Genome-Level Longitudinal Expression of Signaling Pathways and Gene Networks in Pediatric Septic Shock (original) (raw)
Abstract
We have conducted longitudinal studies focused on the expression profiles of signaling pathways and gene networks in children with septic shock. Genome-level expression profiles were generated from whole blood-derived RNA of children with septic shock (n = 30) corresponding to day one and day three of septic shock, respectively. Based on sequential statistical and expression filters, day one and day three of septic shock were characterized by differential regulation of 2,142 and 2,504 gene probes, respectively, relative to controls (n = 15). Venn analysis demonstrated 239 unique genes in the day one dataset, 598 unique genes in the day three dataset, and 1,906 genes common to both datasets. Functional analyses demonstrated time-dependent, differential regulation of genes involved in multiple signaling pathways and gene networks primarily related to immunity and inflammation. Notably, multiple and distinct gene networks involving T cell- and MHC antigen-related biology were persistently downregulated on both day one and day three. Further analyses demonstrated large scale, persistent downregulation of genes corresponding to functional annotations related to zinc homeostasis. These data represent the largest reported cohort of patients with septic shock subjected to longitudinal genome-level expression profiling. The data further advance our genome-level understanding of pediatric septic shock and support novel hypotheses.
INTRODUCTION
Septic shock affects a large number of children in the United States and worldwide (1–3). The morbidity and mortality associated with pediatric septic shock remain unacceptably high and current therapy is relatively limited to prevention (i.e. vaccines), antibiotics, and pediatric intensive care unit (PICU)-based physiological support (3,4). We have suggested that one powerful approach to more comprehensively understand the inherent biological complexity of pediatric septic shock involves translational research at the genomic level (5,6). The feasibility of a genomic approach to increase our understanding of septic shock pathobiology is indicated by a series of recent studies (7–12), including our own initial report (6). Furthermore, a comprehensive, unbiased examination of gene transcripts at the level of the whole genome may identify novel biological mechanisms and therapeutic targets.
In a previous study, we generated an unprecedented, genome-level expression profile dataset of pediatric septic shock specifically focused on a single time point, within the first 24 h of presentation to the PICU (6). The previous dataset demonstrated the feasibility of conducting genome-centered, translational research in critically ill children. In addition, we demonstrated that biologically meaningful microarray data can be generated and analyzed using a readily accessible clinical sample, whole blood-derived RNA.
In the current study, we sought to broaden our initial approximation of the genomic response of children with septic shock. We have generated and analyzed additional microarray data representing two distinct time points during the initial 72 h of presentation to the PICU with septic shock. We have focused our analyses on coordinately regulated genes corresponding to signaling pathways and gene networks. Our overall goal for these studies is to define the longitudinal, genome-level expression profiles that occur in pediatric septic shock as a means of further advancing our understanding of this condition at the translational level.
METHODS
Patients
The study protocol was approved by the individual Institutional Review Boards of each participating institution. Children < ten years of age admitted to the PICU and meeting criteria for septic shock were eligible for the study. Septic shock was defined using pediatric-specific criteria (13). Control patients were recruited from the participating institutions using the following exclusion criteria: a recent febrile illness (within two weeks), recent use of anti-inflammatory medications (within two weeks), or any history of chronic or acute disease associated with inflammation.
Sample and Data Collection
After informed consent, blood samples for RNA isolation were obtained within 24 h of admission to the PICU, heretofore called “day one” of septic shock. An additional sample was obtained for RNA isolation 48 hours after obtaining the day one sample (“day three” of septic shock). Severity of illness at study entry was calculated using the PRISM III score (14), and organ failure was defined using pediatric-specific criteria (13,15,16). Clinical and laboratory data were collected daily while in the PICU. All study patients were followed for 28 days to determine survival. Clinical, laboratory, and biological data were entered and stored using a locally developed, web-based database.
RNA Extraction and Microarray Hybridization
The data and protocols described in this manuscript are deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, GEO Series accession number GSE8121).
Total RNA was isolated from whole blood samples using the PaxGene Blood RNA System (PreAnalytiX, Qiagen/Becton Dickinson, Valencia, CA, USA) according to the manufacturer’s specifications. Microarray hybridization was performed by the Affymetrix Gene Chip Core facility at Cincinnati Children’s Hospital Research Foundation as previously described (Human Genome U133 Plus 2.0 GeneChip, Affymetrix, Santa Clara, CA, USA) (17).
Data Analysis
Analyses were performed using one patient sample per chip. Image files were captured using an Affymetrix GeneChip Scanner 3000. CEL files were subsequently preprocessed using Robust Multiple-Array Average (RMA) normalization using GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA, USA) (18). All signal intensity-based data were used after RMA normalization, which specifically suppresses all but significant variation among lower intensity probe sets (18). All chips were then normalized to the respective median values of controls.
Differences in mRNA abundance between patient samples were determined using GeneSpring GX 7.3 (Agilent Technologies). All statistical analyses used corrections for multiple comparisons. A list of genes differentially regulated on day one of septic shock was generated using sequential statistical and expression filters as follows. We first conducted a two-group ANOVA (Benjamini-Hochberg false discovery rate of five percent) using controls and the day one samples from patients with septic shock as the comparison groups, and all gene probes within the microarray (54,681 gene probes). The list of genes passing this statistical filter was then subjected to an expression filter that selected only the genes having ≥ two-fold expression difference between the median values of controls and patients with septic shock on day one, respectively. Identical sequential and expression filters were used to generate a list of genes differentially regulated on day three of septic shock, using controls and day three septic shock samples as the comparison groups.
Two-dimensional cluster maps were constructed using GeneSpring GX 7.3 (Agilent Technologies). Gene trees are represented in the vertical dimension and condition trees are represented in the horizontal dimension. Gene trees and condition trees are both based on the Distance similarity algorithm. The coloring conventions for all maps are as follows: red intensity correlates with increased gene expression, blue intensity correlates with decreased gene expression, and yellow intensity correlates with no change in gene expression relative to the median of controls.
Gene lists of differentially expressed genes were primarily analyzed using the Ingenuity Pathways Analysis application (IPA, Ingenuity Systems, Redwood City, CA, USA) that provides a tool for discovery of signaling pathways and gene networks within the uploaded gene lists (7). Additional analyses for derivation of functional gene annotations were performed by uploading specific gene expression lists to two distinct web-based applications that allow public access to relational databases of functional gene annotations: D.A.V.I.D. (Database for Annotation, Visualization and Integrated Discovery) (19) and the PANTHER Classification System (Protein Analysis Through Evolutionary Relationships) (20–22). These applications use specific approaches to estimate significance based on non-redundant representations of the microarray chip and to convert the uploaded gene lists to gene lists containing a single value per gene.
