Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination (original) (raw)
Abstract
The T early α (TEA) promoter in the murine Tcra locus generates noncoding transcripts that extend across the 65 kb Jα array. Here, we have analyzed the significance of TEA transcription for Tcra locus regulation through the targeted introduction of a transcription terminator downstream of the TEA promoter. We demonstrate that noncoding transcription driven by this single promoter can instruct both positively and negatively the activity of downstream Jα promoters, and can similarly instruct alterations in Jα chromatin structure and Jα recombination. TEA transcription activates promoters associated with relatively proximal Jα segments and stimulates histone acetylation, histone methylation and chromatin accessibility in this region. In contrast, at more distal locations, TEA transcription inhibits promoter activity through transcriptional interference and suppresses chromatin accessibility. In combination, these effects target initial Vα-to-Jα recombination to TEA-proximal Jα segments and promote the ordered usage of the Jα array. The ability of TEA transcription to coordinate the activity of multiple downstream promoters maximizes the biological potential of the Jα array and diversifies the Tcra repertoire.
Keywords: chromatin remodeling, T-cell receptor, transcriptional interference, V(D)J recombination
Introduction
Noncoding transcription is thought to serve important functions in gene regulation, and to do so through a variety of mechanisms (Mattick and Makunin, 2006; Prasanth and Spector, 2007). In some settings, the noncoding RNA itself is directly involved in gene silencing or activation (Meister and Tuschl, 2004; Feng et al, 2006; Sanchez-Elsner et al, 2006). In other instances, noncoding transcription, rather than the resultant transcript, is thought to serve a regulatory function. Noncoding transcripts traverse regulatory elements, genes and nonregulatory intergenic regions of a variety of complex developmentally regulated loci in mammals (Gribnau et al, 2000; Masternak et al, 2003; Bolland et al, 2004; Rogan et al, 2004; Ho et al, 2006). In these instances, noncoding transcription generally correlates with locus activation and is thought to be involved in the establishment of open chromatin domains. This line of thinking is based, in part, on studies demonstrating that elongating RNA polymerase II (RNA pol II) and associated proteins can disrupt chromatin structure and revise the pattern of histone modifications across transcribed coding regions (Belotserkovskaya et al, 2003; Schwabish and Struhl, 2006; Li et al, 2007). Histone modifications introduced by the process of transcription include acetylation, methylation and ubiquitylation. Histone H4 acetylation can disrupt the overall compaction of nucleosome arrays (Shogren-Knaak et al, 2006), whereas H2B ubiquitylation (Pavri et al, 2006) and H3 K36 methylation (Carrozza et al, 2005) are important for the regulated disassembly and redeposition of nucleosomes that occurs during RNA pol II passage. However, because studies of intergenic transcription have often failed to establish causal relationships, it is unclear whether the detected transcription serves a true regulatory function, and what the function might be. In fact, several examples of intergenic transcription that were thought initially to have roles in chromatin opening have more recently been linked to gene silencing via siRNA (Baguet et al, 2005; Haussecker and Proudfoot, 2005).
Noncoding transcription also has the potential to mediate the repression of genes linked in cis through a process known as transcriptional interference (Shearwin et al, 2005). In one well-studied example, repression of the Saccharomyces cerevisiae SER3 gene is mediated by regulated transcription across SER3 from the upstream SRG1 promoter. SRG1 transcription can suppress the activity of a downstream promoter by interfering with the binding of transcriptional activators to the promoter (Martens et al, 2004). In other examples of transcriptional interference, transcription from an interfering promoter prevents the transition of RNA pol II from initiation to elongation (Callen et al, 2004) or promotes the release of elongating RNA pol II from the DNA (Prescott and Proudfoot, 2002). While these examples implicate the process of transcription in interference with downstream promoters, a transcribed RNA itself suppresses dhfr gene expression by annealing to promoter DNA (Martianov et al, 2007). Recent reports have also suggested that transcriptional interference can repress Drosophila Ubx (Petruk et al, 2006) and the murine β-like globin gene βh0 (Hu et al, 2007). However, as these studies tested for transcriptional interference by promoter deletion rather than transcriptional blockade, a formal distinction between transcriptional interference and promoter competition could not be made. Well-characterized examples of transcriptional interference in higher eukaryotes are rare.
V(D)J recombination functions in developing lymphocytes to assemble the variable region exons of both immunoglobulins and T-cell receptors (TCRs) from germline variable (V), diversity (D) and joining (J) gene segments (Cobb et al, 2006). The recombinase-activating gene-1 (RAG1) and RAG2 proteins initiate this assembly process by introducing a double-strand break between the coding gene segments and flanking recombination signal sequences (RSSs). Components of the nonhomologous end-joining pathway then join coding ends to form coding joints and signal ends to form signal joints. V(D)J recombination is developmentally regulated, at least in part, via changes in the accessibility of chromosomal RSSs to the recombinase machinery (Cobb et al, 2006). Recombinase-accessible chromatin tends to contain hyperacetylated and histone 3 lysine 4 (H3 K4) methylated histones and low levels of DNA methylation. However, a detailed understanding of the chromatin requirements for recombinase access is lacking. The initiation of V(D)J recombination has long been known to coincide with noncoding (or germline) transcription at antigen receptor loci (Yancopoulos and Alt, 1985). However, until recently, the functional relevance of these transcripts had not been established (Abarrategui and Krangel, 2006).
The murine Tcra locus undergoes Vα-to-Jα recombination at the CD4+CD8+ (double positive, DP) stage of thymocyte development, leading to the production of the α chain of the αβTCR. Near the 3′ end of the Tcra locus is an array of 61 Jα segments that occupy a 65-kb region (Krangel et al, 2004). Accessibility of these Jα segments is controlled by the Tcra enhancer (Eα) at the extreme 3′ end of the locus (Sleckman et al, 1997), and two promoters that are activated by Eα (Villey et al, 1996; Hawwari et al, 2005). These promoters, T early α (TEA) at the 5′ end of the Jα array, and Jα49 15 kb downstream of TEA, produce noncoding transcripts that extend 71 and 56 kb, respectively, across the Jα and Cα gene segments. Eα is required for all Vα-to-Jα recombination events, whereas the two promoters are required for the targeting of primary Vα-to-Jα rearrangements to the most 5′ Jα segments. Subsequently, secondary Vα-to-Jα rearrangements can replace primary rearrangements through the use of progressively more 5′ Vα and 3′ Jα segments (Wang et al, 1998; Buch et al, 2002). Studies of TEA-deleted alleles have revealed that TEA controls chromatin structure across a 12-kb region at the 5′ end of the Jα array (Mavieux et al, 2003; Hawwari et al, 2005; Hawwari and Krangel, 2007). These studies also suggested that TEA is required for the activation of several additional promoters associated with the 5′ Jα segments and for the suppression of several Jα promoters located further 3′. However, it was not clear whether these effects are mediated by TEA transcription or by some other structural or functional property of the TEA promoter region. To initially assess the regulatory function of transcription at the Tcra locus, we blocked transcription within the Jα array by introducing a transcription terminator downstream of Jα56 (Abarrategui and Krangel, 2006). Transcriptional blockade suppressed recombination of the downstream Jα53 and Jα52 gene segments, and caused an accompanying decrease in H3 K4 methylation at these sites. In addition, transcriptional blockade activated downstream promoters associated with Jα47, Jα42 and Jα37, indicating that these promoters are normally suppressed through a transcriptional interference mechanism.
