The Edinburgh human metabolic network reconstruction and its functional analysis (original) (raw)
Abstract
A better understanding of human metabolism and its relationship with diseases is an important task in human systems biology studies. In this paper, we present a high-quality human metabolic network manually reconstructed by integrating genome annotation information from different databases and metabolic reaction information from literature. The network contains nearly 3000 metabolic reactions, which were reorganized into about 70 human-specific metabolic pathways according to their functional relationships. By analysis of the functional connectivity of the metabolites in the network, the bow-tie structure, which was found previously by structure analysis, is reconfirmed. Furthermore, the distribution of the disease related genes in the network suggests that the IN (substrates) subset of the bow-tie structure has more flexibility than other parts.
Keywords: bow-tie, human, metabolic network, pathway
Introduction
Many human diseases are caused by or resulted in an abnormal metabolic state such as the high glucose concentration in blood of diabetes patients and the high urine amino-acid level resulted from liver or renal disorders. Metabolic processes are also heavily involved in xenobiotics degradation and drug clearance (Lewis, 2003). Drug safety is often linked to inhibition of metabolic processes (Mogilevskaya et al, 2006). Detecting the unusual level of certain specific metabolites in a patient's blood or urine has long been established as an effective method to identify biomarkers for diagnosing particular diseases (Schwedhelm and Boger, 2003; Schlotterbeck et al, 2006). Recent rapid developments of advanced metabolomics technology is opening up new horizons, as hundreds or even thousands of metabolites can be measured simultaneously, providing a much more comprehensive assessment of a patient's health status (Griffin and Nicholls, 2006; Kell, 2006; Wishart et al, 2007). However, for a better and in-depth understanding of the large amounts of data generated from metabolomics, a complete and high-quality human metabolic network is essential. This network links various metabolites by enzyme catalyzed reactions and thus allows us to discover the genetic mechanism, which causes the abnormal state of metabolites by network analysis, and further kinetic modelling. Then drugs, which could target on so far uncharacterised genes/proteins can be developed for disease treatment.
Although some human metabolic pathways such as glycolysis and urea cycle has been discovered almost a hundred years ago and many pathways have been extensively studied in biomedical journals and textbooks, a complete whole picture of human metabolic network is still missing. Especially the recent development of genome technology requires us to reconstruct the whole network from genome level to better understand the genetic basis of metabolic organization and regulation. A preliminary metabolic network can be computationally reconstructed easily from the gene annotation information (Ma and Zeng, 2003; Romero et al, 2005; Reed et al, 2006). Actually such computational reconstructed networks are available from several metabolic databases such as KEGG and HumanCyc (Kanehisa et al, 2004; Romero et al, 2005). However, the quality and completeness of such networks are often not very high. They need to be carefully investigated and calibrated through the experimental results reported in literature. This is a very labor-intensive and time consuming (somehow endless) manual process. Even for simple microorganisms such as Escherichia coli and Saccharomyces cerevisiae, the high-quality metabolic networks reconstruction often take years to finish (Reed et al, 2003; Duarte et al, 2004). In contrast to these microorganisms, which can use only a few substrates such as glucose to synthesis all the metabolites, human (and other animals) requires many essential nutrients for growth. Moreover, human is a multicell, multi-tissue organism with complex networks of interactions between them. The functions of the cells, tissues and organs are well differentiated and the metabolites are transferred within the body through the blood circulation system. Therefore, the human metabolic network is much more complex than those of microorganisms, and has different structural and functional features, which may be representative for higher organisms, especially animals.
In this paper we report our ongoing work on human metabolic network reconstruction. We have combined genome reconstruction with reconstruction based on literature to obtain a high-quality human metabolic network with more than 2000 metabolic genes and nearly 3000 metabolic reactions (referred as EHMN: Edinburgh Human Metabolic Network, in the following sections). It allows us to have a coherent picture that could be used in different studies as a reference. To better understand the functional organization of the network, we have reorganized the enzyme reactions into about 70 pathways according to their functional relationships. We further compared our network with other available human networks such as HumanCyc (Romero et al, 2005) and the recent reconstruction by Palsson's group (Duarte et al, 2007). A bow-tie connectivity structure is rediscovered from a functional rather than structural point of view, and the distribution of disease related metabolic reactions in the bow-tie structure was investigated.