Derivation of Gene Networks
The IPA algorithm for generating gene networks is as follows. The IPA application maps each gene identifier to its corresponding gene object in the IPA Knowledge Base. These focus genes are then overlaid onto a global molecular network developed from information contained in the Knowledge Base. Networks of these focus genes are then algorithmically generated based on their connectivity. Each individual IPA network has a maximum of 35 focus genes and is assigned a significance score (based on P value) representing the likelihood that the focus genes within the network are found therein by random chance. A high number of focus genes within a dataset leads to a higher network score (equal to the negative exponent of the respective P value such that a score of three corresponds to a P value of 10E-3) (7).
RESULTS
General Subject Data
The current cohort of 30 patients with septic shock and 15 controls were included in a previous publication involving 42 total patients and focused exclusively on day one of septic shock (6). The current cohort represents a subset of those 42 patients having complete microarray data on both day one and day three of septic shock. Summary demographic, clinical, and microbiologic data for the study subjects are provided in Tables 1 and 2. Analysis was performed on 75 individual microarray chips representing 15 controls and 30 patients with septic shock (separate day one and day three samples for each patient with septic shock). The controls were comparable to the patients with septic shock with regard to age, race, and gender. Among the patients with septic shock there were 20 having positive identification of an infecting organism (67 percent) and five deaths (17 percent mortality). Overall, the patients with septic shock were heterogeneous with respect to age, illness severity, gender, race, infectious organisms, and primary sites of infection.
Table 1.
Clinical and Demographic Data for All Subjects
| Controls | Septic Shock | |
|---|---|---|
| Number of individual subjects | 15 | 30 |
| Mean age (years) ± S.D. | 3.1 ± 3.5 | 3.8 ± 3.5 |
| Mean PRISM Score ± S.D. | n/a | 17.9 ± 8.4 |
| Number with comorbidity1 | n/a | 14 (47 percent) |
| Mean number of organ failures ± S.D.2 | n/a | 2.8 ± 1.1 |
| Gender (Male/Female) | 8/7 | 18/12 |
| Race (number) | A.A./Black (6) | A.A./Black (10) |
| Asian (4) | Asian (1) | |
| Caucasian (5) | Unreported (2) | |
| Caucasian (17) |
Table 2.
Microbiology Data for Patients with Septic Shock1
| Organism (number) | Primary source of positive culture (number) |
|---|---|
| Candida albicans (1) | Blood (15) |
| Cytomegalovirus (1) | Lung (3) |
| Enterococcus faecalis (1) | Urine (1) |
| Escherichia coli (1) | Retropharyngeal abscess (1) |
| Streptococcus pyogenes (2) | |
| Streptococcus agalactiae (1) | |
| Herpes simplex I (1) | |
| Influenza B (2) | |
| Klebsiella pneumoniae (1) | |
| Neisseria meningitidis (2) | |
| Ricketssia ricketsii (1) | |
| Staphylococcus aureus (2) | |
| Staphylococcus epidermidis (1) | |
| Streptococcus viridans (2) | |
| Varicella (1) |
Differential Gene Expression on Day One of Pediatric Septic Shock
To begin testing the hypothesis that the genome-level expression profile of pediatric septic shock has a longitudinal component, we generated a list of differentially regulated genes on day one of septic shock using the filtering strategy outlined in the Methods section. The initial statistical filter yielded a list of 21,146 gene probes that were differentially regulated between day one patients with septic shock and controls. The subsequent expression filter yielded a final list of 2,145 gene probes that were differentially regulated between day one patients with septic shock and controls.
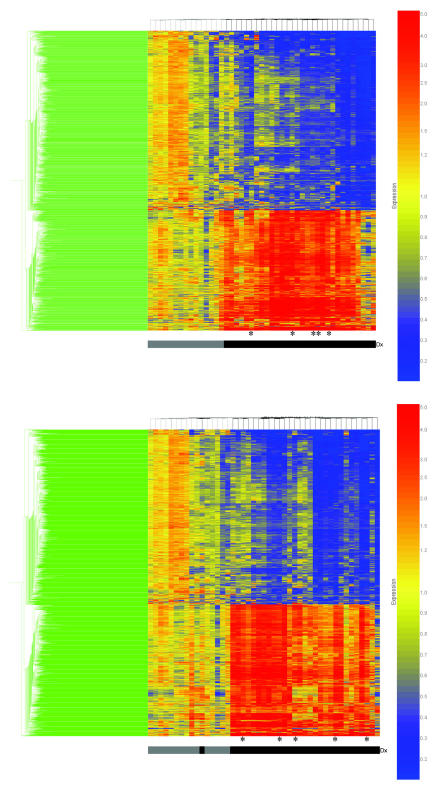
These 2,145 genes were subjected to two-dimensional cluster analysis (Figure 1A). All patients with septic shock clustered together within the right two-third portion of the map in a homogenous manner, thus indicating relatively cohesive gene regulation on day one of pediatric septic shock despite clinical heterogeneity. The homogenous clustering was dependent on a group of genes in the upper portion of the map having decreased expression (1,283 genes) and a group of genes in the lower portion of the map having increased expression (862 genes). Based on the clustering pattern, these gene expression profiles did not appear to be dependent on the class of infecting organism. For example, patients having a viral etiology (indicated by asterisks, bottom of Figure 1A) were distributed throughout the map in a heterogeneous manner. Similar observations regarding clustering patterns were made for patients having gram positive, gram negative bacterial infections, or no documented infection (not shown).
Figure 1.
A. Two dimensional gene cluster map representing 2,145 genes differentially regulated between day one patients with septic shock and control patients (see text for filtering strategy). Individual patients are oriented horizontally and individual genes are oriented vertically. The color-coded bar at the bottom of the map represents controls in gray and the patients with septic shock in black. Asterisks indicate patients having a viral etiology. B. Two dimensional gene cluster map representing 2,504 genes differentially regulated between day three patients with septic shock and control patients (see text for filtering strategy). The same conventions apply for Figure 1B as for that of Figure 1A.
Differential Gene Expression on Day Three of Pediatric Septic Shock
To compare the genome-level expression profiles of day one and day three, we next generated a list of differentially regulated genes on day three. We used the identical, sequential statistical, and expression filters as that described above for the day one analysis, except that we used controls and day three samples from patients with septic shock as the comparison groups. The initial statistical filter yielded a list of 22,403 gene probes that were differentially regulated between day three patients with septic shock and controls. The subsequent expression filter yielded a final list of 2,504 gene probes that were differentially regulated between day three patients with septic shock and controls.
These 2,504 genes then were subjected to two-dimensional cluster analysis (Figure 1B). All patients with septic shock, except one, clustered together within the right two-third portion of the map in a homogenous manner. Similar to the day one map, these data indicate relatively cohesive gene regulation on day three of pediatric septic shock. Again, the homogenous clustering was dependent on a group of genes in the upper portion of the map having decreased expression (1,432 genes) and a group of genes in the lower portion of the map having increased expression (1,072 genes), but independent of the infecting organism class or absence of documented infection.