Here, we introduced a transcription terminator downstream of the TEA promoter to block TEA transcription across the entire length of the Jα array. We found that blockade of TEA transcription led to the silencing of additional Jα promoters immediately downstream of TEA, but to the activation of Jα promoters located further 3′. These pertubations in Tcra transcription were accompanied by dramatic changes in chromatin structure and by the retargeting of Vα-to-Jα recombination events to more 3′ Jα segments. These results demonstrate that the effects of TEA promoter deletion are due entirely to the loss of TEA transcription and that noncoding transcription driven by a single promoter can have both positive and negative influences on downstream promoter activity and chromatin structure.
Results
Blockade of TEA transcription in vivo
To investigate the role of TEA promoter-derived transcription in the regulation of Jα chromatin, we used homologous recombination to introduce a transcription terminator cassette into the Jα array immediately downstream of the TEA exon (Figure 1). Previous work had established this cassette as a highly effective transcription terminator in vivo (Abarrategui and Krangel, 2006). When introduced downstream of Jα56, it caused >99% reduction in steady-state transcript levels as measured by RT–PCR of nuclear RNA and a drop in RNA pol II density to background levels as measured by nuclear run-on.
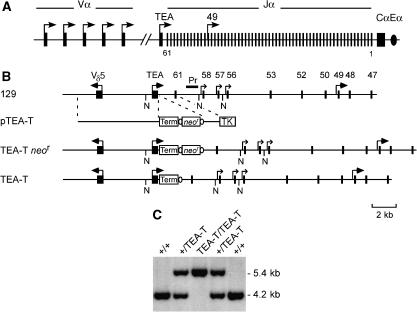
Figure 1.
Tcra locus organization and gene targeting. (A) Tcra locus, including the relative positions of V, J and C gene segments and the TEA exon (filled rectangles), promoters (arrows) and Eα (oval). (B) Strategy for the generation of the TEA-T allele. Wild-type 129/Sv allele (129), targeting construct pTEA-T, targeting intermediate allele TEA-T _neo_r and mutated allele TEA-T. Term, transcriptional terminator; TK, thymidine kinase (open rectangles); ovals, lox P sites. N, _Nco_I sites; Pr, probe. (C) Southern blot of _Nco_I-digested genomic DNA from wild-type (+/+), and targeted (+/TEA-T, TEA-T/TEA-T) mice. Molecular sizes are indicated on the right margin.
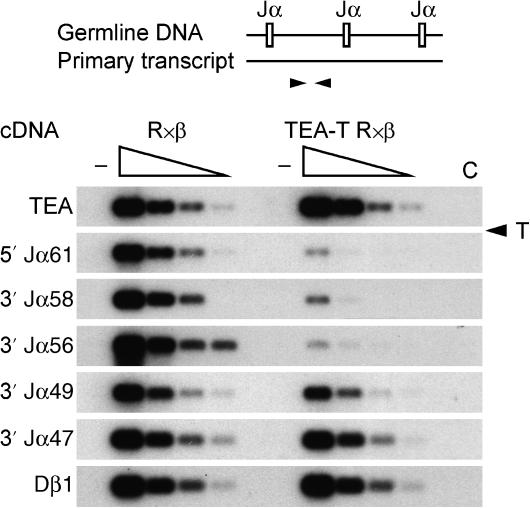
To analyze the effectiveness of the terminator in the blockade of TEA-derived transcription in vivo, mice homozygous for TEA-T alleles (‘TEA-T' mice) were bred onto a _Rag2_−/− × TCRβ transgene (R × β) background (Shinkai et al, 1993). On this background, the absence of Rag2 protein maintains TCR loci in unrearranged configuration, whereas expression of TCRβ permits thymocyte development to proceed to the CD4+CD8+ (DP) stage of development in which the Tcra locus is active. Steady-state transcript levels in thymocytes of R × β and TEA-T R × β mice were analyzed by RT–PCR of nuclear RNA (Figure 2). As expected, transcript levels upstream of the terminator (TEA) were identical on wild-type and TEA-T alleles. In contrast, transcript levels on TEA-T alleles were reduced to ∼4% of wild type immediately downstream of the terminator (5′ Jα61), and to ∼1% of wild type downstream of Jα56. Transcript levels partially recovered downstream of Jα49, and reached wild-type levels downstream of Jα47, presumably as a consequence of promoter activity associated with these Jα segments (Hawwari et al, 2005).
Figure 2.
Transcription termination in TEA-T mice. In the PCR strategy for analysis of transcription termination (top), arrowheads denote PCR primers. Fivefold serial dilutions (wedges) of cDNA from thymocyte nuclear RNA were PCR amplified and detected by Southern blot and hybridization to radiolabeled oligonucleotide probes to detect unspliced products. Dβ1 transcripts served as loading control. Control PCR reactions: −, no reverse transcriptase; C, water. T (with arrowhead), position of the transcription terminator. Results are representative of two independent experiments.
