host mucosa (with epithelium and reactive neutrophils, monocytes, plasma cells, and lymphocytes) by the lower box. (a) All H. pylori cells are identical. Three polymorphic loci are shown. The lateral loci each are subject to phase variation, and each one may either be ON, encoding a full-length protein, or OFF, i.e., translation has been prematurely terminated. The middle locus represents a site in which two major alleles (represented as black and red) are present. Recombination events can permit the total or partial replacement of one locus by the other. In a, all H. pylori cells exert the same effect on the host since they are identical. (b) A single H. pylori population is present, but now individual cells are polymorphic with respect to the lateral loci. This H. pylori population will develop a different equilibrium with the host than does the population represented in a, because of its altered expression from the lateral loci. The altered host response in turn creates a different environment that selects for phenotypes different from those for a. The individual cells are thus in equilibrium with one another, and the relative proportion of one to another will be selected by the host signals. In the absence of exogenous perturbation, this model can move toward a stable equilibrium, but independent changes in the environment or periodic changes (selection) affecting the bacterial population may then move the equilibrium into new territory. (c) Two H. pylori populations are present that are recombining with one another. The dynamic equilibrium between host and the microbial population parallels that for b, except that the recombinational events change the overall equilibrium. With recombination, the variety of potential interactions expands greatly and allows a highly fluid response to host conditions. This polymorphic population can optimize overall fitness to the particular host, while remaining subject to the host selective pressures as transmitted by its signals in response to the bacterial signals. This model permits ongoing fine-adjustment of adaptation in multiple local niches.
A more complex condition occurs when two or more different H. pylori strains colonize the same stomach (Figure 1c), because recombination generates new genotypes that may outcompete their parental and sibling strains. If each bacterial cell independently signals the host, and even a localized host response potentially affects all bacterial cells in the locality, new more fit individuals might have substantial effects on the equilibrium even before their selective advantage is fully realized. This dynamic condition allows for the creation of population structures that can optimize the use of resources in any host.
One potential cost of this system to H. pylori diversity — and hence to the resilience of the population in the face of sudden shifts in the host environment — is that recombination can create a new, much more fit genotype that will overwhelm the mixed population. Such a selective sweep, by definition, eliminates the diversity that might be useful in future years to create new individuals if and when conditions change (i.e., when transmitted to a new host). Although evolution is not anticipatory, barriers to recombination, as occur with strain-specific restriction endonucleases (29), might nevertheless be selected because they slow gene exchange in populations. That some R-M genes contain mono- or dinucleotide repeats (20, 21) suggests that they are contingency genes (see Bayliss et al., this Perspective series, ref. 27) and illustrates another potential level of regulation of recombination and genome evolution. Restriction barriers between H. pylori strains cocolonizing an individual stomach may modulate the frequency and intensity of selective sweeps and may have been selected over the long term to maximize the diversity of the population colonizing individual hosts (29).
The cag island as an example of polymorphisms affecting equilibrium and clinical outcome
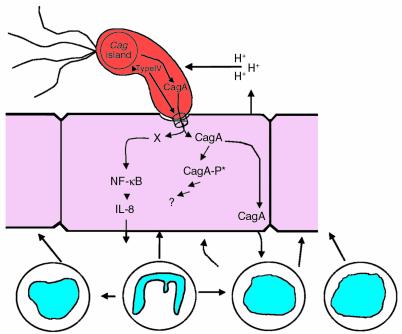
The H. pylori cag island is a 35- to 40-kb genetic element that probably entered the genome after H. pylori became a species, and contains homologs to genes of other bacterial species encoding type IV secretion systems (20, 33–35). One substrate for this system now is known; using the type IV system, cag+ strains inject the CagA protein into host cells (10) (Figure 2). CagA undergoes tyrosine phosphorylation by a host-cell kinase, and the new phosphoprotein alters the physiology of the affected cells.
Figure 2.
Role of the cag island in regulating gastric acidity. The conditions of a feedback system regulating both host response and bacterial transcription are potentially fulfilled by the interaction between cag+ strains and humans. Gastric acidity (represented by H+ ions) induces transcription of cagA (40). The type IV secretion system in the cag island (20) exports the cagA product into gastric epithelial cells, where it undergoes tyrosine phosphorylation (-P*) by a host-cell kinase (10). Although the exact role of the CagA phosphoprotein is not curreVntly known, signaling of the host by cag+ strains induces an increased leukocytic cellular response, as well as proinflammatory cytokines, including IL-1β (42). Because such inflammatory mediators can reduce gastric acidity, at least in part by their direct effects on the specialized epithelial cells involved in acid secretion, the _H. pylori_–host system shifts toward a new equilibrium value (lowered acidity, thus lowering cagA transcription). Thus all the components necessary for a simple “thermostat” have been refined. However, higher degrees of regulation can be added to this simple negative-feedback (homeostatic) model. Humans are polymorphic for IL-1β expression; thus host status helps determine the level of the equilibrium, which has important disease consequences (41). Genetic variation in the cag island may offer H. pylori populations one means to respond to long-term or local environmental changes. Because H. pylori are naturally competent, genetic determinants lost by some members of the population can be regained through transformation. DNA also may be exchanged via conjugation. Thus, metastable cag islands in a population of H. pylori cells may serve as high-gain “thermostats,” governing both H. pylori and host response dynamics.