Comparison of Day One and Day Three Datasets
The 2,145 genes differentially regulated on day one of septic shock and the 2,504 genes differentially regulated on day three of septic shock were compared by Venn analysis (not shown). This analysis demonstrated that there were 239 unique genes in the day one dataset, 598 unique genes in the day three dataset, and 1,906 genes common to both the day one and day three datasets. These data demonstrate that while a large number of genes are differentially regulated compared with controls, the majority of these genes are consistently up- or downregulated over the initial three days of septic shock. Conversely, smaller sets of genes are uniquely regulated either early (day one) or later (day three) during septic shock. Thus, over the initial three-day course of pediatric septic shock, the longitudinal genome-level expression profile has a temporal expression component and a relatively constant expression component.
Signaling Pathway Analysis Focused on Upregulated Genes
To derive biological meaning from the datasets representing increased gene expression on day one and day three of pediatric septic shock, we conducted a comparison analysis using the IPA application (7). We individually uploaded the 862 genes upregulated on day one and the 1,072 genes upregulated on day three to the IPA application, and focused the comparative analytical output on upregulation of genes associated with signaling pathways.
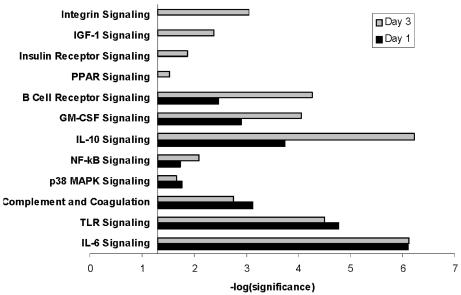
As shown in Figure 2, this analysis demonstrated that the two datasets contained genes relevant to 12 distinct signaling pathways (individual gene lists for each pathway are provided in the supplementary data). Five of the 12 signaling pathways had a similar level of increased gene expression on day one and day three (interleukin-6 signaling, Toll-like receptor signaling, complement and coagulation, p38 MAP kinase signaling, and NF-κB signaling), as defined by having respective significance values within one log-fold range. Three of the 12 signaling pathways had a higher level of increased gene expression on day three, compared with day one (interleukin-10 signaling, granulocyte macrophage colony stimulating factor signaling, and B cell receptor signaling), as defined by having a respective significance value difference > one log-fold. Four of the 12 signaling pathways did not have significant upregulation on day one, but were significantly upregulated on day three (PPAR signaling, insulin receptor signaling, IGF-1 signaling, and integrin signaling), as defined by having a respective significance value > 1.3 (i.e. -log (P value = 0.05) = 1.3). These data indicate that in pediatric septic shock, the longitudinal pattern of increased gene expression relevant to signaling pathways is heterogeneous, with some pathways being equally upregulated on day one and day three, some pathways having a higher degree of upregulation on day three relative to day one, and some pathways having exclusive upregulation on day three.
Figure 2.
Comparison analysis of the signaling pathways corresponding to the 862 genes upregulated on day one and the 1,072 genes upregulated on day three of pediatric septic shock. The analysis is derived from the Ingenuity Pathways Analysis application (see text for details). The y-axis represents the -log (significance). For illustrative purposes, the x-axis crosses the y-axis at the 1.3 threshold of significance (i.e. -log (P value = 0.05) = 1.3). Significance is directly proportional to the number of genes within the data set that corresponds to the respective canonical signaling pathway, and inversely proportional to the number of genes in the entire data set uploaded to the Ingenuity Pathways Analysis application.
Signaling Pathway Analysis Focused on Downregulated Genes
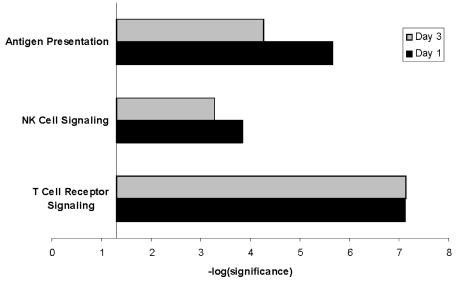
To derive biological meaning from the gene lists representing decreased gene expression on day one and day three of pediatric septic shock, we again conducted an analysis using the IPA application in an identical manner to that described above, except that we individually uploaded the 1,283 genes downregulated on day one and the 1,432 genes downregulated on day three to the IPA application. As shown in Figure 3, this analysis demonstrated that both gene lists contained genes relevant to three distinct signaling pathways (individual gene lists for each pathway are provided in the supplementary data). Two of the three signaling pathways had a similar degree of downregulation on day one and day three (T cell receptor signaling and NK cell signaling), as defined by having respective significance values within one log-fold range. In contrast, the signaling pathway involving antigen presentation was downregulated to a greater degree on day one, compared with day three, as defined by having a respective significance value difference > one log fold. These data indicate that the downregulated genes on day one and day three of pediatric septic shock conform to a more limited number of signaling pathways relative to that of the respective upregulated genes. Notably, however, the limited number of signaling pathways associated with the downregulated genes correspond to T cell function and antigen presentation.
Figure 3.
Comparison analysis of the signaling pathways corresponding to the 1,072 genes downregulated on day one and the 1,432 genes downregulated on day three of pediatric septic shock. The analysis is derived from the Ingenuity Pathways Analysis application (see text for details). The y-axis represents the -log (significance). For illustrative purposes, the x-axis crosses the y-axis at the 1.3 threshold of significance (i.e. -log (P value = 0.05) = 1.3). Significance is directly proportional to the number of genes within the data set that corresponds to the respective canonical signaling pathway, and inversely proportional to the number of genes in the entire data set uploaded to the Ingenuity Pathways Analysis application.
Gene Network Analysis Focused on Upregulated Genes
To gain further insight regarding the biological meaning of genes upregulated on day one and day three, we focused the subsequent analysis on the presence of gene networks (see Methods section) (7). The datasets containing the 862 upregulated day one genes and the 1,072 upregulated day three genes were individually uploaded to the IPA application. The network analysis focused on upregulated genes yielded eight highly significant gene networks associated with either the day one or day three datasets (Tables 3A and 3B). In each case, the maximum 35 focus genes that comprised each of the networks were present in the respective upregulated datasets such that all eight upregulated networks had significance scores ≥ 47. Three unique gene networks were identified within the day one upregulated dataset (Table 3A) and five unique gene networks were identified within the day three upregulated dataset (Table 3B).
Table 3A.
Gene Networks Upregulated Uniquely on Day One
| Network and Central Node1 | Ingenuity Annotations2 | Panther Classifications3 Pathway (P value) Biological Process (P value) Molecular Function (P value) | D.A.V.I.D. Functional Annotation4 Category Term P value |
|---|---|---|---|
| Network 1 MAPK14 | Inflammatory disease Infectious disease Cell-cell signaling | Oxidative stress response (3.3E-8) Immunity and defense (1.3E-12) Protein kinase (5.3E-5) | GOTERM_BP_ALL Response to pest, pathogen or parasite 1.9E-14 |
| Network 2 STAT3 | Immune response Cellular movement Hematologic system development | Inflammation mediated by chemokine and cytokine signaling (2.2E-7) Immunity and defense (2.6E-13) Cytokine receptor (1.8E-9) | SP_PIR_KEYWORDS Signal 6.6E-10 |
| Network 3 LYN | Cell-cell signaling Tissue morphology Cellular movement | PDGF signaling pathway (1.9E-7) Immunity and defense (1.4E-13) Signaling molecule (8.0E-6) | GOTERM_BP_ALL Response to wounding 1.1E-7 |
Table 3B.