Reconstruction of the global network
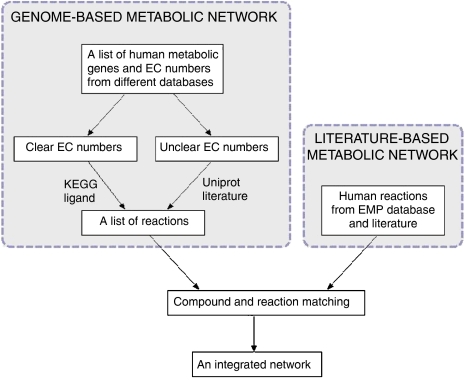
The main processes for the reconstruction of the human network are shown in Figure 1. The first step is the reconstruction of the network solely based on the human gene annotation information. This network is called the genome-based network. This step can be automated and thus the genome-based network can be easily updated with the new annotation information in the databases. Unfortunately, the human gene contents in online databases are often very different. For example, there are more than 38 000 human genes in the NCBI EntrezGene database (Maglott et al, 2007), but only about 27 thousand in HGNC (Eyre et al, 2006). Moreover, about 3000 genes in HGNC are not in EntrezGene. Therefore, it is very important to integrate information from different databases to get a more complete enzyme gene list for the reconstruction. In our reconstruction, we mainly obtained the enzyme annotation from KEGG, Uniprot and HGNC. Information from NCBI EntrezGene (Maglott et al, 2007), Ensembl (Hubbard et al, 2007) and Genecard (Safran et al, 2002) databases are also included to provide more complete crosslinks between gene (protein) IDs in different databases and validate the enzyme gene annotation.
Figure 1.
Processes for reconstruction of the high-quality human metabolic network.
Special attention was paid on the genes with unclear EC numbers such as 1.-.-.-. The existence of the unclear EC numbers is because only a chemically well-characterized enzyme is assigned an EC number by IUBMB. In the post-genome era, this process is far behind the function annotation of genes, which is mainly based on the DNA sequence. For example, in the UniProt database there are more than 800 proteins annotated with an unclear EC numbers (Wu et al, 2006). For these genes we could not get the reactions catalyzed by its encoded enzymes through the EC numbers. The reactions can only be added directly from the function annotation part in Uniprot and many genes need to be manually examined in literature.
Another problem in the reconstruction of the genome-based network is the somehow ambiguous relationships between EC numbers and reactions in the reaction databases. A protein in human may not catalyze a reaction, which is catalyzed by a protein with same EC number in other organisms. For example, the GBA3 gene in human codes for cytosolic beta-glucosidase, which have an EC number 3.2.1.21, whereas in other organisms, proteins with this EC number also function as cellobiase catalyzing the degradation of cellulose. This degradation reaction apparently does not occur in human. Unfortunately there is still no automatic way to obtain the human-specific EC number–reaction relationships. We first used the KEGG ligand database to generate the reaction list, because it is one of the most complete metabolic reaction databases including more than 7000 reactions (Goto et al, 2002). Then the reactions were manually checked to exclude non-human reactions.