TEA transcription regulates J_α_ promoter activity
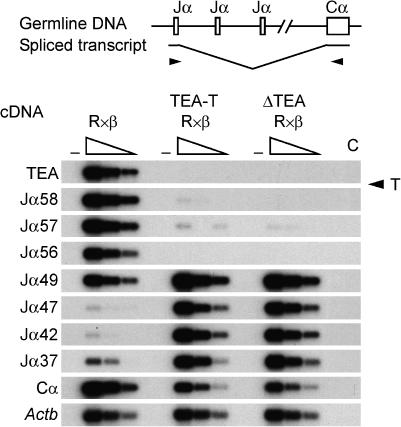
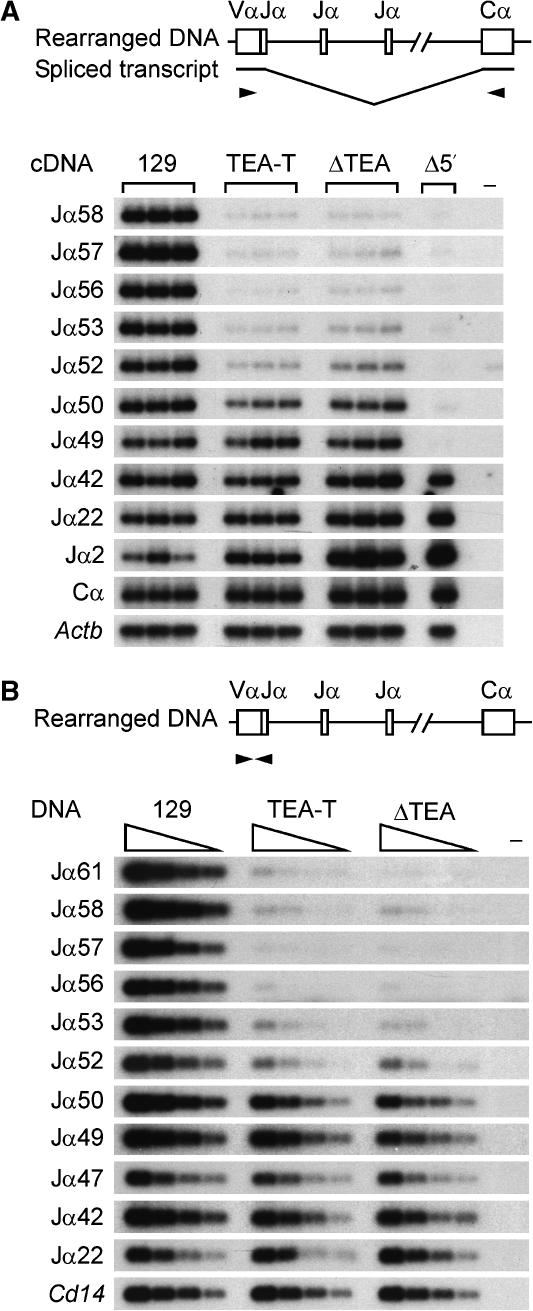
The low steady-state transcript levels across the 5′ Jα region suggested that, as we have previously described for ΔTEA alleles (Hawwari et al, 2005), the activity of promoters associated with the Jα58, Jα57 and Jα56 gene segments might be compromised on TEA-T alleles. Primary transcripts that initiate at TEA or at Jα gene segments extend across the entire Jα array to Cα, where they are terminated and polyadenylated (Villey et al, 1997). Mature transcripts are typically generated by RNA splicing between a donor sequence 3′ of either the TEA exon or the first Jα segment in the primary transcript, and a splice acceptor 5′ of the first Cα exon. Therefore, as one measure of the activation status of Jα promoters in the absence of TEA transcripts, we compared the levels of mature Jα-Cα transcripts produced by wild-type and TEA-T alleles by semiquantitative RT–PCR (Figure 3). As expected, mature TEA-Cα transcripts were eliminated on TEA-T alleles due to the transcriptional block imposed by the terminator cassette. Notably, Jα58, Jα57 and Jα56-Cα transcripts were also undetectable, suggesting that the blockade of TEA transcripts prevents the normal activation of these promoters.
Figure 3.
Jα region germline transcription in TEA-T mice. PCR and Southern blot analysis of threefold serial dilutions of thymocyte cDNA to detect spliced germline Jα transcripts. TEA-Cα, Jα-Cα and total Cα products were detected by Southern blot with a radiolabeled Cα probe. Amplification of Actb controlled for cDNA loading. Control PCR reactions: −, no reverse transcriptase; C, water. Results are representative of two independent experiments.
We previously showed that either TEA promoter deletion or the insertion of a transcription terminator downstream of Jα56 causes derepression of promoters located in the central portion of the Jα array (Jα47, Jα45, Jα42 and Jα37) (Hawwari et al, 2005; Abarrategui and Krangel, 2006; Hawwari and Krangel, 2007). Consistent with this, Jα49-Cα transcripts were increased threefold, and Jα47, Jα42 and Jα37-Cα transcripts were increased tenfold on TEA-T alleles as compared to wild-type alleles (Figure 3). These results corroborate our previous data indicating that central Jα promoters are normally suppressed through a transcriptional interference mechanism (Abarrategui and Krangel, 2006).
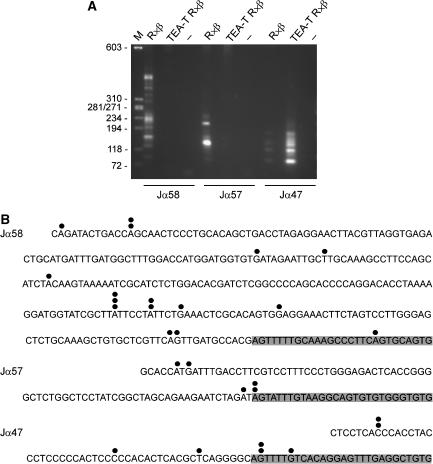
The above analysis is ambiguous with respect to the true initiation sites of the transcripts detected by RT–PCR. As transcripts initiated at TEA span the entire Jα array on wild-type alleles, the identified Jα58, Jα57 and Jα56-Cα transcripts might reflect alternatively spliced forms of transcripts that initiate at the TEA promoter. Similarly, as transcripts that initiate at the upregulated Jα49 promoter on TEA-T alleles span the central Jα segments, the detected Jα47, Jα42 and Jα37-Cα transcripts might reflect initiation at Jα49. To eliminate this ambiguity, we mapped Jα58, Jα57 and Jα47 transcription start sites by 5′ RACE using total RNA from thymocytes isolated from R × β and TEA-T R × β mice (Figure 4A). This approach allows the specific amplification of mature, capped, full-length transcripts within the total mRNA pool. PCR amplification of 5′ RACE products of R × β thymocytes identified a heterogeneous array of initiation sites extending from ∼100 to ∼500 bp upstream of Jα58 and also identified two main initiation sites upstream of Jα57. These transcripts were not detected in TEA-T R × β thymocytes. On the contrary, transcripts initiating upstream of Jα47 were detected at low abundance in R × β thymocytes, but were highly upregulated in TEA-T R × β thymocytes. Sequencing of cloned 5′ RACE products confirmed that the detected transcripts initiated at multiple sites upstream of Jα segments, and for all three gene segments identified a fraction of transcripts that initiated within the RSS (Figure 4B). We also independently verified transcription start sites by primer extension of thymocyte RNA from R × β and TEA-T R × β mice (Supplementary Figure 1). Taken together, these results demonstrate that transcripts initiating at TEA simultaneously function to activate a set of relatively proximal downstream Jα promoters and to repress a set of more distal Jα promoters.
Figure 4.