Gene Networks Upregulated Uniquely on Day Three
| Network and Central Node(s)1 | Ingenuity Annotations2 | Panther Classifications3 Pathway (P value) Biological Process (P value) Molecular Function (P value) | D.A.V.I.D. Functional Annotation4 Category Term P value |
|---|---|---|---|
| Network 1 FOS and IL1A | Immune response Connective tissue disorders Inflammatory disease Immunity and defense | Inflammation mediated by chemokine and cytokine signaling (6.0E-5) Cytokine receptor (1.5E-09) | GOTERM_BP_ALL Defense response 7.7E-11 |
| Network 2 MAPK1 | Cellular movement Hematologic system Development Immune response | Toll receptor signaling pathway (4.0E-10) Proteolysis (2.9E-12) Serine protease (3.6E-9) | SP_PIR_KEYWORDS Signal 1.3E-9 |
| Network 3 SP1 | Inflammatory disease Hematologic system development Immune response | Plasminogen activating cascade (3.3E-4) Immunity and defense (3.2E-8) Complement component (3.8E-5) | SP_PIR_KEYWORDS Direct protein sequencing 2.7E-9 |
| Network 4 STAT3 and STAT5 | Cellular movement Cell morphology Cellular function and maintenance | PDGF signaling pathway (1.4E-10) Signal transduction (1.2E-16) Non-receptor tyrosine protein kinase (2.6E-09) | SP_PIR_KEYWORDS Phosphorylation 2.2E-14 |
| Network 5 NFKB1A | Cell death Cancer Hematologic disease | Insulin/IGF pathway-protein kinase B signaling pathway (1.7E-7) Apoptosis (2.5E-8) Membrane traffic protein (1.3E-3) | GOTERM_MF_ALL Protein binding 1.4E-9 |
The functional annotations assigned to each upregulated network by the IPA application are provided in Tables 3A and 3B. Because these functional annotations are relatively generic, we attempted to derive more detailed biological meaning from these eight upregulated networks by uploading the individual network gene lists to the D.A.V.I.D. database (19) and the PANTHER classification system (20–22). As shown in Tables 3A and 3B, the D.A.V.I.D.-based analysis of these networks also yielded relatively generic functional annotations. In contrast, the PANTHER-based analysis yielded more specific signaling pathways, molecular functions, and biological processes within each of the upregulated networks.
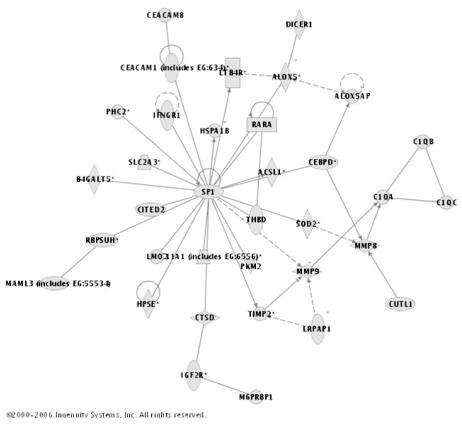
A representative upregulated gene network identified within the day three dataset is provided in Figure 4. This gene network is centered on Sp1 transcription factor (SP1) and the genes within the network correspond to the plasminogen-activating cascade, immunity and defense, and the complement system (PANTHER-based classifications, Table 3B). Figures of the other upregulated networks listed in Tables 3A and 3B, a network legend, and the corresponding gene lists are provided in the supplementary data. Collectively, these data demonstrate that a relatively large proportion of the genes upregulated on day one and day three of pediatric septic shock are functionally connected in the form of biologically relevant gene networks. In addition, the up-regulated networks have a temporal expression pattern in that there are unique networks corresponding to the day one and day three data sets, respectively.
Figure 4.
Representative gene network within the 1,072 genes upregulated on day three of pediatric septic shock. The network is approximately centered on a gene node corresponding to Sp1 transcription factor (SP1). The genes within the network correspond functionally to the plasminogen activating cascade, immunity and defense, and complement component (based on the Panther Classification System). See text for network derivation. A network legend and the entire gene list contributing to the network are provided in the supplementary data.
Gene Network Analysis Focused on Downregulated Genes
In this analysis we focused on gene networks associated with downregulated genes. The analysis was conducted in an identical manner to that described above, except that we individually uploaded the datasets corresponding to the 1,283 downregulated day one genes and the 1,432 downregulated day three genes to the IPA application. The network analysis focused on downregulated genes yielded six gene networks associated with either the day one or day three datasets (Tables 4A and 4B). All six downregulated networks had significance scores ≥ 44 and within each network the maximum 35 focus genes were present in the respective datasets that were used to generate the networks. Three unique gene networks were identified within the day one downregulated dataset (Table 4A) and three unique gene networks were identified within the day three downregulated dataset (Table 4B).
Table 4A.
Gene Networks Downregulated Uniquely on Day One
| Network and Central Node1 | Ingenuity Annotations2 | Panther Classifications3 Pathway (P value) Biological Process (P value) Molecular Function (P value) | D.A.V.I.D. Functional Annotation4 Category Term P value |
|---|---|---|---|
| Network 1 BCL25 | Hematologic system development Immune system development Lymphatic system development | T cell activation (2.2E-12) T cell mediated immunity (7.4E-14) Immunoglobulin receptor family member (1.9E-10) | SP_PIR_KEYWORDS T cell 1.4E-16 |
| Network 2 HLA6 | RNA posttranscriptional modification Cell-cell signaling Immune response | T cell activation (4.0E-7) MHCII-mediated immunity (6.9E-19) MHC antigen (2.0E-15) | KEGG_PATHWAY Antigen processing and presentation 2.2E-12 |
| Network 3 TCF3 | Hematologic system development Immune system development Lymphatic system development | Wnt signaling pathway (1.2E-2) mRNA transcription regulation (3.2E-8) Other transcription factor (1.1E-5) | GOTERM_BP_ALL Regulation of cellular process 3.9E-9 |
Table 4B.
