HCAR and MCAR: The human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses (original) (raw)
Abstract
The subgroup C of the adenoviruses (Ad) and the group B coxsackieviruses (CVB) are structurally unrelated viruses that are known to compete for an unidentified cell surface receptor. We now describe the isolation of cDNAs from human and mouse that encode the human CVB and Ad2 and 5 receptor (HCAR) and the mouse CVB Ad2 and 5 receptor (MCAR). Both are 46-kDa glycoproteins whose primary amino acid sequences are highly homologous. Structurally, HCAR and MCAR appear to be transmembrane proteins that contain two extracellular immunoglobulin-like domains and therefore belong to this superfamily. Transfection of either of these cDNA molecules into receptor-negative NIH 3T3 cells conferred susceptibility to CVB infection and permitted the expression of β-galactosidase from a recombinant Ad5 vector. In addition, HCAR and MCAR mRNAs could be detected on Northern blots of oligo(dT)-selected RNA from receptor-positive HeLa cells and TCMK-1 as well as several tissues of human and mouse origin that are known to be targets for Ad and CVB infections. Finally, Western blots using antibodies that inhibit virus binding to either the human or mouse CVB receptors detected 46-kDa proteins in HCAR- and MCAR-transfected cells, respectively. Taken together, these results confirm that the isolated cDNAs encode the receptors for the subgroup C Ad and CVB.
The ability of animal viruses to infect host cells is dependent on the presence of an appropriate cellular receptor. In general, viruses of different families do not compete for binding to a common receptor. However, adenovirus (Ad) serotypes 2 (Ad2) and 5 (Ad5) and the parental group B coxsackieviruses (CVB) are human pathogens that obviously share a common receptor although they belong to divergent virus families (1). The Ad are DNA viruses that contain fiber proteins protruding from the 5-fold vertices of an icosahedral capsid. It is the terminal portion of the fiber known as the “knob” that is responsible for receptor binding (2). In contrast, the CVB are RNA viruses that lack fiber structures and are presumed to attach to cells through insertion of the receptor into a “canyon” on the surface of the virus (3). Interestingly, although these viruses use a common receptor they do not exhibit a similar host range (4). The CVB are known to infect a variety of organs, including the brain, intestines, heart, pancreas, and lungs, whereas the Ad primarily infect the intestines and lungs (4, 5). Therefore, these differences must be due to restrictions within the Ad life cycle subsequent to receptor attachment. We demonstrate here that transfection of either HCAR or MCAR cDNA into receptor-negative NIH 3T3 cells is sufficient to confer susceptibility to subgroup C Ad and CVB infection.
MATERIALS AND METHODS
cDNA Library Screening.
Five micrograms of twice oligo- (dT)-selected TCMK-1 RNA was used to construct a cDNA library in the λ ZAP Express vector using the ZAP Express cDNA synthesis kit (Stratagene). A library of 4 × 105 primary clones was amplified once in the XL1-Blue MRF′ strain of Escherichia coli. The library was plated at a density of 5 × 104 pfu/150 mm NZY agar plate and overlaid with nitrocellulose filters that had been wet in sterile distilled water containing 10 mM IPTG (isopropyl thio-β-d-galactoside) after incubation for 2.5 h at 42°C. The plates were transferred to 37°C for 8 h before the filters were removed and washed once in Tris-buffered saline to remove agarose and bacteria before being assayed with anti-p46 (6) antiserum using the 5-bromo-4-chloro-3-indolyl phosphate _p_-toluidine salt /nitroblue tetrazolium detection system.
Northern Blots.
Analysis of cellular RNA was conducted by transfer to nylon membranes after electrophoretic separation through a 1.2% formaldehyde agarose gel according to published procedures (7). Hybridizations were performed in 50% formamide, 1% SDS and 5× standard saline phosphate/EDTA (SSPE; 0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), for 18–24 h at 42°C. Membranes were washed once in 2× SSPE, 0.1% SDS at room temperature, and twice in 0.1× SSPE, 0.1% SDS at 42°C for 30 min after hybridization.
Transfections.