Jα58, Jα57 and Jα47 promoter activity in TEA-T mice. (A) Agarose gel electrophoresis of 5′ RACE products. M, molecular size markers; −, water. (B) Identification of transcription start sites by sequencing of 5′ RACE products. Start sites are identified by filled circles and the number of circles over a particular nucleotide indicates the number of times that 5′ RACE clone was isolated. Shading identifies the RSS.
Altered J_α_ chromatin structure in TEA-T mice
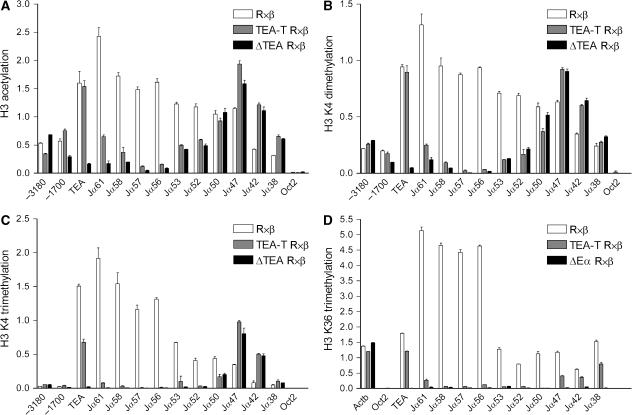
To understand the effects of transcriptional blockade on Jα chromatin structure and function, we used chromatin immunoprecipitation to monitor several histone modifications that are characteristic of active transcription units (Figure 5). On wild-type alleles, diacetylation of histone H3 was highest at the 5′ end of the Jα array, and gradually decreased toward the 3′ Jα segments, roughly paralleling the gradient of RNA pol II density along the Jα array (Abarrategui and Krangel, 2006) (Figure 5A). On ΔTEA alleles, H3 acetylation was dramatically reduced from TEA through Jα56 and then gradually increased starting at Jα53 to reach wild-type levels at Jα50. In contrast to ΔTEA alleles, TEA-T alleles revealed wild-type H3 acetylation at TEA. H3 acetylation then diminished gradually across the Jα segments just downstream of the terminator, reaching the level observed on ΔTEA alleles by Jα57. The disparity in acetylation between TEA-T and ΔTEA alleles at 5′ Jα segments implies a transcription-independent mechanism for the spreading of this histone modification over several kb from the activated TEA promoter. In support of this idea, H3 diacetylation also diminished gradually upstream of the TEA promoter on TEA-T alleles. On the other hand, the dramatic reductions in acetylation at sites 3′ of the terminator on TEA-T as compared to wild-type alleles are solely attributable to blockade of transcriptional elongation. Within the affected portion of the Jα array, the decline in H3 acetylation at Jα61 should simply reflect the loss of transcriptional readthrough, whereas the declines at Jα58, Jα57 and Jα56 might reflect the combined effects of diminished transcriptional readthrough and the loss of promoter activity at these sites. At central Jα segments, the ΔTEA and TEA-T alleles displayed parallel increases in histone acetylation at Jα47, Jα42 and Jα38 as compared to wild-type, consistent with the activation of central Jα promoters on both mutant alleles.
Figure 5.
Jα region histone modifications in TEA-T mice. Analysis of H3 acetylation (A), H3 K4 di- and trimethylation (B, C) and H3 K36 trimethylation (D) by chromatin immunoprecipitation. −3180 and −1700 indicate nucleotide positions upstream of the TEA transcript start site; TEA is positioned 300 nucleotides downstream of the start site. Oct2, negative control. Actb (exon 4), positive control. Data represent the mean±s.e.m. of triplicate PCR. Results are representative of two independent experiments.
Similar analyses of dimethylation and trimethylation at H3 K4 revealed profiles that were almost identical to those for H3 acetylation (Figure 5B and C). The sole exception was that H3 K4 trimethylation declined sharply both 5′ and 3′ of the TEA exon on TEA-T alleles. This implies that H3 K4 trimethylation at promoter distal sites can only be acquired through a transcription-dependent mechanism.
Histone H3 K36 methylation is associated with transcriptional elongation (Xiao et al, 2003) and has been shown to recruit the Rpd3S histone deacetylase complex to create a repressive environment that suppresses spurious transcriptional initiation within coding regions (Carrozza et al, 2005). We reasoned that reduced transcriptional readthrough across central Jα segments on TEA-T alleles might result in lower H3 K36 trimethylation and a more relaxed chromatin environment that would be permissive for central Jα promoter function. On wild-type alleles, H3 K36 trimethylation was moderate at the TEA exon, was substantially elevated at Jα61 through Jα56 and then decreased significantly at Jα53 and across the central Jα segments (Figure 5D). As expected, this modification was undetectable on ΔEα alleles. On TEA-T alleles, H3 K36 trimethylation was dramatically reduced from immediately downstream of the terminator through Jα50, indicating that its deposition is strictly dependent on transcriptional elongation. Moreover, although H3 K36 trimethylation was detectable at Jα47, Jα42 and Jα38, the level of trimethylation was only about half of that on wild-type alleles. Increased histone acetylation and promoter activity at central Jα segments on TEA-T alleles might therefore occur as a consequence of this reduction.
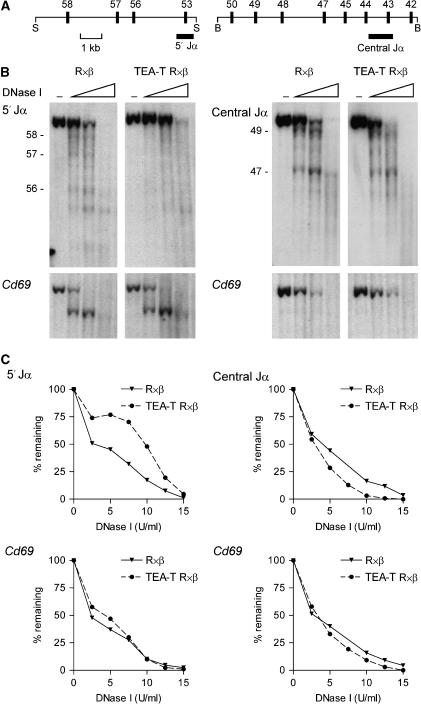
Altered J_α_ accessibility in TEA-T mice
We directly examined the influence of TEA transcription on Jα chromatin accessibility by measuring the sensitivity of Jα chromatin to DNaseI digestion (Figure 6). Thymocyte nuclei from R × β and TEA-T R × β mice were incubated with increasing amounts of DNaseI, and purified DNA was then digested with restriction enzymes, blotted and hybridized to radiolabeled control or Jα probes (Figure 6A and B). We detected no difference in DNaseI sensitivity of the control Cd69 gene between the two samples, indicating that digestion conditions were well matched (Figure 6B, bottom). Nevertheless, as compared to wild-type alleles, on TEA-T alleles the parental _Spe_I fragment detected by the 5′ Jα probe was more resistant to DNaseI digestion, whereas the parental _Bam_HI fragment detected by the central Jα probe was more sensitive (Figure 6B, top). These conclusions were confirmed by quantitative analysis of an independent experiment (Figure 6C). Thus, transcriptional blockade leads to reduced chromatin accessibility across 5′ Jα segments and increased accessibility across central Jα segments, consistent with the changes in promoter activity and histone modifications at these sites. In addition, blockade of TEA transcription led to the loss of three hypersensitive sites corresponding to the Jα58, Jα57 and Jα56 promoters on TEA-T alleles (Figure 6B, left panels). This suggests a defect in stable transcription factor loading in the absence of readthrough transcription. Nevertheless, at central Jα segments, the major hypersensitive sites at Jα49 and Jα47, as well as other minor hypersensitive sites, were detected on both wild-type and TEA-T alleles (Figure 6B, right panels). This suggests that central Jα promoters may be at least partially loaded with transcription factors on wild-type alleles even though their transcription is repressed.