Gene networks Downregulated Uniquely on Day Three
| Network and Central Node(s)1 | Ingenuity Annotations2 | Panther Classifications3 Pathway (P value) Biological Process (P value) Molecular Function (P value) | D.A.V.I.D. Functional Annotation4 Category Term P value |
|---|---|---|---|
| Network 1 BCL25 | Hematologic system development Immune system development Lymphatic system development | T cell activation (1.7E-10) T cell mediated immunity (5.3E-14) Immunoglobulin receptor family member (8.3E-9) | SP_PIR_KEYWORDS T cell 1.8E-16 |
| Network 2 IL8 and GATA3 | Hematologic system development Immune system development Lymphatic system development | B cell activation (6.3E-6) Granulocyte-mediated immunity (9.8E-5) Guanyl-nucleotide exchange factor (9.2E-4) | GOTERM_BP_ALL Defense response 1.7E-7 |
| Network 3 HLA6 | RNA posttranscriptional modification Cell-cell signaling Immune response | T cell activation (4.5E-4) MHCII-mediated immunity (4.0E-14) MHC antigen (4.5E-11) | KEGG_PATHWAY Antigen processing and presentation 1.4E-13 |
The functional annotations assigned to each downregulated network by the IPA application are provided in Tables 4A and 4B. Similar to that of the upregulated gene networks, these functional annotations are relatively generic. Accordingly, we again uploaded the individual network gene lists to both the D.A.V.I.D. database and the PANTHER classification system. As shown in Tables 4A and 4B, both the D.A.V.I.D.- and PANTHER- based analyses yielded functional annotations related to T cell- and antigen presentation (MHC)-related biological processes. In addition, the day three-specific network approximately centered on IL8 (CXCL8) and GATA was notable for a B cell-related functional annotation (Table 4B).
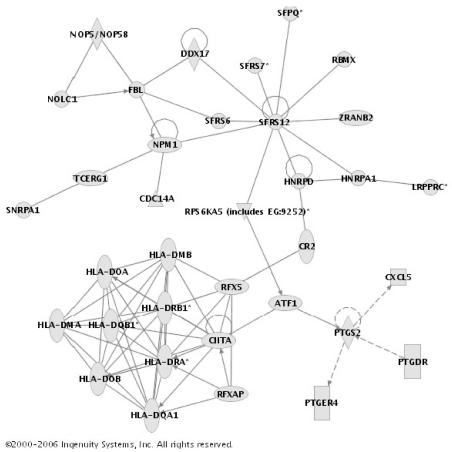
A representative gene network identified within the day one dataset is provided in Figure 5. This gene network contains eight individual gene nodes corresponding to human leukocyte antigens (HLA), and the genes within the network correspond functionally to T cell activation, MHC II-mediated immunity, and MHC antigens. Figures of the other networks listed in Table 4A and 4B, and the corresponding gene lists are provided in the supplementary data. Collectively, these data demonstrate that a relatively large proportion of the genes downregulated on day one and day three of pediatric septic shock are functionally connected by their uniform identification within networks of biologically related genes. In addition, the longitudinal expression patterns of these networks indicate large scale and persistent downregulation of genes directly involved in lymphocyte- and MHC-related biological processes.
Figure 5.
Representative gene network within the 1,283 genes downregulated on day one of pediatric septic shock. The gene network contains eight individual gene nodes corresponding to the major histocompatibility complex. The genes within the network correspond functionally to T cell activation, MHCII-mediated immunity, and MHC antigen (based on the Panther Classification System). See text for network derivation. A network legend and the entire gene list contributing to the network are provided in the supplementary data.
Absolute Lymphocyte Counts and Differential Gene Expression
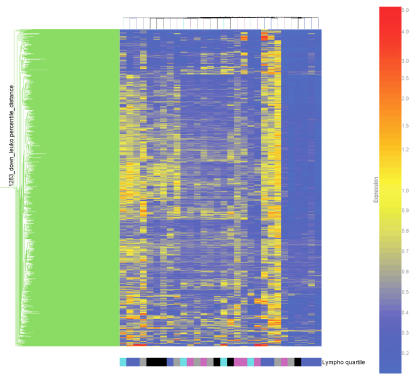
As shown in Table 5, the study patients had mean or median absolute lymphocyte counts at the lower range of normal on both day one and day three (23). This observation raises the possibility that the data shown in Tables 4A and 4B, indicating large scale downregulation of genes involved in T cell function, are an artifact of relative lymphopenia, perhaps related to apoptosis, rather than a reflection of a genuine change in gene expression. To test this possibility, we grouped the patients with septic shock based on absolute lymphocyte count quartiles, and conducted two-dimensional cluster analyses focused on genes that were downregulated on either day one or day three. As shown in Figure 6, patients within each of the four quartiles clustered in a heterogeneous manner (Distance similarity measurement) throughout the map representing the day one downregulated genes. This heterogeneous pattern persisted when we applied either a Pearson Correlation or a Standard Correlation as the similarity measurement for the condition tree (data not shown). Similar observations were made with regard to the genes downregulated on day three (data not shown). Collectively, these data indicate that our observations regarding downregulation of genes relevant to T cell function are not likely to be artifacts of relative lymphopenia as there was no correlation between absolute lymphocyte counts and gene downregulation.
Table 5.
Absolute Leukocyte Numbers (× 103 per mm3) for Patients with Septic Shock.
| Mean ± SEM | Median | 25th percentile | 75th percentile | |
|---|---|---|---|---|
| Neutrophils | ||||
| Day1 | 10.2 ± 2.0 | 6.7 | 2.6 | 15.3 |
| Day 3 | 10.5 ± 1.1 | 8.6 | 7.0 | 13.8 |
| Lymphocytes1 | ||||
| Day 1 | 2.7 ± 0.6 | 1.7 | 1.1 | 3.0 |
| Day 3 | 2.4 ± 0.5 | 1.8 | 1.0 | 3.0 |
| Monocytes | ||||
| Day 1 | 0.9 ± 0.2 | 0.4 | 0.2 | 1.2 |
| Day 3 | 0.7 ± 0.1 | 0.6 | 0.3 | 1.0 |
Figure 6.
Two dimensional gene cluster map focused on the 1,283 downregulated genes on day one of septic shock (see text for filtering strategy). Individual patients are oriented horizontally and individual genes are oriented vertically. The color-coded bar at the bottom of the map represents the patients with septic shock grouped by quartiles of absolute lymphocyte counts (derived from Table 5) as follows: (turquoise) 1st quartile, (blue) 2nd quartile, (pink) 3rd quartile, (grey) 4th quartile; and (black) absolute lymphocyte count data not available.
Persistence of Zinc-Related Functional Annotations
We previously reported that day one of pediatric septic shock is characterized by large scale downregulation of genes having functional annotations related to zinc homeostasis (6). Here we conducted an analysis to determine if gene ontologies related to zinc homeostasis are persistently downregulated during the initial three days of pediatric septic shock. The analysis was conducted by uploading the individual lists of genes down-regulated on day one and day three to the D.A.V.I.D. database. As shown in Table 6, day one was characterized by large scale downregulation of genes corresponding to zinc and metal functional annotations as previously reported (6), and this pattern of downregulation persisted into day three. In fact, downregulation of gene ontologies related to zinc and metal homeostasis appears to be an even more robust feature on day three of pediatric septic shock as demonstrated by the respective day one and day three P values.
Table 6.