NIH 3T3 cells (3 × 105) were seeded into 35-mm plates 24 h before transfection. Forty-five minutes before transfection, a solution was prepared by mixing 2 μg of supercoiled plasmid diluted in 100 μl of DMEM (GIBCO) and 7 μl of Lipofectamine (GIBCO) diluted in 100 μl of DMEM. After incubation at room temperature, 800 μl of DMEM was added, and the solution was overlaid onto plates that were washed twice with DMEM. The plates were incubated for 5 h at 37°C in 5% CO2 before the solution was removed, and 2 ml of DMEM containing 10% fetal calf serum was added. The plates were incubated for an additional 18 h at 37°C. Cells were used for experiments 18–24 h posttransfection.
Virus Assays.
Twenty hours after transfection, 35-mm plates containing 5 × 105 cells were washed twice and 200 μl of DMEM containing 1 × 107 pfu of picornaviruses added. Plates were incubated for 90 min at room temperature before being washed four times with 2 ml of DMEM. Plates were overlaid with 5 ml of DMEM/10% fetal calf serum, and 24 h later frozen and thawed three times. Cellular debris was removed by centrifugation, and dilutions of the resulting supernatants quantitated by plaque assay (8). The Ad vector construct was kindly provided by Robert Schneider at New York University Medical Center. It contains the β-galactosidase under a cytomegalovirus (CMV) promoter inserted in the E1 region of Ad5. The plasmids used for virus construction are described in ref. 9.
Western Blots.
Cells were solubilized in 1% Triton X-100, 1% deoxycholate, and analyzed by SDS/10% PAGE using the discontinuous buffer system (10). After transfer to poly(vinylidene difluoride) membrane supports, blots were probed with either 125 ng/ml RmcB or a 1:1000 dilution of anti-p46 antibody for 1 h at room temperature. Immunoreactivity was determined after incubation with horseradish peroxidase conjugated secondary antibodies and the ECL Western Blotting Detection System (Amersham).
Immunofluorescence.
Transfectants (1 × 106) were removed from plastic dishes with 0.05 mM EDTA and washed once in 1× PBS. The washed cells were resuspended in 100 μl of 1× PBS containing 2 μg of RmcB and incubated for 1 h at 4°C. Cells were washed three times with 1× PBS and incubated for 30 min at 4°C in 100 μl of 1× PBS containing 0.35 μg of a goat anti-mouse fluorescein isothiocyanate-conjugated antibody. Fluorescence was monitored using a Zeiss Axioplan fluorescent microscope.
RESULTS
Isolation of cDNA.
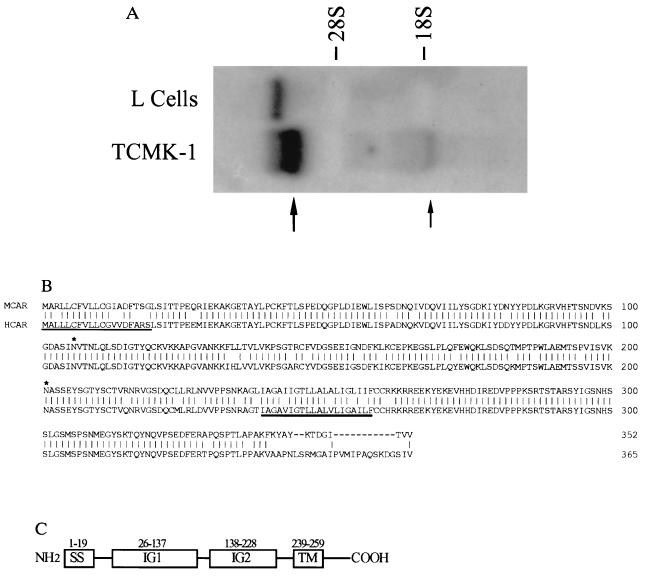