Figure 6.
Jα region accessibility in TEA-T mice. (A) Strategy for Southern blot analysis of Jα region DNase I sensitivity using a 5′ Jα probe in _Spe_I (S) digests and a central Jα probe in _Bam_HI (B) digests. (B) Thymocytes were incubated with DNaseI (5, 10, 15 U) and Southern blots of _Spe_I (left panels) and _Bam_HI (right panels) digested genomic DNA samples were analyzed by hybridization to the indicated Jα probes (top panels). Blots were then stripped and reanalyzed by hybridization to a control Cd69 probe (bottom panels) to insure equal DNase I digestion and sample loading. Jα segments are indicated on the left. –, no DNaseI. Data are representative of two independent experiments (C) In a third experiment, Southern analysis was quantified for loss of the parental _Spe_I (left panels) and _Bam_HI (right panels) fragments using a phosphorimager. Blots were sequentially analyzed with Jα and Cd69 probes.
TEA transcription controls the targeting of primary rearrangements
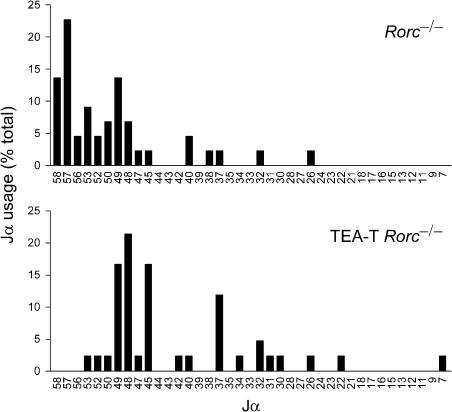
We hypothesized that the changes in promoter activity, histone modifications and DNaseI sensitivity at the 5′ and central Jα segments would translate into differences in RAG protein recognition and cleavage on wild type and TEA-T alleles. To study the role of TEA transcription in the targeting of initial Vα-to-Jα recombination events in DP thymocytes, we introduced the TEA-T allele onto the _Rorc_−/− background (Sun et al, 2000). DP thymocytes from mice deficient for the transcription factor RORγ are short lived and can support initial, or primary Vα-to-Jα rearrangement events, but not subsequent, or secondary rearrangement events (Guo et al, 2002). We examined Jα usage in thymocytes of _Rorc_−/− and TEA-T _Rorc_−/− mice by using Vα8 and Cα primers in RT–PCR to amplify the mature transcripts of Vα8-to-Jα rearranged alleles. Sequence analysis of the amplified products identified the distribution of Jα segments used (Figure 7). Consistent with previous work, primary rearrangements in _Rorc_−/− thymocytes were targeted to Jα segments that were immediately downstream of the TEA promoter (Jα58–Jα52), and that surrounded the Jα49 promoter (Jα50–Jα48) (Guo et al, 2002; Hawwari et al, 2005). However, in thymocytes of TEA-T _Rorc_−/− mice, primary rearrangements to the most 5′ Jα segments were almost completely abolished (54.5% in _Rorc_−/− versus 0.5% in TEA-T _Rorc_−/− at Jα58–Jα52), whereas primary rearrangements to central Jα segments were substantially increased (18.2% in _Rorc_−/− versus 54.8% in TEA-T _Rorc_−/− at Jα47–Jα22). Therefore, transcriptional blockade retargets primary Vα-to-Jα rearrangements in a fashion that is fully consistent with its effects on Jα promoter activity, histone modifications and chromatin accessibility. These results demonstrate that TEA transcription is required to focus initial Vα-to-Jα rearrangements to the 5′ end of the Jα array.
Figure 7.
Primary Jα usage in TEA-T mice. Vα8-Cα RT–PCR products of _Rorc_−/− (top) and TEA-T _Rorc_−/− (bottom) mice were cloned and sequenced for analysis of Jα usage. Data represent the percentage of clones using the indicated Jα segments. Number of clones analyzed: 44 _Rorc_−/− and 42 TEA-T _Rorc_−/−.
Reduced TCR_α_ repertoire in TEA-T mice
We addressed the effect of transcriptional blockade on the steady-state Tcra repertoire using two different approaches. We first analyzed Jα usage in mature Tcra transcripts in thymocytes of wild-type, TEA-T and ΔTEA mice. Following RT–PCR using Vα8 and Cα primers, PCR products were blotted and detected by hybridization with radiolabeled Jα probes (Figure 8A). Mice with a genomic deletion encompassing TEA to Jα49 (Δ5′) were similarly analyzed to provide a specificity control (Hawwari et al, 2005). This analysis revealed a dramatic reduction in usage of Jα58 to Jα52 on TEA-T alleles that was equivalent to the reduction on ΔTEA alleles. Nevertheless, in both instances, Jα usage increased to near wild-type levels at Jα49 and more 3′ Jα segments, a likely consequence of Jα49 and downstream promoter activity. Finally, we more directly examined Vα-to-Jα rearrangements by PCR of thymocyte genomic DNA using Vα8 and various Jα primers (Figure 8B). Consistent with the cDNA analysis, genomic rearrangements of 5′ Jα segments (Jα61-to-Jα52) were dramatically reduced in TEA-T thymocytes and these reductions were equivalent to those in ΔTEA thymocytes. We conclude that transcription driven by the TEA promoter is critical for the generation of a robust Tcra repertoire.
Figure 8.
Jα use and genomic rearrangements in TEA-T mice. (A) Southern blot of Vα8-to-Cα RT–PCR products prepared from three mice each of the indicated strains and one mouse lacking the 15 kb TEA-Jα49 genomic region (Δ5′). Jα use was determined by hybridization to radiolabeled Jα or Cα oligonucleotide probes. Analysis of Actb controlled for cDNA loading. (−) control PCR lacking cDNA. Data are representative of two independent experiments. (B) Southern blot of twofold serially diluted genomic DNA samples amplified with Vα8 and different Jα primers. PCR products were detected using radiolabeled Jα oligonucleotide probes. Analysis of Cd14 controlled for DNA loading. (−) control PCR lacking DNA. Data are representative of two independent experiments.