The second step of the reconstruction is to refine the genome-based network based on information from literature. Fortunately, thanks to the EMP database (personal communication with EMP projects Inc.), we already have a literature based human metabolic network available. The EMP network is a compound centric network reconstructed mainly based on information from literature. It contains more than a thousand compounds and nearly 2000 reactions. We then can compare and integrate the two networks together to obtain a more complete and high-quality human metabolic network. However, this integration process is very time consuming mainly due to the different compound and reaction nomenclature systems used in the two datasets. The reactions and compounds in the genome-based network are mainly from the KEGG ligand database, while EMP has its own nomenclature for reactions and compounds. In order to check if two reactions from the two networks are the same, we need to check if all the compounds in the reaction equations are the same. A straightforward way to match the compounds in different databases is to compare them by name. However, even though both KEGG and EMP databases have a synonym list for each compound, the total number of matching compounds is only about 500 of the more than 2000 compounds in the two networks. This is a surprising result considering that both are human metabolic networks. We have tried different methods to obtain more compound matching relationships such as using synonyms in other compound databases such as PubChem (Wheeler et al, 2006) and ChEBI (Brooksbank et al, 2005) to match the compounds in EMP and KEGG, matching compounds by structure and allow fuzzy matching between a generic compound and their specific compounds (for example D-glucose and alpha D-glucose). Unfortunately only about 300 new matching relationships were obtained through these methods. Based on the matched compounds, we found about 700 matching reactions between the genome-based network and the EMP network, by checking if two reactions have the same reactants. These matching reactions allow us to compare the two networks at a higher pathway level. If a pathway in EMP contains one or more matching reactions with a KEGG pathway, then these two pathways will be functionally linked. We can then compare the two pathways to see if they have some unfound matching reactions or a reaction in one pathway complements a gap in another pathway. This pathway consolidation process can only be done by manual examination of the reactions in the pathways and visual inspection of the pathway maps. However, it is an important step to improve and maintain the quality of the reconstructed network, and for us to better understand the biological function of the large-scale network.
Pathway organization of the human metabolic network
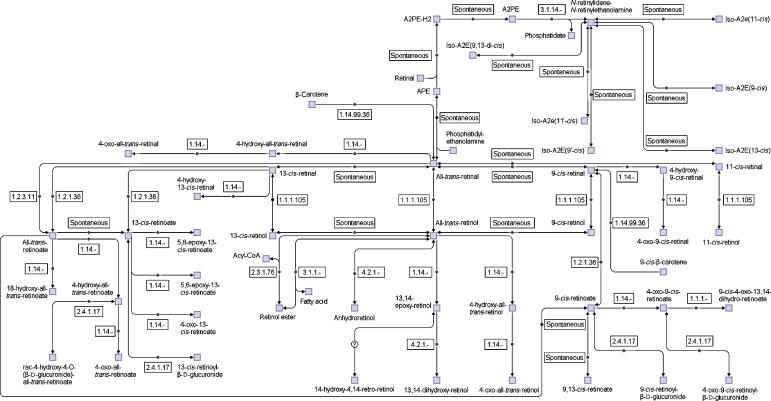
As described above, the pathway organization of reactions is an important step in the network reconstruction. However, in comparing the pathways in EMP and KEGG, we found they are organized very differently. In EMP there are more than 300 metabolic pathways for the human metabolic network. Almost 100 of them are very small, containing three or fewer reactions. Moreover, in the pathway maps there is no link to other pathways shown. Therefore, it is very difficult to gain a whole picture of the human metabolic network from so many small pathways. In KEGG, all the reactions from different organisms are organized into about a hundred metabolic pathways. The problems with the KEGG pathways are the following: (1) it is not human specific. In certain pathways only one or two isolated human reactions exist; (2) there is a high overlap between pathways. For example, there are many overlap reactions in the pathways of glutamate metabolism, urea cycle, arginine and proline metabolism. Grouping these pathways as a big pathway would show the functional relationships between the reactions in them much better; (3) the mass flow between a substrate and a product in the pathways is not as clear as those in EMP or Biocyc (Karp et al, 2005). To address these problems, we decided to define a new set of human-specific pathways which are (1) human specific; (2) less overlap between pathways; (3) large enough to include functional related small pathways; (4) including links to other pathway to get a better overview of functional connectivity between pathways. Basically, the small pathways in EMP and KEGG were grouped on the basis of their functional relationships. Some original pathways may also be separated into different new pathways. Altogether, 2823 reactions are included in the network and reorganized into 66 pathways, with the number of reactions between 5 and 142 (there are also more than 300 isolated reactions in the network). The retinol (vitamin A) pathway is shown in Figure 2 as an example. There are much more reactions in our pathways than in the corresponding KEGG pathways. More pathway maps and the whole set of pathways in SBML format can be seen in the Supplementary files (the Supplementary data set and Supplementary figures, the network and the pathways in SBML format are also available at http://wwwtest.bioinformatics.ed.ac.uk/wiki/PublicCSB/EHMN). The users can directly open the SBML files in CellDesigner (Kitano et al, 2005) or other softwares to generate an automatic layout for the pathways. As described in previous studies, the currency metabolites often cause trouble in graph layout of metabolic pathways because they tend to link all the metabolites in a short path. Therefore in the SBML files for the pathways we include only the main compounds in the ‘listofreactants' and ‘listofproducts' section. This makes it possible to quickly generate clear and nice pathway maps from the SBML files.