To isolate a receptor molecule, we used the classical approach of screening a λ phage expression cDNA library with antiserum directed against a mouse CVB-binding protein (anti-p46) (6). This antibody will prevent infection with CVB in mouse cells and specifically identify a 46-kDa protein from virus-susceptible mouse cells (6). From a transformed mouse kidney cell (TCMK-1) library, a 1-kilobase (kb) cDNA clone (RTMCAR-4) was isolated that hybridized in Northern blots with 6-kb and 1.4-kb RNAs from TCMK-1 cells that were not found in receptor-negative mouse L cells (11) (Fig. 1A). The band observed in the L cells may be due to DNA contamination, because it was not observed in poly(A)-selected RNA (Fig. 4A). blast searches of the GenBank EST database using the nucleotide sequence of RTMCAR-4 revealed two IMAGE consortium clones from newborn human melanocytes (265680) and pancreas islet cells (328668) cDNA libraries that were significantly homologous to RTMCAR-4. These findings suggested that the human clones may encode a homologue of RTMCAR-4. To proceed, clone 265680 was purchased from the American Type Culture Collection, and the nucleotide and deduced amino acid sequence compared with that of RTMCAR-4. Based upon best-fit alignments, both 265680 and RTMCAR-4 contained single open reading frames that exhibited 67% amino acid identity. Clone 265680 was slightly larger, containing an additional 34 amino acids N terminal of the protein encoded by RTMCAR-4. However, neither clone appeared to contain the entire coding sequence, because their open reading frames (that lacked starting methionine codons) began immediately at the 5′ end of each clone. Therefore, primers for amplification of the 5′ ends were designed specifically for either RTMCAR-4 or clone 265680 and used in the PCR to amplify fragments of 450 bp and 280 bp from TCMK-1 and HeLa cell cDNA libraries, respectively. These sequences were sufficiently long to provide a starting ATG codon and encode the additional amino acids necessary to complete the open reading frames. The final length of the cDNA from mouse was around 1.4 kb and around 2.4 kb from the human cells. Although the coding regions of the two cDNAs are similar, the human clone contains 1.2 kb of 3′ untranslated sequence.
Figure 1.
(A) Unique RNAs are found in receptor-expressing TCMK-1 cells. Total RNA from TCMK-1 or mouse L cells was probed with a 32P-labeled fragment of RTMCR-4 to reveal differentially expressed 6-kb (large arrow) and 1.4-kb (small arrow) RNAs. (B) Amino acid alignment of HCAR and MCAR. The deduced amino acid sequences of the cloned HCAR and MCAR cDNAs were aligned using the GeneWorks software program. Vertical lines, amino acid identities; dashes, gaps in the alignment. The positions of the potential signal peptide (thin underline), transmembrane region (thick underline), and N-linked glycosylation sites (∗) are shown. (C) A domain model for the CAR proteins. IG1 and IG2, immunoglobulin domains; SS, signal sequence; TM, transmembrane-spanning region.
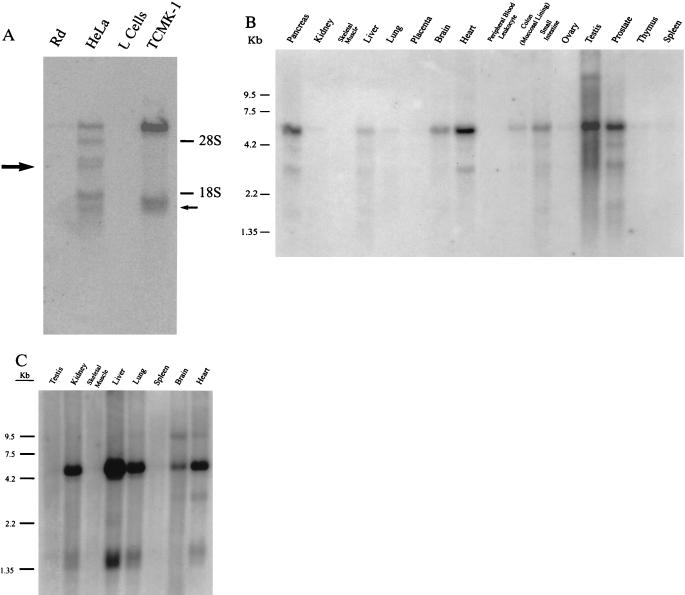
Figure 4.