Functional Annotations related to zinc, downregulated on both day one and day three. The analysis is based on the default parameters in D.A.V.I.D.
| Category | Term | number of genes day one (day three) | P value day one (day three) |
|---|---|---|---|
| SP_PIR_KEYWORDS | zinc-finger | 118 (143) | 2.9E-16 (3.0E-22) |
| SP_PIR_KEYWORDS | zinc | 135 (159) | 3.0E-16 (1.4E-20) |
| SP_PIR_KEYWORDS | metal-binding | 147 (176) | 1.1E-12 (2.4E-17) |
| GOTERM_MF_ALL | zinc ion binding | 149 (172) | 5.9E-11 (1.1E-12) |
All supplementary materials are available online at molmed.org
DISCUSSION
These data represent the most comprehensive analysis of longitudinal gene expression in pediatric septic shock to date. As relatively descriptive data, they are subject to interpretation. Below we provide our pathophysiology-based interpretation of the expression data, being cognizant of alternative interpretations.
The genome-level expression profile during the initial three days of pediatric septic shock is complex and heterogeneous. Between four and five percent of the genes encompassing the entire array were differentially expressed (upregulated or downregulated) on either day one or day three, respectively, compared with controls. Among the differentially expressed genes, 11 percent of the day one differentially expressed genes were unique to day one, and 24 percent of the day three differentially expressed genes were unique to day three. Thus, the genome-level expression profile during the initial three days of pediatric septic shock is characterized by a predominant constant component and a relatively smaller temporal component. The genes contributing to the smaller temporal component, however, appear to be biologically significant in that they contribute to differential expression of signaling pathways and gene networks known to be associated with inflammatory disease states.
The day one and day three datasets contain multiple genes contributing to signaling pathways. Many of the pathways identified within these data sets have been linked previously by the experimental literature to septic shock pathobiology (3,24,25). While the demonstration of some of these pathways does not necessarily represent novel concepts, their presence in the datasets greatly strengthens both the validity and relevance of the overall dataset. For example, we were intrigued to find that day three (but not day one) of pediatric septic shock is characterized by upregulation of genes that participate in insulin-like growth factor (IGF)-, insulin receptor-, and PPAR-signaling. These data indicate an important role for insulin-related biology/metabolism in pediatric septic shock, and are made all the more intriguing by the recent advancements, by Van den Berghe and colleagues, regarding the use of intensive insulin therapy in critically ill adult surgical patients (26).
The primary novelty of the signaling pathway-related data lies in the temporal dependence of pathway expression. Several of the upregulated signaling pathways were consistently upregulated from day one to day three. More than half of the upregulated pathways, however, were expressed in a time-dependent manner. In fact, some of the signaling pathways were exclusively expressed on day three. These data could have implications for the timing and duration of future therapeutic strategies centered on any one of these pathways.
The importance of timing and duration of therapy in critically ill patients is well illustrated in the recent follow-up study of intensive insulin therapy in critically ill adult medical patients by Van den Berghe and colleagues (27). This study involving 1,200 critically ill adults demonstrated that intensive insulin therapy conferred a survival benefit only for patients having an intensive care unit length of stay ≥ three days. In contrast, mortality was greater in patients receiving intensive insulin therapy and having an intensive care unit length of stay < three days. These data indirectly support our observations regarding time-dependent expression of signaling pathways related to insulin biology/metabolism.
Using a pathway-based analytical tool (IPA), the day one and day three datasets were found to contain multiple genes corresponding to distinct networks. The biological importance of these networks is broadly supported at two levels. First, all of the networks selected for presentation had exceptionally high significance scores based on the IPA algorithm. Second, many of the functional annotations corresponding to the networks are biologically consistent with the signaling pathways shown in Figures 3 and 4. Innate immunity and inflammation are the predominant themes of the upregulated gene networks, and these observations are again consistent with some of the current paradigms in the septic shock literature (3,24,25). The network expression patterns were time-dependent, as demonstrated by unique gene networks being expressed on day one and day three, respectively. Thus, the temporal nature of network expression and the implied functionality may also have implications for the timing and duration of future therapeutic strategies.
The most novel feature derived from the network-based analysis is the down-regulation of gene networks related to T cell immunity and the MHC antigen presentation pathway. Importantly, this observation does not appear to be an artifact of relative lymphopenia when analyzed in the context of total lymphocyte counts. The strength of this observation is indicated by the multiple gene networks having these particular annotations (T cell function and the MHC antigen presentation pathway), and the ability to consistently derive these functional annotations using two distinct gene annotation/ontology databases (i.e. D.A.V.I.D., and PANTHER) that are independent from the IPA database. In addition, the analyses focused on discovery of signaling pathways also yielded “T cell receptor signaling” and “antigen presentation” as being predominant themes among the group of genes down-regulated on day one and day three. Collectively, these data indirectly suggest that pediatric septic shock may have a strong component of immune dysfunction secondary to decreased T cell function and antigen presentation.
Our conceptual framework of septic shock pathobiology has evolved over the last decade to include the concepts of immune suppression and “immunoparalysis.” Whereas septic shock has been viewed traditionally as being a reflection of hyperinflammation, it is now thought that septic shock also has a strong, perhaps predominant, anti-inflammatory component that can be manifest as functional immune suppression and the relative inability to effectively clear infection (24,28,29). Our current observation that genes corresponding to interleukin-10 signaling, a major anti-inflammatory pathway (30), are temporally upregulated in this patient cohort, well supports this anti-inflammatory concept.
Monocyte deactivation related to decreased MHC gene mRNA expression and decreased surface expression of MHC molecules has been demonstrated in patients with septic shock (31–34). Regarding lymphocyte dysfunction, Heidecke and colleagues demonstrated that adult patients with intraabdominal infections and septic shock had defective T cell proliferation and T cell-dependent cytokine secretion, consistent with an-ergy/immune suppression (35). Felmet and colleagues identified prolonged lymphopenia and apoptosis-associated depletion of lymphoid organs as independent risk factors for the development of nosocomial infections and multiple organ failure in critically ill children (36). Animal studies have documented the requirement of an intact T cell system to adequately combat a septic challenge (29,37). More recently, animal-based experiments have demonstrated that experimental septic shock is characterized by widespread T cell apoptosis and that preventing T cell apoptosis positively impacts the outcome of experimental sepsis (38–45). Importantly, the concept of T cell apoptosis in human sepsis has been indirectly corroborated by autopsy studies (36,46). Thus, while formal studies of T cell function in pediatric septic shock remain to be conducted, our current data support the concept of immune suppression secondary to T cell dysfunction at the transcriptome level and represent a first step toward demonstrating proof of principle on a clinical genomics scale.
Our previous report indicated that day one of pediatric septic shock is characterized by a large scale downregulation of genes that play a direct role in zinc homeostasis or are functionally dependent on zinc homeostasis (6). We have corroborated this observation and now demonstrate its persistence well into day three of pediatric septic shock. In fact, the current data suggest that the perturbation of genes related to zinc homeostasis is even more profound on day three than on day one. The functional significance of these observations remains to be directly tested and elucidated. However, given the importance of zinc homeostasis for T cell function (47–53), coupled with our observations suggesting large scale downregulation of T cell-related genes, the overall data provide the basis for biologically plausible and testable hypotheses surrounding zinc homeostasis and lymphocyte dysfunction in pediatric septic shock.