Figure 2.
An example pathway of the reconstructed human metabolic network: the retinol (vitamin A) pathway. Compared with corresponding KEGG pathway (map00830), our pathway contains more reactions.
In the process of pathway reorganizing, we noticed that many reactions, especially those related with complex lipid metabolism, are missing in the genome-based network, where the reactions are mainly from KEGG ligand database. For example, the reactions related with omega-3 and omega-6 fatty acid (two essential nutrients for human) metabolism, mono-unsaturated fatty acid metabolism and epoxyeicosatrienoic acids (EETs) metabolism are almost completely missing in KEGG. Due to the great structural variance of complex lipids, the total number of lipid metabolites is more than 8000 (Fahy et al, 2005) and most of them exist in the human metabolic network. Therefore, even though we already added many lipid pathways from literature, the network is still far from complete. A comprehensive database on lipids and their relating enzymes in various organisms has been developed by the LIPID MAPS Consortium (Fahy et al, 2005; Cotter et al, 2006). Based on information in this database and other resources, more lipid-related pathways can be added in the future version of our database.
We further compared our network with another computationally reconstructed network in HumanCyc (version 10.6) (Romero et al, 2005). There are 996 reactions in the database, and among them 766 are catalyzed by enzymes. This is only half of the number of reactions in our database. We extracted 976 EC numbers from HumanCyc and compared them with those in our database. We found 151 EC numbers are in HumanCyc but not in our database. We then checked the reactions and proteins corresponding to these EC numbers, aiming to add new reactions to our database. Surprisingly, we found that 116 of the 151 new EC numbers were without any coding gene, but added by the pathway hole filling algorithm used in Pathologic method for the computational reconstruction of metabolic networks in Biocyc (Karp et al, 2005). However, many of them are in pathways where many reactions are without any gene. For example, in the dTDP-L-rhamnose biosynthesis I pathway, only the reaction catalyzed by 4.2.1.46 is encoded by a human gene. The other three reactions catalyzed by 1.1.1.133, 2.7.7.24 and 5.1.3.13 are all added to complement the pathway. There is even no literature related with these reactions in human. Therefore, we decided not to include these reactions in our network. For the other 35 EC numbers, we examined their corresponding genes and checked how these genes are annotated in other databases and literature. We found that 24 EC numbers unique in HumanCyc are because of wrong annotation of the genes in HumanCyc. For example, among the three genes encoding 2.4.1.87, gta actually functions as a galactosyltransferase activator, CDC2L2 is a galactosyltransferase-associated protein kinase, ENSG00000165196 has already been removed in the latest ENSEMBL database (Hubbard et al, 2007). For the other 10 EC numbers, four have no reaction or a protein modification reaction, which is currently not included in our network. Therefore, we only need to add reactions for six EC numbers from HumanCyc. Actually some of the reactions are already in our reconstruction, but with a different EC number. Altogether nine reactions were added from HumanCyc. A complete list of the manually examined EC numbers unique in HumanCyc can be seen in Supplementary Table 1. The comparative analysis between our network and HumanCyc indicates from one aspect the importance of integrating information from different databases for network reconstruction, and from another side, the importance of human curation for improving the quality of the computationally reconstructed network.