Discussion
Effects of TEA transcription on J_α_ promoter activity
In the present study, we directly analyzed the function of noncoding TEA transcription by introducing a transcription terminator downstream of the TEA promoter at the 5′ end of the Jα array. This approach offered a unique opportunity to discriminate the effects of promoter activation from those depending on transcription per se. We showed that TEA transcription controls chromatin structure and determines the transcriptional profile of the Jα array by promoting the activation of immediately downstream Jα promoters and repressing the activity of more distal promoters. It is remarkable that noncoding transcription initiated at a single promoter has opposing effects on different downstream promoters in the Jα array. However, the basis for these differential effects is unclear. One possibility is that the distance between TEA and downstream promoters is the critical parameter that distinguishes activation from suppression, perhaps as a consequence of histone modifications that vary across the length of the transcription unit. Alternatively, differential responsiveness of downstream promoters might rely on intrinsic differences in promoter strength or transcription factor binding.
Positive effects of TEA transcription were restricted to the Jα58, Jα57 and Jα56 promoters. Previous studies of TEA-deleted alleles had suggested that TEA was necessary for transcripts initiating at these Jα gene segments (Hawwari et al, 2005; Hawwari and Krangel, 2007), and two types of mechanisms had been considered. First, the TEA promoter might act as a physical organizer of the 5′ end of the Jα array. For example, a physical interaction between TEA and Eα might be required to bring the distant Eα to the vicinity of the Jα58, Jα57 and Jα56 promoters, so that it could stimulate their activation. Alternatively, transcription initiated at TEA might itself be regulatory. Recapitulation of the ΔTEA phenotype on TEA-T alleles that contain an active TEA promoter offers unambiguous support for a transcription-dependent mechanism.
TEA transcription could influence 5′ Jα promoter activity if the synthesized RNA itself provided an effector function essential for promoter activation. As an example, a noncoding RNA was shown to mediate recruitment of the transcriptional activator Dlx-2 to the Dlx-5/6 enhancer in mice (Feng et al, 2006). However, this mechanism seems unlikely to stimulate Jα58, Jα57 and Jα56 promoter activity, because there is no obvious sequence conservation near the initiation sites of the three promoters. Alternatively, activation could occur as a consequence of chromatin remodeling events that are linked to transcriptional elongation. Notably, the 5′ end of the Jα array displays a high density of RNA pol II and elevated levels of several histone modifications that are associated with active and accessible chromatin, including H3 acetylation and H3 K4 di- and trimethylation (Abarrategui and Krangel, 2006). Blockade of TEA transcription led to a dramatic reduction of these histone modifications and resulted in a general decrease in accessibility of 5′ Jα segments. The observed changes in chromatin structure at Jα61 must reflect direct effects of transcriptional blockade, as Jα61 lacks its own promoter. However, the decreases in chromatin modifications at Jα58, Jα57 and Jα56 segments on TEA-T alleles are likely to reflect both direct effects of blocked transcription and indirect effects of the loss of promoter activity at these sites. Either the specific histone marks introduced by TEA transcription, or the general increase in chromatin accessibility that they provide, could play an important role in activator binding to the Jα58, Jα57 and Jα56 promoters on wild-type alleles. This would be consistent with the reduced DNaseI hypersensitivity at these promoters on TEA-T alleles. Of note, Myc was recently been found to bind with high affinity in vivo only to binding sites located in regions characterized by high-level H3 acetylation and H3 K4 and K79 methylation (Guccione et al, 2006).
Negative effects of TEA transcription were evident in the central region of the Jα array. Transcriptional interference is generally believed to depend on the process of transcription without any direct involvement of the synthesized RNA (Shearwin et al, 2005). However, it was recently shown that the G/C rich dhfr promoter could be repressed in trans by an RNA that spanned the promoter (Martianov et al, 2007). We do not favor an RNA-mediated mechanism for transcriptional interference at the Jα47, Jα42 and Jα37 promoters, because transcripts initiated at Jα49 also run through the Jα47, Jα42 and Jα37 promoters on TEA-T alleles, but do not repress their activation.
Transcriptional interference by TEA may occur as a consequence of repressive chromatin marks that are introduced by transcriptional elongation across central Jα segments. Although we documented differences in the distribution of H3 K36 trimethylation across the Jα array on wild-type and TEA-T alleles, any role for this modification in transcriptional interference at central Jα promoters remains uncertain. Alternatively, transcriptional interference by TEA may occur because elongating RNA pol II physically interferes with the initiation of transcription at central Jα promoters. The fact that interference occurs with no loss of DNaseI hypersensitivity at Jα47 suggests that it may affect a step that is downstream of initial activator binding. Regardless, any explanation incorporating physical interference by elongating polymerase must address why similar interference is not observed at upstream promoters. We suggest that promoter activation or repression in the Jα array might ultimately reflect the balance between two competing effects, transcriptional interference and transcription-dependent chromatin remodeling. Transcriptional interference might have a suppressive influence on downstream promoters along the entire length of the Jα array. However, this suppressive effect might be counterbalanced at TEA-proximal Jα promoters by potent chromatin remodeling dependent on TEA transcription. The net effect would be conducive to promoter function in the TEA-proximal region, but suppressive to promoter function at greater distances from TEA. We suggest that the opposing influences of transcription documented here are likely to be of general significance, as studies of the human and murine transcriptomes indicate the frequencies of overlapping transcription units to be very high (Carnicini et al, 2005; Kimura et al, 2006).
Effects of TEA transcription on J_α_ gene segment recombination
Previous studies of TEA promoter-deleted mice showed that TEA is required to promote primary rearrangement of 5′ Jα gene segments and to suppress primary rearrangement of central Jα segments (Hawwari et al, 2005; Hawwari and Krangel, 2007). Here, we found that both these effects depend on TEA transcription. Several studies have shown that promoter elements stimulate V(D)J recombination by providing short-range accessibility to nearby RSSs (Sikes et al, 2002; Cobb et al, 2006; Oestreich et al, 2006). In addition, we recently demonstrated that transcription per se can provide long-range accessibility and permit the rearrangement of gene segments that are located at some distance from a promoter (Abarrategui and Krangel, 2006). Our results suggest that TEA transcription regulates both types of accessibility at the mouse Tcra locus. For example, accessibility at Jα61 depends exclusively on transcription from the TEA promoter situated 1.7 kb upstream. However, accessibility at Jα58, Jα57 and Jα56 is likely to depend both on the long-range accessibility delivered by TEA transcription and by the short-range accessibility provided by local promoters in response to transcriptional readthrough from TEA.