Many of the limitations inherent to the data in our previous report are applicable to the current data and have been discussed previously (6), including the possibility that our observations are confounded by differences in the white blood cell populations. Similar to the previous report, we have not found any correlation between our current observations and differential white blood cell counts (at the level of neutrophils, monocytes, and total lymphocytes). We are cognizant, however, that differences in T cell subset numbers (i.e. CD4 cells, CD8 cells, etc.) could account for some of our observations regarding T cell receptor signaling and antigen presentation. Thus, we again maintain that our observations are reasonably representative of genuine genomic responses, rather than sampling artifact related to whole blood-derived RNA.
Two interrelated limitations, more specific to the current data, warrant further discussion. First, the data are descriptive and limited to two time points. The data are, however, unprecedented in scope by addressing the problem of clinical pediatric septic shock using a translational approach at the level of the entire genome. Second, the data require functional validation in the form of directly testing the hypotheses derived from the current observations. We are currently in the process of designing a validation, follow-up study in which we will formally measure lymphocyte function in a large cohort of children with septic shock.
These limitations aside, the current data represent an unprecedented first approximation of longitudinal, genome-level responses of pediatric septic shock. The data demonstrate: 1) the complex nature of this genomic response; 2) a predominance of constant, coordinated gene regulation over the first three days of illness; 3) a relatively small but biologically relevant temporal component of coordinated gene regulation; 4) large scale downregulation of genes corresponding to T cell function and MHC-mediated antigen presentation; and 5) persistent downregulation of a large subset of genes related to zinc homeostasis. Collectively, the data support novel hypotheses that can be tested readily at both the experimental and translational levels.
Supplemental Data
ACKNOWLEDGMENTS
Additional Genomics of Pediatric SIRS/Septic Shock Investigators: Robert J. Freishtat, M.D., M.P.H., (Children’s National Medical Center, Washington, D.C.); Mary Ann Tagavilla, M.D. (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH); Julie Simon, R.N. (Children’s Hospital and Research Center Oakland, Oakland, CA); Carey Roth Bayer, Ed.D., R.N. (The Children’s Hospital of Philadelphia, Philadelphia, PA); Joseph Hess, R.N. (Penn State Children’s Hospital, Hershey, PA); Margaret Winkler, M.D. (The University of Alabama at Birmingham, Birmingham, AL); Robert Fitzgerald, M.D. (Devos Children’s Hospital, Grand Rapids, MI); Gwenn McLaughlin, M.D. (Jackson Memorial Hospital, Miami, FL); Cheri Landers, M.D. (Kentucky Children’s Hospital, Lexington, KY); Gary Kohn, M.D. (Morristown Memorial Hospital, Morristown, NJ); Paul Checchia, M.D. (St. Louis Children’s Hospital, St. Louis, MO); Jose Gutierrez, M.D. (Pediatric Critical Care of Arizona, Phoenix, AZ); Nick Anas, M.D. (Children’s Hospital of Orange County, Orange, CA) and Steve Shane, M.D. (Washoe Medical Center, Reno, NV). Supported by a grant from the National Institute of General Medical Sciences (RO1 GM064619), and The Amanda Kanowitz Foundation (http://www.amandakfoundation.org).
Footnotes
REFERENCES
- 1.Watson RS, Carcillo JA, Linde-Zwirble WT, Clermont G, Lidicker J, Angus DC. The epidemiology of severe sepsis in children in the United States. Am J Respir Crit Care Med. 2003;167:695–701. doi: 10.1164/rccm.200207-682OC. [DOI] [PubMed] [Google Scholar]
- 2.Watson RS, Carcillo JA. Scope and epidemiology of pediatric sepsis. Pediatr Crit Care Med. 2005;6:S3–5. doi: 10.1097/01.PCC.0000161289.22464.C3. [DOI] [PubMed] [Google Scholar]
- 3.Shanley TP, Hallstrom C, Wong HR. Sepsis. In: Fuhrman BP, Zimmerman JJ, editors. Pediatric Critical Care Medicine. Mosby; St. Louis: 2006. pp. 1474–93. [Google Scholar]
- 4.Carcillo JA, Fields AI. Clinical practice parameters for hemodynamic support of pediatric and neonatal patients in septic shock. Crit Care Med. 2002;30:1365–78. doi: 10.1097/00003246-200206000-00040. [DOI] [PubMed] [Google Scholar]
- 5.Shanley TP, Wong HR. Molecular genetics in the pediatric intensive care unit. Crit Care Clin. 2003;19:577–94. doi: 10.1016/s0749-0704(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 6.Wong HR, et al. Genome level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–55. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 8.Chinnaiyan AM, et al. Molecular signatures of sepsis: multiorgan gene expression profiles of systemic inflammation. Am J Pathol. 2001;159:1199–209. doi: 10.1016/S0002-9440(10)62505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb JP, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102:4801–6. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prucha M, et al. Expression profiling: toward an application in sepsis diagnostics. Shock. 2004;22:29–33. doi: 10.1097/01.shk.0000129199.30965.02. [DOI] [PubMed] [Google Scholar]
- 11.Pachot A, Lepape A, Vey S, Bienvenu J, Mougin B, Monneret G. Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol Lett. 2006;106:63–71. doi: 10.1016/j.imlet.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Ramilo O, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–77. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein B, Giroir B, Randolph A. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6:2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 14.Pollack MM, Patel KM, Ruttimann UE. The Pediatric Risk of Mortality III—Acute Physiology Score (PRISM III-APS): a method of assessing physiologic instability for pediatric intensive care unit patients. J Pediatr. 1997;131:575–81. doi: 10.1016/s0022-3476(97)70065-9. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson JD, Pollack MM, Ruttimann UE, Glass NL, Yeh TS. Outcome of pediatric patients with multiple organ system failure. Crit Care Med. 1986;14:271–4. doi: 10.1097/00003246-198604000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Proulx F, Fayon M, Farrell CA, Lacroix J, Gauthier M. Epidemiology of sepsis and multiple organ dysfunction syndrome in children. Chest. 1996;109:1033–7. doi: 10.1378/chest.109.4.1033. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann N, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–74. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 20.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–52. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas PD, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malatack JJ, Blatt J, Penchansky L. Hematology and oncology. In: Zittelli BJ, Davis HW, editors. Atlas of Pediatric Physical Diagnosis. Mosby; St. Louis: 1997. pp. 305–41. [Google Scholar]