During the review process of the paper, another high-quality human metabolic network reconstructed by Palsson's group (referred as HMN-P below) was published (Duarte et al, 2007). We obtained their data from the BiGG database and compared with our network (EHMN). At the gene level, EHMN contains 2322 genes from different databases, HMN-P contains 1496 genes mainly from EntrezGene (actually all genes have EntrezGene ID). The common part is 1069 genes. At enzyme level, in HMN-P only less than half of the genes are assigned EC numbers (total EC numbers less than 500 including unclear EC numbers). In EHMN, all the genes have clear or unclear EC number because we start the reconstruction from such genes. The total number of ECs is more than 800 (excluding unclear ECs). One may argue that in HMN-P ECs are not used to link genes with reactions. However, as a widely used standard for representing metabolic reactions, introducing EC number in the network can greatly simplify the comparative analysis of metabolic networks for that the direct comparison of reaction equations is very difficult due to compound synonyms. At the metabolite level, EHMN contains 2671 compounds, and 1769 of them can be found in KEGG database. HMN-P has 1469 compounds and about a half of them linked to KEGG. For the non-KEGG compounds, in HMN-P only one compound name is given. This makes it difficult to find a matching compound in other databases. As stated previously, we have developed a compound database with synonyms, structure information and IDs in different databases (in Supplementary data set). At the reaction level, HMN-P contains more reactions than EHMN (3731 versus 2823). However, there are 1189 transport reactions and 457 exchange reactions, which are not considered in EHMN because the subcellular location information is still not included. Furthermore, there are 290 repeat reactions in HMN-P, which are the same reaction but in different compartments. Therefore, the number of reactions comparable with EHMN is just 1795. Because of the intrinsic complexity of human cell, it is very difficult to place the reactions into a small number of compartments. Actually we have collected protein location information from different databases and have identified hundreds of cellular locations. We are working on it to develop a GO (Gene Ontology) (Ashburner et al, 2000)-based hierarchically compartmented human network for the next release.
Functional analysis of the human metabolic network and the bow-tie structure
Unlike most microorganisms, which can use simple substrates to produce all the metabolites required for its growth, human requires many essential nutrients obtained directly from the food to maintain a health physiological state. The typical essential nutrients include 10 amino acids, omega-3 and omega-6 fatty acids and various vitamins. To verify the essentiality of these metabolites, we manually examined the pathways related with them and found that there is no pathway for the synthesis of these metabolites from the central metabolites. For a more systematic analysis of the metabolic capacity of the human metabolic network, we start from the central metabolites (in glycolysis, pentose phosphate pathway and TCA cycle pathway) and classify other metabolites as exchangeable (pathways for both the synthesis from central metabolites and the degradation to central metabolites exist), degradable (only degradation pathways exist), synthesizable (only synthesis pathways exist) and isolated (no pathway from/to central metabolites) based on pathway analysis. We found that many of the essential nutrients fall into the isolated metabolites and some essential amino acids are degradable. This metabolite classification is quite similar with the bow-tie structure of metabolic networks discovered previously based on graph analysis (Ma and Zeng, 2003). In the bow-tie structure, all the metabolites in the giant strong component (GSC) can convert to each other (equal to the exchangeable metabolites), the metabolites in the IN subset can convert to metabolites in GSC (degradable), those in the OUT subset can be produced from metabolites in GSC (synthesizable) and those in the isolated subset are not connected with GSC. Here we rediscovery the bow-tie structure by functional analysis of the metabolic network. Because graph is a simplified representation of metabolic networks, which lose some structure information (for example a multiple substrate multiple product reaction just be represent as several links with one substrate and one product), if the bow-tie structure found by graph analysis represents the true biological organization principle of metabolic network is still an open question. Here, based on a functional analysis of the high-quality human metabolic network, we confirmed the bow-tie structure from a biological point of view. Due to the simplification in the graph conversion of a metabolic network, the exact position of certain metabolites in the bow-tie may be different for the two different approaches. However, as a system level macroscopic structure, the bow-tie is true from both structural and biological aspects. This fact strengthens the hypothesis that bow-tie is an important organization principle for complex systems to be robust and flexible (Csete and Doyle, 2004; Kitano, 2004).
Based on the classification of the metabolites, the reactions can also be classified into four subsets forming a bow-tie structure similar to the bow-tie of the reaction graphs of metabolic networks (Ma et al, 2004). The reactions occur between the exchangeable metabolites are in the GSC. The reactions in a pathway from the degradable metabolites to the exchangeable metabolites form the IN subset. Correspondingly, the reactions in a pathway from the exchangeable metabolites to the synthesizable metabolites form the OUT subset. All the other reactions are in the isolated subset. The bow-tie classification of the reactions is more interesting because the reactions are linked to the genes and proteins, which are the main functional regulation units in the cell. The full classification of the reactions can be seen in the Supplementary file.