The exact mechanism by which the long-range accessibility is delivered through the process of transcription is not known. Specific histone modifications introduced by transcription are likely to contribute, either by modulating the packing of the nucleosome arrays or by recruiting additional remodeling activities that are in turn important for accessibility (Hassan et al, 2001; Belotserkovskaya et al, 2003; Pavri et al, 2006; Shogren-Knaak et al, 2006; Taverna et al, 2006; Wysocka et al, 2006). Notably, RAG2 was recently shown to contain a functionally important PHD domain (Elkin et al, 2005). Because PHD domains can confer binding to trimethylated H3 K4, this transcription-dependent histone modification could play a direct role in recruiting the recombinase to transcribed RSSs. Alternatively, recombination could be more directly stimulated by transcriptional elongation through any of several mechanisms. A direct interaction between RNA pol II and RAG proteins might deliver the recombinase directly to transcribed RSSs. Moreover, if RAG proteins were to travel with RNA pol II in this fashion, they might then take advantage of the transient nucleosome disassembly that accompanies elongation to access the underlying RSSs (Belotserkovskaya et al, 2003; Schwabish and Struhl, 2006). Finally, we cannot rule out the possibility that the synthesized transcript itself might help to recruit RAG proteins to RSSs. Regardless, the work presented here extends our previous data (Abarrategui and Krangel, 2006) and emphasizes that transcription is likely to function broadly as a regulator of accessibility for V(D)J recombination. Understanding precisely how transcription does so will be an important goal of future research.
Materials and methods
Generation of TEA-T mice
Homology arms were PCR amplified using pfu Ultra (Stratagene) and PCR fragments were verified by sequencing. The long and short arms extend from nucleotide 12 960 to nucleotide 19 676 and from nucleotide 19 677 to nucleotide 20 884, respectively of GeneBank accession number M64239. The transcription terminator, composed of polyadenylation sites followed by a lac operator array, has been described elsewhere (Abarrategui and Krangel, 2006). Homology regions were cloned into the pLNTK vector containing neo r and thymidine kinase selection markers driven by the phosphoglycerate kinase promoter. The TC1 129S6/SvEvTAc embryonic stem cell line was transformed by electroporation with the _Not_I-linearized targeting vector. Drug-resistant embryonic stem cell clones were first screened by PCR and correctly targeted clones were then verified by Southern blot using a Cδ fragment to probe _Kpn_I-digested genomic DNA, and a Jα58 fragment to probe _Pvu_II-digested genomic DNA. Chimeric mice were bred with mice expressing a Cre recombinase transgene under the control of the cytomegalovirus promoter to obtain germline transmission and neo r deletion. The TEA-T allele was bred onto _Rag2_−/− × TCRβ transgene (Shinkai et al, 1993) (R × β) and _Rorc_−/− (Sun et al, 2000) backgrounds. The wild-type Tcra allele carried by R × β mice was of 129 origin. Mice used for analysis were negative for the Cre recombinase. All mice were used in accordance with protocols approved by the Duke University Animal Care and Use Committee (Durham, NC).
Semiquantitative RT–PCR analysis of germline transcripts
Analysis of unspliced germline transcripts was performed on thymocyte nuclear RNA by semiquantitative RT–PCR as described previously (Abarrategui and Krangel, 2006), with the additional primers 5′TEA-F (5′-AGCTTTGACATCCCTGTCTTCC-3′), 5′TEA-R (5′-TTCTTACCAGTGTGCTCCAGGG-3′), 3′TEA-F (5′-ACCTTCCCAGTGAGCAATTTGC-3′), 3′TEA-R (5′-ACTCCGTGTTCCTCAGAGAGGC-3′). Spliced germline transcripts were analyzed in total RNA by RT–PCR as described previously (Abarrategui and Krangel, 2006).
5′ RACE
5′ RACE analysis was performed on total thymocyte RNA using the GeneRacer™ Kit (Invitrogen) according to the manufacturer′s instructions. Gene specific 3′ primers Jα58 (5′-GTTCTGGATGTCTG GACTCACTGTG-3′), Jα57 (5′-GTTCTGGATGTATGAGCTCACTGTC-3′) and Jα47 (5′-GTTCTGGATGTGAGGTCTGACTCTC-3′), which partially overlap the first Cα exon, were used for PCR amplification of cDNA products. PCR products were resolved on 2% agarose gels, were cloned using a TOPO TA Cloning® Kit (Invitrogen) and were analyzed by sequencing.
Chromatin immunoprecipitation
Immunoprecipitation of mononucleosomes using anti-diacetylated H3 (Upstate Biotechnology, 06-599), anti-dimethylated H3 K4 (Upstate Biotechnology, 07-030), anti-trimethylated H3 K4 (Abcam, ab8580) and control rabbit-IgG (R&D Systems, ab-105-c) antibodies was performed as described previously (Hawwari et al, 2005). Immunoprecipitations using anti-trimethylated H3 K36 (Abcam, ab9050) were performed as described elsewhere (Bannister et al, 2005). Immunoprecipitated and input material were quantified using a Roche LightCycler and a FastStart DNA Master Syber Green I kit (Roche) by real-time PCR. For immunoprecipitations using anti-diacetylated H3, anti-dimethylated H3 K4 and anti-trimethylated H3K4, analysis of β2-microglobulin was used to normalize the ratios of bound/input in different samples. Analysis of Actb exon 3 was used to normalize anti-trimethylated H3 K36 immunoprecipitations. Primer sequences are provided in Supplementary Table 1.
DNase I sensitivity assay
Sensitivity of thymocytes to DNaseI digestion was assayed as described elsewhere (Boyes and Felsenfeld, 1996) with a few modifications. Briefly, thymocytes from a whole thymus were isolated and incubated for 5 min on ice with 5 ml of 0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM Na2EDTA, pH 7.4, followed by addition of 20 ml of PBS. Thymocytes were then washed with Hanks buffer, resuspended in 0.15 M sucrose, 80 mM KCl, 30 mM HEPES, pH 7.4, 5 mM MgCl2, 0.5 mM CaCl2 and 0.067 mg/ml lysolecithin at 107 cells/ml, and were incubated for 5 min on ice. Thymocytes (3 × 107) of each genotype were then incubated with DNaseI for 10 min on ice. Digestions were stopped by bringing the reactions to 10 mM EDTA pH 8, 0.4 mg/ml Proteinase K and 0.4% SDS, followed by an overnight incubation at 37°C. Genomic DNA was then isolated by successive phenol, phenol/chloroform and chloroform extractions, followed by ethanol precipitation. _Spe_I or _Bam_HI digests were then fractionated by 0.7% agarose gel electrophoresis, transferred to a nylon membrane and hybridized to either a 5′Jα or a central Jα probe. To insure equivalent DNaseI digestion of different samples, the blots were stripped and re-hybridized to a Cd69 probe. The primers used to generate probes were: 5′Jα-F (5′-TGGGACCA GAAACCAGGGAG-3′), 5′Jα-R (5′-TTTCTGGCCTGGGCTACATCC-3′), central Jα-F (5′-AGAGAAGGCATCTTCTAGAGAAGTCC-3′), central Jα-R (5′-CTCCAAATCGTGGGGCATTGTTGT-3′), _Cd69_-F (5′-GGTAAAATTGTGAAGTTCCTGTGC-3′), and _Cd69_-R (5′-GTTAGGTGAAGTGGGCTTGG-3′). Quantitative analysis of blots was accomplished using a Typhoon 9410 Variable Mode Imager (Amersham Biosciences).