HCAR and MCAR mRNA is expressed in a variety of tissues. (A) Hybridizing mRNAs of 6 kb and 1.4 kb can be detected in poly(A)+ Northern blots of TCMK-1 cells, but not mouse L cells. In contrast, six species of mRNAs are detected in HeLa cells that range in size from 6 kb to 1.2 kb that are not found in receptor-negative rhabdomyosarcoma (Rd) cells. The sizes of the RNAs that correlate with the size of the isolated HCAR (large arrow) and MCAR (small arrow) cDNAs are shown. (B) Human multiple tissue Northern blots were purchased from CLONTECH and hybridized with a 32P-labeled fragment that corresponded to the open reading frame of HCAR. Hybridizing sequences are similar in size to those detected in HeLa cells. (C) Mouse multiple tissue Northern blots (CLONTECH) were hybridized with a 32P-labeled fragment that corresponded to the open reading frame of MCAR. Hybridizing sequences are similar in size to those detected in TCMK-1 cells.
The deduced full length of the putative HCAR protein is composed of 365 amino acids totaling around 40 kDa. MCAR is 352 amino acids long with a molecular mass of around 39 kDa. These molecules are 83% identical with conservative amino acid substitutions found at most of the differing positions (Fig. 1B). The only dramatic difference between the molecules is a sequence divergence at the carboxyl terminus. Both proteins contain potential leader sequences, transmembrane domains, and two N-linked glycosylation sites. Structurally they contain two disulfide-bonded loops (amino acids 35–130 and amino acids 155–220), which resemble the characteristic V and C2 domains of the immunoglobulin superfamily (Fig. 1C) (12). However, no significant homologies have been found during blast searches between sequences present in the SWISSPROT database and HCAR or MCAR.
Receptor Function.
To determine whether the HCAR and MCAR cDNAs truly encoded functional receptors, they were inserted into pBK-CMV vectors (Stratagene), transfected into NIH 3T3 cells, and transfectants assayed for virus production or reporter gene expression. The strain of NIH 3T3 that we have used for expression of growth-arresting genes (13) does not express the MCAR mRNA and cannot be infected with subgroup C Ad (not shown). The plasmid pRTHR, which contains a 1.1-kb HCAR cDNA that lacks the 3′ untranslated region, or pRTMR, which contains the complete 1.4-kb MCAR cDNA, allowed production of 102- to 103-fold higher titers of infectious CVB3 and CVB4 than cells transfected with plasmid alone (pBK-CMV). As shown in Table 1, the background level after transfection with control plasmid only refers to surviving virus from the inoculum. The infectability was specific for CVB, because no increase in titer was observed when another picornavirus family member, poliovirus T1, was used (Table 1).
Table 1.
Production of plaque-forming units after virus infection of transfected NIH 3T3 cells
| Plasmid* | Virus yield, plaque-forming units† | ||
|---|---|---|---|
| CVB3 | CVB4 | Poliovirus | |
| pRTHR | 2.92 × 107 | 1.7 × 106 | 2.65 × 104 |
| pRTMR | 4.22 × 106 | 6.0 × 105 | 2.60 × 104 |
| pBK-CMV | 2.25 × 104 | 1.5 × 103 | 2.27 × 104 |
In addition, pRTHR and pRTMR, but not plasmid control transfectants, stained positively for β-galactosidase 16 h after incubation with a recombinant Ad5 expressing β-galactosidase from the CMV promoter (Fig. 2A). HCAR and MCAR are specific Ad-binding proteins, because 35S-Ad3 (group B Ad), which does not compete with group C Ad for the same receptor (14), exhibits 5-fold less binding to transfectants than 35S-Ad2. The binding of labeled virus could be competed with cold virus, as previously described (1). Thus, these results confirm that HCAR and MCAR are the receptors for Ad2, A5, and the CVB.
Figure 2.
(A) pRTHR and pRTMR confer susceptibility to Ad5 entry. Only (A) TCMK-1 positive controls or NIH 3T3 cells transfected with either (B) pRTHR or (C) pRTMR stained for β-galactosidase expression after incubation with a CMV-β-galactosidase recombinant Ad5 vector at a multiplicity of infection of around 50. (D) pBK-CMV controls showed no reactivity after incubation under identical conditions. (B) HCAR expression is localized to the exterior of the cell. Bright staining was observed over the entire plasma membrane in unfixed NIH 3T3 cells transfected with pRTHR and labeling transfectants with RmcB and a fluorescein isothiocyanate-labeled goat anti-mouse antibody (A). B represents pBK-CMV control cells.
Expression of the Receptor.