For the human network, we found that the scale of the isolated subset is often very large (more than one fourth of the whole network). We investigated the metabolites and reactions in the isolated subsets and found that many of them are actually not truly isolated, but can be produced from metabolites in the IN subset and may have important physiological function. As mentioned previously, human requires many essential nutrients for growth. These nutrients are essential because they are used for producing certain metabolites with important physiological functions. For example, the aromatic amino acids are precursors for monoamine hormones and neuron transmitters. These signal metabolites can bind to different protein receptors and then regulate the amount and activity of proteins to change the physiological state. Because many such signal molecules are produced from essential nutrients, which are in the IN subset of the bow-tie, they should be in the isolated subset of the bow-tie structure. To distinguish these metabolites from the real isolated metabolites, we generate a new subset called ‘OUT2' to include the metabolites synthesized from the metabolites in the ‘IN' subset. Correspondingly, an ‘IN2' subset, which contains the metabolites for producing metabolites in ‘OUT' is also added. Therefore a six-subset modified bow-tie structure of the human metabolic network is produced as shown in Figure 3. The numbers of reactions in the six subsets are shown in Table I.
Figure 3.
The bow-tie structure of the human metabolic network.
Table 1.
The bow-tie structure of human metabolic network and diseases
| Subsets | No. of reactions | No. of reactions with encoding genes | No. of disease-related reactions | Percentage of disease-related reactions |
|---|---|---|---|---|
| Total | 2823 | 2314 | 2215 | 95.72% |
| GSC | 605 | 550 | 535 | 97.27% |
| IN | 330 | 283 | 258 | 91.17% |
| OUT | 984 | 788 | 777 | 98.60% |
| IN2 | 12 | 12 | 8 | 66.67% |
| OUT2 | 257 | 201 | 194 | 96.52% |
| Isolated | 635 | 480 | 443 | 92.29% |
A main objective of human metabolic network analysis is to see how it is related with human disease. More than 10 000 human genes (half of the whole genome) have been reported to be related with one or more human diseases in the OMIM database (Hamosh et al, 2005). In the human metabolic network, we found that 2215 (of 2823) reactions are catalyzed by enzymes, which are coded by disease related genes. If we exclude the reactions, which are spontaneous or with an unknown gene, the proportion of disease related reactions is even higher at 95% (2215 of 2314 reactions with encoding genes). This surprisingly high value raises a question to us: what does network robustness really mean from a biological aspect? As the most complex organism on the earth, human is expected to have a very robust metabolic network. Actually previous studies on biological network truly suggest that as scale free networks, metabolic networks are robust against random errors from a structural point of view (Jeong et al, 2000). However, the result that most reactions are linked with disease genes indicates that the human metabolic network is fragile. We analyzed the distribution of the disease related reactions in the bow-tie structure and the result is shown in Table I. Interestingly the proportion of disease-related reactions in the IN subset is much less than that in the OUT subset, implying that the reactions leading to metabolic products are more fragile than the reactions for the degradation of various substrates. This result looks unusual but understandable from biochemistry. The key function of the central pathways (glycolysis and TCA cycle) is to produce energy and precursors for biosynthesis. Most of the bioproducts can be synthesized from a number of common metabolite precursors in the central pathways, and most of the substrates are also converted to these precursors first for further conversion. Therefore, theoretically one substrate is enough to produce all the necessary products, if it can be converted to these precursors. The existence of multiple pathways for multiple substrates just provides more flexibility to the organism and thus blocking one pathway is unlikely to damage the organism. In contrast, a product is synthesized in an organism often because it has some unique function important for the organism (as a structure molecule or a signal molecule). Hence, a failure in a product synthesis pathway can make the whole system organized improperly, causing a disease. One step further, we may hypothesize that the organization of the bio-products and their synthesis pathways from the common precursors rather than the substrates and their degradation pathways determine the feature of a biosystem. Back to the bow-tie structure, the metabolites and reactions in the Out subset may better define a biosystem than the metabolites and reactions in the IN subset. Further studies on comparative analysis of the metabolic networks of different organisms are needed to validate this hypothesis.