Not all H. pylori strains contain the cag island; this dichotomy represents the most important differentiation among H. pylori strains found to date. In addition, there is a strong association between the presence of the cag island and several other genotypes (and their resultant phenotypes) that also relate to the interaction of H. pylori cells with their host. Strains that are cag+ are significantly more likely than cag– strains to possess the s1 allele of _vac_A, which encodes a secreted molecule that induces vacuole formation in epithelial cells (36). Similarly, cag+ strains also are more likely to express the _bab_A product, which governs adherence to Lewisb on gastric epithelial cells (37), and to express Lewis antigens as part of their LPS (37). That these properties are strongly associated with one another, but are not mechanistically dependent on the function of the cag island, suggests that the underlying genes may be considered as components of a coadapted gene complex. The cag– strains probably have a different lifestyle, since they typically also have much reduced Lewisb adherence and are not toxigenic. The coexistence of cag+ and cag– cells in the same host (19) suggests that they occupy different niches, where they would be subject to different selection pressures. Because of their close linkage, it is difficult to dissect the role of each polymorphism (cag/vac/bab) with relation to human disease, but studies based on CagA can serve as a proxy for the general phenomenon.
We and others have used serologic responses to the CagA protein to identify persons carrying a cag+ strain and to probe the relationship between carriage of such a strain and risk for disease. In Western populations, persons carrying cag+ strains have an enhanced risk for developing atrophic gastritis, noncardia gastric adenocarcinoma, and peptic (especially duodenal) ulceration, compared with those with cag– strains (38). Conversely, they are at lower risk for having gastroesophageal reflux disease, Barrett’s esophagus, and adenocarcinoma of the esophagus (5, 39). These observations indicate trends toward virulence or protection, not absolutes. Diversity in cag islands and differences in background H. pylori genotypes, in host, and in environmental cofactors all can further modulate disease risk. In Asian populations, since nearly all persons carry cag+ strains, the relationship of cag with disease is more difficult to establish.
Why do cag+ strains tend to be more virulent? A concept that is emerging is that they are more interactive with the host (39) and live in a different equilibrium than do cag– strains (15). For example, both in vivo and in vitro, cag+ cells induce higher levels of the proinflammatory cytokines IL-1β and IL-8. That such effects are specific for cag island genes has been shown by mutation studies. Interestingly, while mutation in each of several type IV pathway genes ablates the enhanced IL-8 induction, deletion of another such gene, the _virD4_-homolog carried on the Cag pathogenicity island, has the opposite effect in at least one strain (40). Such studies suggest that by differential expression of particular cag island genes, H. pylori can modulate its proinflammatory signals to the hosts. That cag+ and cag– strains can coexist in the same host, that all or part of the island can be deleted, and that H. pylori strains can recombine suggest that a population of H. pylori cells may use the extent of its _cag_-positivity as a type of “thermostat” to regulate its response to specific stimuli over the course of colonization of a particular host (Figure 1c). The stimuli to which the cag system is responding are not all known.
One hypothesis is that H. pylori uses the cag system to help regulate lumenal pH, which is a critical factor for all bacteria in the gastric milieu. Since transcription of _cag_A is increased at low pH (41), injection of the CagA protein into the epithelium (10) may be a means to limit acid stress (Figure 2). This model of coevolved and linked signaling between microbe and host illustrates a functioning thermostat for regulating physiologic phenomena of consequence to both parties. H. pylori has relatively few of the now-classical two-component signal-transduction regulatory systems that many other bacteria rely on to respond to environmental stress (20). Thus, we speculate that the thermostatic regulation of gene expression might at least in part be based on mutation (insertion/deletion as well as smaller point mutation changes), implying that H. pylori often uses mutation rather than conventional physiologic regulatory systems to adapt to changes in its niche. As one example, size variation in the CagA protein due to insertion or deletion of repetitive DNA sequences affects potential tyrosine phosphorylation sites, which then may alter the physiology (Figure 2) of the affected epithelial cells (10). Similarly, mutations leading to frameshifts (and their reversion) in type IV genes would affect Cag delivery by a proportion of the H. pylori population.