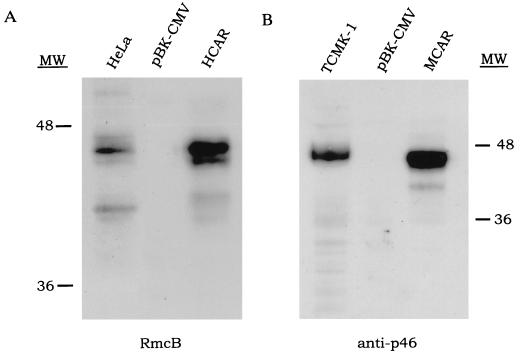
The ability of the viruses to infect NIH 3T3 cells indicated that the receptors were being properly expressed. Immunofluorescence using the well defined anti-human CVB receptor monoclonal antibody (RmcB), which can block infection in human cells of CVB viruses (15), revealed a bright staining on the surface of suspended HCAR transfectants (Fig. 2B), confirming that receptors were localized on the external side of the plasma membrane. Western blots of detergent-solubilized cell extracts detected two proteins of around 46 kDa and 44 kDa from HCAR transfectants corresponding to the size of the polypeptides in the HeLa cell positive control (although the latter had much less of the 44-kDa protein), whereas the empty plasmid pBK-CMV transfectants did not produce any signal (Fig. 3). When the rat anti-p46 (6) antiserum was used to probe MCAR transfectant extracts, a very strong signal was also observed at 46 kDa. In both cases it appears that the translated receptor proteins have undergone posttranslational processing, increasing their size from around 40 kDa to 46 kDa. The doublet observed with HCAR may be attributable to the 46-kDa molecule having been glycosylated at both of its N-linked sites, whereas the 44-kDa molecules may be glycosylated at only a single site. The HCAR antibody did not react with the MCAR moiety and vice versa, as previously reported (15).
Figure 3.
HCAR and MCAR are detectable by CVB receptor-specific antibodies. Forty-six-kilodalton proteins are visible in HCAR- and MCAR-transfected cells that are absent in pBK-CMV transfected cells. Similar sized proteins (≈46 kDa) are also detectable in HeLa cell and TCMK-1 cell positive controls. Immunodetection was performed using RmcB (A) (15) or anti-p46 (B) (6).
Northern blots of poly(A)+ RNA probed with fragments from the coding regions of HCAR and MCAR identified RNAs of around 6 kb and 1.4 kb from TCMK-1 cells, and a range of RNAs from around 6 kb to 1.3 kb from Hela cells (Fig. 4A). Neither fragment hybridized to RNAs of the respective receptor-negative cells of human (rhabdomyosarcoma) or murine (L cell) origin. We currently are assessing the nature of the larger mRNAs to determine if they are incompletely processed transcripts or RNAs that are related to the receptors. Nonetheless, the sizes of the identified cDNAs correlate with sizes of RNAs detected by the hybridization (Fig. 4A), indicating they are truly representative of the parental RNA species.
Receptor RNA in Different Tissues.
When poly(A)+ RNA from multiple human tissues were probed with HCAR in Northern blots, the pancreas, brain, heart, small intestine, testis, and prostate had the highest amount of HCAR message (Fig. 4B). The liver and lung had small amounts of HCAR RNA, whereas no signal could be detected in kidney, placenta, peripheral blood leukocytes, thymus, and spleen. In comparison, the highest level of MCAR RNA was observed in mouse liver (Fig. 4C). Other mouse organs that expressed detectable levels were the kidney, heart, lung, and brain. The expression of HCAR and MCAR at the RNA level appears to correlate well with previous data of target organs infected by the CVB (5, 16).
DISCUSSION
We have demonstrated here that previously unidentified human and mouse cDNAs encode for proteins that serve as the receptors for the subgroup C Ad and the CVB. Transfection of either cDNA into receptor-negative cells resulted in measurable increases in CVB titers of 102- to 103-fold. In addition, β-galactosidase activity could be detected 16 h after infection with a recombinant Ad5 vector expressing the β-galactosidase gene from a CMV promoter. Although it is possible that the molecules encoded by the HCAR and MCAR cDNAs may activate an endogenous mouse cell receptor, assays with the human CVB receptor specific RmcB monoclonal antibody prove this is not the case. It is known that RmcB will block infection of human cells but not mouse cells by CVB (15). Therefore, the positive RmcB immunofluorescent staining of HCAR-transfected cells, along with the detection of a 46-kDa protein in RmcB Western blots indicates that the ligand for RmcB (which must be of human origin) is being expressed. Collectively, these results, together with the presence of detectable mRNAs in virus susceptible cells, confirm the function of HCAR and MCAR.