J_α_ usage
Primary Jα usage in Vα8 transcripts was analyzed by RT–PCR followed by sequencing as described previously (Hawwari et al, 2005). Steady-state Jα usage in Vα8 transcripts was analyzed by RT–PCR followed by hybridization with Jα probes as described previously (Hawwari et al, 2005). Vα8-to-Jα rearrangements were analyzed using Vα8 and Jα primers in genomic PCR as described previously (Abarrategui and Krangel, 2006).
Supplementary Material
Supplementary data
Acknowledgments
We thank E Oltz, B Sleckman, Y Zhuang and H Kondilis for critical review of the manuscript. Supported by the National Institutes of Health Grant GM41052 (to MSK).
References
- Abarrategui I, Krangel MS (2006) Regulation of T cell receptor-α gene recombination by transcription. Nat Immunol 7: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Baguet A, Sun X, Arroll T, Krumm A, Bix M (2005) Intergenic transcription is not required in Th2 cells to maintain histone acetylation and transcriptional permissiveness at the Il4-Il13 locus. J Immunol 175: 8146–8153 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2005) Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE (2004) Antisense intergenic transcription in V(D)J recombination. Nat Immunol 5: 630–637 [DOI] [PubMed] [Google Scholar]
- Boyes J, Felsenfeld G (1996) Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J 15: 2496–2507 [PMC free article] [PubMed] [Google Scholar]
- Buch T, Rieux-Laucat F, Forster I, Rajewsky K (2002) Failure of HY-specific thymocytes to escape negative selection by receptor editing. Immunity 16: 707–718 [DOI] [PubMed] [Google Scholar]
- Callen BP, Shearwin KE, Egan JB (2004) Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell 14: 647–656 [DOI] [PubMed] [Google Scholar]
- Carnicini P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest ARR, Zavolan M, Davis MJ, Wilming LG, Aidinis V et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM (2006) Accessibility control of V(D)J recombination. Adv Immunol 91: 45–109 [DOI] [PubMed] [Google Scholar]
- Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, Gozani OP (2005) A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem 280: 28701–28710 [DOI] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD (2006) The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20: 1470–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell 5: 377–386 [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B (2006) _Myc_-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 8: 764–770 [DOI] [PubMed] [Google Scholar]
- Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW (2002) Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat Immunol 3: 469–476 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL (2001) Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Proudfoot NJ (2005) Dicer-dependent turnover of intergenic transcripts from the human β-globin gene cluster. Mol Cell Biol 25: 9724–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A, Bock C, Krangel MS (2005) Regulation of the T cell receptor-α gene assembly by a complex hierarchy of germline Jα promoters. Nat Immunol 6: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A, Krangel MS (2007) Role for rearranged variable gene segments in directing secondary T cell receptor α recombination. Proc Natl Acad Sci USA 104: 903–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Liebhaber SA, Cooke NE (2006) Locus control region transcription plays an active role in long-range gene activation. Mol Cell 23: 365–375 [DOI] [PubMed] [Google Scholar]
- Hu X, Eszterhas S, Pallazzi N, Bouhassira EE, Fields J, Tanabe O, Gerber SA, Bulger M, Engel JD, Groudine M, Fiering S (2007) Transcriptional interference among the murine β-like globin genes. Blood 109: 2210–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T et al. (2006) Diversification of transcriptional modulation: large scale identification and characterization of putative alternative promoters of human genes. Genome Res 16: 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel MS, Carabana J, Abarrategui I, Schlimgen R, Hawwari A (2004) Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/δ locus. Immunol Rev 200: 224–232 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W (2003) Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat Immunol 4: 132–137 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15 (Spec No 1): R17–R29 [DOI] [PubMed] [Google Scholar]
- Mavieux L, Villey I, de Villartay JP (2003) TEA regulates local TCR-Jα accessibility through histone acetylation. Eur J Immunol 33: 2216–2222 [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM (2006) Regulation of TCRβ gene assembly by a promoter/enhancer holocomplex. Immunity 24: 381–391 [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A (2006) Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the ‘genome complexity' conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ (2002) Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA 99: 8796–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan DF, Cousins DJ, Santangelo S, Ioannou PA, Antoniou M, Lee TH, Staynov DZ (2004) Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc Natl Acad Sci USA 101: 2446–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F (2006) Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science 311: 1118–1123 [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K (2006) Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell 22: 415–422 [DOI] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB (2005) Transcriptional interference—a crash course. Trends Genet 21: 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Koyasu S, Nakayama K, Murphy KM, Loh DY, Reinherz EL, Alt FW (1993) Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science 259: 822–825 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM (2002) Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc Natl Acad Sci USA 99: 12309–12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW (1997) Function of the TCR-α enhancer in αβ and γδT cells. Immunity 7: 505–515 [DOI] [PubMed] [Google Scholar]
- Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR (2000) Requirement for RORγ in thymocyte survival and lymphoid organ development. Science 288: 2369–2373 [DOI] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24: 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP (1996) Defect in rearrangement of the most 5′ TCR-Jα following targeted deletion of T early α (TEA): implications for TCR α locus accessibility. Immunity 5: 331–342 [DOI] [PubMed] [Google Scholar]
- Villey I, Quartier P, Selz F, de Villartay JP (1997) Germ-line transcription and methylation status of the TCR-Jα locus in its accessible configuration. Eur J Immunol 27: 1619–1625 [DOI] [PubMed] [Google Scholar]
- Wang F, Huang CY, Kanagawa O (1998) Rapid deletion of rearranged T cell antigen receptor (TCR) Vα-Jα segment by secondary rearrangement in the thymus: role of continuous rearrangement of TCR α chain gene and positive selection in the T cell repertoire formation. Proc Natl Acad Sci USA 95: 11834–11839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, Alt FW (1985) Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40: 271–281 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data