This model also encourages analysis of interactions of H. pylori cell populations with individuals in the polymorphic human population. For example, humans are polymorphic in the IL1β promoter region, which controls synthesis of IL-1β, a proinflammatory cytokine (42). IL1β is known to inhibit gastric acidity, and recent studies show that certain alleles are associated with increased risk of gastric cancer (42). Persons carrying cag+ strains have enhanced transcription of IL-1β (43), which, by lowering acidity, may permit better adaptation of the H. pylori population to the stomach, as well as accounting in part for the enhanced gastric cancer risk associated with these strains. Whether these differences are due to the cag island per se, or reflect the entire complex of coadapted genes, is unknown. However, we hypothesize that the cag interaction with the host is a model for a “linked signal” equilibrium system, which can undergo genetic change (Figure 1c) to maximize versatility of the population of cells, and that the particular equilibria developed affect risk of disease in the host. Equivalent considerations should apply to other virulence-associated polymorphic loci including _vac_A, _bab_A, _ice_A, and _oip_A. Risk of disease, then, is the summation of the effects on the “thermostat” by the polymorphic nature of the bacterial population, the individual host phenotypes, and the environmental exposures that affect the interaction.
Conclusions
H. pylori is a medically important bacterium, central to the pathogenesis of important upper gastrointestinal illnesses. Understanding its polymorphisms and their effects on interactions with hosts will permit greater understanding of these important diseases. In addition to the polymorphisms related to the cag island, other polymorphic loci that appear clinically relevant have been identified.
The _H. pylori_–human intercourse also is a model for interactions between persisting well-adapted microbes and their hosts. Sufficient analytical tools now exist, including genome sequences, measurable phenotypes, practical animal models, and mathematical modeling, to allow discovery of the fundamental biological basis of H. pylori persistence. This model, facilitated by _H. pylori_’s undisputed mastery of its niche, without substantial microbial competition for decades, may develop as one of the most significant rewards for studying H. pylori.
Acknowledgments
This work was supported in part by RO1 DK53707, RO1 GM63270, and P30 DK2574 from the NIH, and by the Medical Research Service of the Department of Veterans Affairs. We thank members of our research groups, and the many colleagues worldwide whose findings have contributed to the development of views presented here. We regret that space limitations precluded citing many important papers and apologize for these omissions.
References
- 1.Parsonnet J. Incidence of Helicobacter pylori infection. Aliment Pharmacol Ther. 1995;9(Suppl.):45–51. [PubMed] [Google Scholar]
- 2.Blaser MJ. Helicobacters are indigenous to the human stomach: duodenal ulceration is due to changes in gastric microecology in the modern era. Gut. 1998;43:721–727. doi: 10.1136/gut.43.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kersulyte D, et al. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosunen TU, et al. Helicobacter antibodies in 1973 and 1994 in the adult population of Vammala, Finland. Epidemiol Infect. 1997;119:29–34. doi: 10.1017/s0950268897007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser MJ. The changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999;179:1523–1530. doi: 10.1086/314785. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M.J. 2000. The amphibiotic relationship of Helicobacter pylori to humans. In Helicobacter pylori: basic mechanisms to clinical cure 2000. R.H. Hunt and G.N.J. Tytgat, editors. Kluwer Academic Publishers. Dordrecht, The Netherlands. 25–30.