Supplementary Material
Supplementary Table 1
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary data set 1
Supplementary data set 2
Supplementary data set 3
Supplementary data set 4
Supplementary data set 5
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooksbank C, Cameron G, Thornton J (2005) The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res 33: D46–D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter D, Maer A, Guda C, Saunders B, Subramaniam S (2006) LMPD: LIPID MAPS proteome database. Nucleic Acids Res 34: D507–D510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csete M, Doyle J (2004) Bow ties, metabolism and disease. Trends Biotechnol 22: 446–450 [DOI] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Herrgard MJ, Palsson BO (2004) Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 14: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ (2006) The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res 34: D319–D321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA (2005) A comprehensive classification system for lipids. J Lipid Res 46: 839–861 [DOI] [PubMed] [Google Scholar]
- Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M (2002) LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res 30: 402–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JL, Nicholls AW (2006) Metabolomics as a functional genomic tool for understanding lipid dysfunction in diabetes, obesity and related disorders. Pharmacogenomics 7: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33: D514–D517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K et al. (2007) Ensembl 2007. Nucleic Acids Res 35: D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL (2000) The large-scale organization of metabolic networks. Nature 407: 651–654 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N (2005) Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res 33: 6083–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell DB (2006) Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discov Today 11: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Kitano H (2004) Biological robustness. Nat Rev Genet 5: 826–837 [DOI] [PubMed] [Google Scholar]
- Kitano H, Funahashi A, Matsuoka Y, Oda K (2005) Using process diagrams for the graphical representation of biological networks. Nat Biotechnol 23: 961–966 [DOI] [PubMed] [Google Scholar]
- Lewis DF (2003) Human cytochromes P450 associated with the phase 1 metabolism of drugs and other xenobiotics: a compilation of substrates and inhibitors of the CYP1, CYP2 and CYP3 families. Curr Med Chem 10: 1955–1972 [DOI] [PubMed] [Google Scholar]
- Ma HW, Zeng AP (2003) Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics 19: 270–277 [DOI] [PubMed] [Google Scholar]
- Ma HW, Zeng AP (2003) The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics 19: 1423–1430 [DOI] [PubMed] [Google Scholar]
- Ma HW, Zhao XM, Yuan YJ, Zeng AP (2004) Decomposition of metabolic network based on the global connectivity structure of reaction graph. Bioinformatics 20: 1870–1876 [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T (2007) Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 35: D26–D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilevskaya E, Demin O, Goryanin I (2006) Kinetic model of mitochondrial Krebs cycle: unraveling the mechanism of salicylate hepatotoxic effects. J Biol Phys 32: 245–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Famili I, Thiele I, Palsson BO (2006) Towards multidimensional genome annotation. Nat Rev Genet 7: 130–141 [DOI] [PubMed] [Google Scholar]
- Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD (2005) Computational prediction of human metabolic pathways from the complete human genome. Genome Biol 6: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, Ben-Dor U, Esterman N, Rosen N, Peter I, Olender T, Chalifa-Caspi V, Lancet D (2002) GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics 18: 1542–1543 [DOI] [PubMed] [Google Scholar]
- Schlotterbeck G, Ross A, Dieterle F, Senn H (2006) Metabolic profiling technologies for biomarker discovery in biomedicine and drug development. Pharmacogenomics 7: 1055–1075 [DOI] [PubMed] [Google Scholar]
- Schwedhelm E, Boger RH (2003) Application of gas chromatography-mass spectrometry for analysis of isoprostanes: their role in cardiovascular disease. Clin Chem Lab Med 41: 1552–1561 [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD et al. (2006) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 34: D173–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G et al. (2007) HMDB: the Human Metabolome Database. Nucleic Acids Res 35: D521–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B (2006) The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res 34: D187–D191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary data set 1
Supplementary data set 2
Supplementary data set 3
Supplementary data set 4
Supplementary data set 5