New interest in understanding how two different viruses use the same receptor molecule has been revived 20 years after the initial observation that Ad2, Ad5, and the CVB share a common receptor (1). Emphasis has been given to understanding the pathogenesis of the CVB due to the severe diseases they cause such as, among others, myocarditis (5). Expression of the cellular receptor obviously influences what tissues are most susceptible to infection and by what routes the virus spreads through the body. In more recent times, however, identification of this receptor has been sparked by a need to define target organs for Ad gene therapy. The Ad do not have a broad host range like the CVB and must encounter other restrictions in the virus life cycle after receptor attachment (4). However, the expression of the HCAR RNA in several tissues indicates that Ad gene therapy may have a strong application in a variety of organs. The two molecules described herein must have other cellular functions than that of a virus receptor. Studies currently are underway to determine the normal cellular functions of these molecules to determine if they influence virus infectivity. Preliminary data based on sequence comparison indicate the HCAR locus to be on human chromosome 21, and not on chromosome 19 as previously proposed (17). Isolation of the genomic sequences for both HCAR and MCAR may provide insight into the regulation of their expression, which may be useful when considering possible targets for Ad gene therapy or devising strategies to inhibit Ad and CVB infectivity.
ABBREVIATIONS
Ad
adenovirus
CVB
group B coxsackieviruses
CMV
cytomegalovirus
HCAR
human CVB and Ad2 and 5 receptor
MCAR
mouse CVB Ad2 and 5 receptor
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. U90715U90715 and U90716U90716 for the MCAR and HCAR sequences, respectively).
References
- 1.Lonberg-Holm K, Crowell R L, Philipson L. Nature (London) 1976;259:679–681. doi: 10.1038/259679a0. [DOI] [PubMed] [Google Scholar]
- 2.Xia D, Henry L J, Gerard R D, Deisenhofer J. Structure (London) 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]
- 3.Muckelbauer J K, Kremer M, Minor I, Diana G, Dutko F J, Groarke J, Pevear D C, Rossman M G. Structure (London) 1995;3:653–667. doi: 10.1016/s0969-2126(01)00201-5. [DOI] [PubMed] [Google Scholar]
- 4.Shenk T. In: Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath J P, Roizman B, Straus S E, editors. Philadelphia: Lippincott-Raven; 1996. pp. 2112–2137. [Google Scholar]
- 5.Melnick J L. In: Virology. Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath J P, editors. New York: Raven; 1990. pp. 549–600. [Google Scholar]
- 6.Xu R, Mohanty J G, Crowell R L. Vir Res. 1995;35:323–340. doi: 10.1016/0168-1702(94)00100-q. [DOI] [PubMed] [Google Scholar]
- 7.Davis L G, Dibner M D, Battey J F. Basic Methods in Molecular Biology. New York: Elsevier Science; 1986. [Google Scholar]
- 8.Crowell R L, Syverton J T. J Exp Med. 1961;113:419–435. doi: 10.1084/jem.113.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doria M, Klein N, Lucito R, Schneider R J. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Hsu K-H L, Crowell R L. J Virol. 1989;63:3105–3108. doi: 10.1128/jvi.63.7.3105-3108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams A F, Barclay A N. Annu Rev Immun. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 13.Peverali F A, Ramqvist T, Saffrich R, Pepperkok R, Barone M V, Philipson L. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Defer C, Belin M-T, Caillet-Boudin M-L, Boulanger P. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu K-H L, Lonberg-Holm K, Alstein B, Crowell R L. J Virol. 1988;62:1647–1652. doi: 10.1128/jvi.62.5.1647-1652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalldorf G, Melnick J L. In: Viral and Rickettsial Infections of Man. Horsfall F L, Tamm I, editors. Philadelphia: Lippincott; 1965. pp. 474–512. [Google Scholar]
- 17.Couillin P, Boue A, Rebourcet R, Van Cong N. Pathol Biol. 1976;24:195–203. [PubMed] [Google Scholar]