- 7.Akopyanz NS, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achtman M, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 9.Boren T, Normark S, Falk P. Helicobacter pylori: molecular basis for host recognition and bacterial adherence. Trends Microbiol. 1994;2:221–228. doi: 10.1016/0966-842x(94)90626-2. [DOI] [PubMed] [Google Scholar]
- 10.Odenbreit S, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Ge Z, Rasko DA, Taylor DE. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol Microbiol. 2000;36:1187–1196. doi: 10.1046/j.1365-2958.2000.01934.x. [DOI] [PubMed] [Google Scholar]
- 12.Pérez-Pérez GI, Shepherd VL, Morrow JD, Blaser MJ. Activation of human THP-1 and rat bone marrow-derived macrophages by Helicobacter pylori lipopolysaccharide. Infect Immun. 1995;63:1183–1187. doi: 10.1128/iai.63.4.1183-1187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wirth H-P, Yang M, Peek RM, Tham KT, Blaser MJ. Helicobacter pylori Lewis expression is related to the host Lewis phenotype. Gastroenterology. 1997;113:1091–1098. doi: 10.1053/gast.1997.v113.pm9322503. [DOI] [PubMed] [Google Scholar]
- 14.Blaser MJ. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology. 1992;102:720–727. doi: 10.1016/0016-5085(92)90126-j. [DOI] [PubMed] [Google Scholar]
- 15.Blaser MJ, Kirschner D. Dynamics of Helicobacter pylori colonization in relation to the host response. Proc Natl Acad Sci USA. 1999;96:8359–8364. doi: 10.1073/pnas.96.15.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Putsep K, Branden CI, Boman HG, Normark S. Antibacterial peptide from Helicobacter pylori. Nature. 1999;398:671–672. doi: 10.1038/19439. [DOI] [PubMed] [Google Scholar]
- 17.Mattsson A, Lonroth H, Quiding-Jarbrink M, Svennerholm AM. Induction of B cell responses in the stomach of Helicobacter pylori-infected subjects after oral cholera vaccination. J Clin Invest. 1998;102:51–56. doi: 10.1172/JCI22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothenbacher D, Blaser MJ, Bode G, Brenner H. An inverse relationship between gastric colonization by Helicobacter pylori and diarrheal illnesses in children: results of a population-based cross-sectional study. J Infect Dis. 2000;182:1446–1449. doi: 10.1086/315887. [DOI] [PubMed] [Google Scholar]
- 19.Kersulyte D, Chalkauskas H, Berg DE. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 20.Tomb J-F, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 21.Alm RA, et al. Genome-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 22.Kleanthous H, Clayton CL, Tabaqchali S. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-positive bacteria. Mol Microbiol. 1991;5:2377–2389. doi: 10.1111/j.1365-2958.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 23.Kersulyte D, Akopyants NS, Clifton SW, Roe BA, Berg DE. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene. 1998;223:175–186. doi: 10.1016/s0378-1119(98)00164-4. [DOI] [PubMed] [Google Scholar]
- 24.Guttman DS, Dykuizen DE. Detecting selective sweeps in naturally occurring Escherichia coli. Genetics. 1994;138:993–1003. doi: 10.1093/genetics/138.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois A, et al. Individual host specificity of Helicobacter pylori strains and host responses in experimentally challenged non-human primates. Gastroenterology. 1999;116:90–96. doi: 10.1016/s0016-5085(99)70232-5. [DOI] [PubMed] [Google Scholar]
- 26.Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 27.Bayliss CD, Field D, Moxon ER. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J Clin Invest. 2001;107:657–662. doi: 10.1172/JCI12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuipers EJ, et al. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart in the same host. J Infect Dis. 2000;181:273–282. doi: 10.1086/315173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Q, Morgan RD, Roberts RJ, Blaser MJ. Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc Natl Acad Sci USA. 2000;97:9671–9676. doi: 10.1073/pnas.97.17.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome: restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 31.Milkman R, et al. Molecular evolution of the Escherichia coli chromosome. V. Recombination patterns among strains of diverse origin. Genetics. 1999;153:539–554. doi: 10.1093/genetics/153.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peek RM, Jr, et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531–544. [PubMed] [Google Scholar]
- 33.Tummuru MKR, Sharma SA, Blaser MJ. Helicobacter pylori picB, a homolog of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol. 1995;18:8670–8676. doi: 10.1111/j.1365-2958.1995.18050867.x. [DOI] [PubMed] [Google Scholar]
- 34.Censini S, et al. cag, a pathogenicity island of Helicobacter pylori encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akopyanz NS, et al. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 36.Cover TL. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 37.Ilver D, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 38.Wirth H-P, Yang M, Karita M, Blaser MJ. Expression of the human cell surface glycoconjugates Lewis X and Lewis Y by Helicobacter pylori isolates is related to cagA status. Infect Immun. 1996;64:4598–4605. doi: 10.1128/iai.64.11.4598-4605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaser, M.J. 1998. The interaction of cag+Helicobacter pylori strains with their hosts. In Helicobacter pylori: basic mechanisms to clinical cure 1998. R.H. Hunt and G.N.J. Tytgat, editors. Kluwer Academic Publishers. Dordrecht, The Netherlands. 27–32.
- 40.Crabtree JE, Kersulyte D, Li SD, Lindley IJ, Berg DE. Modulation of Helicobacter pylori induced interleukin-8 synthesis in gastric epithelial cells mediated by cag PAI encoded VirD4 homologue. J Clin Pathol. 1999;52:653–657. doi: 10.1136/jcp.52.9.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karita M, Tummuru MKR, Wirth H-P, Blaser MJ. Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect Immun. 1996;64:4501–4507. doi: 10.1128/iai.64.11.4501-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Omar E, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 43.Peek RM, et al. Heightened inflammatory response and cytokine expression to cagA+Helicobacter pylori strains. Lab Invest. 1995;73:760–770. [PubMed] [Google Scholar]