- 24.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–50. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 25.Abraham E. Why immunomodulatory therapies have not worked in sepsis. Intensive Care Med. 1999;25:556–66. doi: 10.1007/s001340050903. [DOI] [PubMed] [Google Scholar]
- 26.van den Berghe G, et al. Intensive insulin therapy in the critically ill patients. N Engl J Med. 2001;345:1359–67. doi: 10.1056/NEJMoa011300. [DOI] [PubMed] [Google Scholar]
- 27.Van den Berghe G, et al. Intensive insulin therapy in the medical ICU. N Engl J Med. 2006;354:449–461. doi: 10.1056/NEJMoa052521. [DOI] [PubMed] [Google Scholar]
- 28.Munford RS, Pugin J. Normal responses to injury prevent systemic inflammation and can be immunosuppressive. Am J Respir Crit Care Med. 2001;163:316–21. doi: 10.1164/ajrccm.163.2.2007102. [DOI] [PubMed] [Google Scholar]
- 29.Mahidhara R, Billiar TR. Apoptosis in sepsis. Crit Care Med. 2000;28:N105–13. doi: 10.1097/00003246-200004001-00013. [DOI] [PubMed] [Google Scholar]
- 30.Shanley TP, Schmal H, Friedl HP, Jones ML, Ward PA. Regulatory effects of intrinsic IL-10 in IgG immune complex-induced lung injury. J Immunol. 1995;154:3454–60. [PubMed] [Google Scholar]
- 31.Volk HD, et al. Monocyte deactivation-rationale for a new therapeutic strategy in sepsis. Intensive Care Med. 1996;22(Suppl 4):S474–81. doi: 10.1007/BF01743727. [DOI] [PubMed] [Google Scholar]
- 32.Docke WD, et al. Monocyte deactivation in septic patients: restoration by IFN-gamma treatment. Nat Med. 1997;3:678–81. doi: 10.1038/nm0697-678. [DOI] [PubMed] [Google Scholar]
- 33.Monneret G, et al. The anti-inflammatory response dominates after septic shock: association of low monocyte HLA-DR expression and high interleukin-10 concentration. Immunol Lett. 2004;95:193–8. doi: 10.1016/j.imlet.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Le Tulzo Y, et al. Monocyte human leukocyte antigen-DR transcriptional downregulation by cortisol during septic shock. Am J Respir Crit Care Med. 2004;169:1144–51. doi: 10.1164/rccm.200309-1329OC. [DOI] [PubMed] [Google Scholar]
- 35.Heidecke CD, et al. Selective defects of T lymphocyte function in patients with lethal intraabdominal infection. Am J Surg. 1999;178:288–92. doi: 10.1016/s0002-9610(99)00183-x. [DOI] [PubMed] [Google Scholar]
- 36.Felmet KA, Hall MW, Clark RS, Jaffe R, Carcillo JA. Prolonged lymphopenia, lymphoid depletion, and hypoprolactinemia in children with nosocomial sepsis and multiple organ failure. J Immunol. 2005;174:3765–72. doi: 10.4049/jimmunol.174.6.3765. [DOI] [PubMed] [Google Scholar]
- 37.Hotchkiss RS, et al. Overexpression of Bcl-2 in transgenic mice decreases apoptosis and improves survival in sepsis. J Immunol. 1999;162:4148–56. [PubMed] [Google Scholar]
- 38.Wesche DE, Lomas-Neira JL, Perl M, Chung CS, Ayala A. Leukocyte apoptosis and its significance in sepsis and shock. J Leukoc Biol. 2005;78:325–37. doi: 10.1189/jlb.0105017. [DOI] [PubMed] [Google Scholar]
- 39.Chung CS, Song GY, Lomas J, Simms HH, Chaudry IH, Ayala A. Inhibition of Fas/Fas ligand signaling improves septic survival: differential effects on macrophage apoptotic and functional capacity. J Leukoc Biol. 2003;74:344–51. doi: 10.1189/jlb.0102006. [DOI] [PubMed] [Google Scholar]
- 40.Chang KC, et al. Multiple triggers of cell death in sepsis: death receptor and mitochondrial-mediated apoptosis. Faseb J. 2007;21:708–19. doi: 10.1096/fj.06-6805com. [DOI] [PubMed] [Google Scholar]
- 41.Unsinger J, Herndon JM, Davis CG, Muenzer JT, Hotchkiss RS, Ferguson TA. The role of TCR engagement and activation-induced cell death in sepsis-induced T cell apoptosis. J Immunol. 2006;177:7968–73. doi: 10.4049/jimmunol.177.11.7968. [DOI] [PubMed] [Google Scholar]
- 42.Schwulst SJ, et al. Agonistic monoclonal antibody against CD40 receptor decreases lymphocyte apoptosis and improves survival in sepsis. J Immunol. 2006;177:557–65. doi: 10.4049/jimmunol.177.1.557. [DOI] [PubMed] [Google Scholar]
- 43.Hotchkiss RS, et al. TAT-BH4 and TAT-Bcl-xL peptides protect against sepsis-induced lymphocyte apoptosis in vivo. J Immunol. 2006;176:5471–7. doi: 10.4049/jimmunol.176.9.5471. [DOI] [PubMed] [Google Scholar]
- 44.Hotchkiss RS, Osmon SB, Chang KC, Wagner TH, Coopersmith CM, Karl IE. Accelerated lymphocyte death in sepsis occurs by both the death receptor and mitochondrial pathways. J Immunol. 2005;174:5110–8. doi: 10.4049/jimmunol.174.8.5110. [DOI] [PubMed] [Google Scholar]
- 45.Scumpia PO, et al. Increased natural CD4+CD25+ regulatory T cells and their suppressor activity do not contribute to mortality in murine polymicrobial sepsis. J Immunol. 2006;177:7943–9. doi: 10.4049/jimmunol.177.11.7943. [DOI] [PubMed] [Google Scholar]
- 46.Hotchkiss RS, et al. Apoptotic cell death in patients with sepsis, shock, and multiple organ dysfunction. Crit Care Med. 1999;27:1230–51. doi: 10.1097/00003246-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 47.Failla ML. Trace elements and host defense: recent advances and continuing challenges. J Nutr. 2003;133:1443S–7S. doi: 10.1093/jn/133.5.1443S. [DOI] [PubMed] [Google Scholar]
- 48.Fraker PJ, King LE. Reprogramming of the immune system during zinc deficiency. Annu Rev Nutr. 2004;24:277–98. doi: 10.1146/annurev.nutr.24.012003.132454. [DOI] [PubMed] [Google Scholar]
- 49.Ibs KH, Rink L. Zinc-altered immune function. J Nutr. 2003;133:1452S–6S. doi: 10.1093/jn/133.5.1452S. [DOI] [PubMed] [Google Scholar]
- 50.King LE, Osati-Ashtiani F, Fraker PJ. Apoptosis plays a distinct role in the loss of precursor lymphocytes during zinc deficiency in mice. J Nutr. 2002;132:974–9. doi: 10.1093/jn/132.5.974. [DOI] [PubMed] [Google Scholar]
- 51.Kitamura H, et al. Toll-like receptor-mediated regulation of zinc homeostasis influences dendritic cell function. Nat Immunol. 2006;7:971–7. doi: 10.1038/ni1373. [DOI] [PubMed] [Google Scholar]
- 52.Rink L, Haase H. Zinc homeostasis and immunity. Trends Immunol. 2007;28:1–4. doi: 10.1016/j.it.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Shankar AH, Prasad AS. Zinc and immune function: the biological basis of altered resistance to infection. Am J Clin Nutr. 1998;68:447S–63S. doi: 10.1093/ajcn/68.2.447S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.