Gene silencing mechanisms mediated by Aubergine–piRNA complexes in Drosophila male gonad (original) (raw)
Abstract
Genetic studies have shown that Aubergine (Aub), one of the Piwi subfamily of Argonautes in Drosophila, is essential for germ cell formation and maintaining fertility. aub mutations lead to the accumulation of retrotransposons in ovaries and testes, and Stellate transcripts in testes. Aub in ovaries associates with a variety of Piwi-interacting RNAs (piRNAs) derived from repetitive intergenic elements including retrotransposons. Here we found that Aub in testes also associates with various kinds of piRNAs. Although in ovaries Aub-associated piRNA populations are quite diverse, piRNAs with Aub in testes show a strong bias. The most abundant piRNAs were those corresponding to antisense transcripts of Suppressor of Stellate [_Su(Ste)_] genes known to be involved in Stellate gene silencing. The second most abundant class was made up of those from chromosome X and showed strong complementarity to vasa transcripts. Immunopurified Aub–piRNA complexes from testes displayed activity in cleaving target RNA containing sequences complementary to Stellate and vasa transcripts. These results provide the first biochemical insights into gene silencing mechanisms mediated by Aub and piRNAs in fly testes.
Keywords: Aubergine, piRNA, Stellate, RNA silencing, germlines
INTRODUCTION
RNA silencing is an evolutionarily conserved physiological process involved in regulating gene expression, defending against viruses, and maintaining genome stability (Tomari and Zamore 2005; Zaratiegui et al. 2007). RNA interference (RNAi) (Fire et al. 1998) is the most typical of such processes and causes gene silencing by cleaving (slicing) the target mRNAs in a sequence-dependent manner (Tomari and Zamore 2005). A molecule rendering sequence specificity is a 21–23 nucleotide (nt) long RNA, termed short interfering RNA (siRNA) (Tomari and Zamore 2005).
microRNAs (miRNAs) are a large group of endogenous small RNAs (21–23 nt in length) encoded in the genome of a variety of organisms (Ambros 2004; Du and Zamore 2005; Kloosterman and Plasterk 2006). miRNA also functions in RNA silencing, as does siRNA, a process through which the expression levels of genes involved in various developmental and metabolic processes are modulated. Besides their functional commonality, siRNA and miRNA show strong similarities even in maturation events. Both are processed from a longer precursor by an RNase III enzyme (Dicer), unwound, and then loaded onto a member of the Argonaute family of proteins (Carmell et al. 2002; Parker and Barford 2006). After maturation and association with Argonautes, they ultimately exert their effects in RNA silencing as guide molecules to specify the RNA targets to be silenced.
Members of the Argonaute family are defined by the presence of the PAZ and PIWI domains (Carmell et al. 2002). In Drosophila, five genes—AGO1, AGO2, AGO3, Piwi, and Aubergine (Aub)—exist in the genome as members of the Argonaute family. AGO1 and AGO2 are expressed ubiquitously through development, whereas the other three are only expressed in germline cells (Williams and Rubin 2002). The latter are grouped as the Piwi subfamily of Argonautes because they show strong sequence similarities to each other.
AGO1 and AGO2 in Drosophila function in gene silencing through binding with miRNA and siRNA, respectively (Okamura et al. 2004; Tomari et al. 2004; Miyoshi et al. 2005). Recent studies have shown that the structure of a double-stranded small RNA intermediate strongly influences its partitioning between AGO1- and AGO2-RISC (RNA-induced silencing complex) (Forstemann et al. 2007; Tomari et al. 2007). siRNA-loaded AGO2 functions in RNAi as Slicer and is directly responsible for cleaving a target completely complementary to siRNA (Miyoshi et al. 2005). AGO2 is also involved in RISC formation by slicing the passenger strand within the siRNA duplex (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Kim et al. 2006; Leuschner et al. 2006). Although AGO1 is thought to function in miRNA-mediated translational repression, like AGO2 it also possesses Slicer activity (Miyoshi et al. 2005).
Piwi, one of the Piwi subfamily proteins in Drosophila, is an essential factor in germline stem cell (GSC) self-renewal in both males and females (Cox et al. 1998, 2000; Szakmary et al. 2005). piwi mutations impact transposon mobility (Sarot et al. 2004; Kalmykova et al. 2005); this most likely results from the deleterious effects of transposon activation. Indeed, recent studies have shown that Piwi in ovaries is associated with repeat-associated siRNAs (rasiRNAs) derived from a variety of repetitive intergenic elements such as retrotransposons (Saito et al. 2006; Vagin et al. 2006; Brennecke et al. 2007). Their longer length distinguishes rasiRNAs (24–30 nt) from siRNAs and miRNAs (Aravin et al. 2003). Recombinant Piwi protein produced in Escherichia coli is able to exhibit Slicer activity in vitro (Saito et al. 2006), and Piwi is localized in the nucleus (Cox et al. 2000; Saito et al. 2006; Brennecke et al. 2007). Taken together, it can be postulated that Piwi functions, at least in part, like Slicer in silencing repetitive/transposable genes through its association with rasiRNAs in the nucleus, and thus protects the genome from invasive elements. rasiRNAs in Drosophila are also called piRNAs (Brennecke et al. 2007; Zamore 2007); we use the latter term hereinafter.
Genetic studies have revealed that Aub is required for pole cell formation (Harris and Macdonald 2001) and for activating RNAi during Drosophila oocyte maturation (Kennerdell et al. 2002). Aub is also involved in silencing retrotransposons in the germline (Vagin et al. 2004, 2006; Savitsky et al. 2006) and Stellate genes in testis through the homologous Suppressor of Stellate [_Su(Ste)_] repeats on chromosome Y from which Su(Ste) piRNAs are derived (Aravin et al. 2004; Vagin et al. 2006). Aub is required for accumulation of Su(Ste) piRNAs (Aravin et al. 2004; Vagin et al. 2006). Mutations in Aub cause male sterility, which is directly attributable to the failure in silencing the repetitive Stellate locus. Aub in ovaries is associated with piRNAs originating mainly from retrotransposon antisense transcripts (Vagin et al. 2006; Gunawardane et al. 2007). A large-scale study identifying small RNAs associated with Aub in ovary has recently been reported (Brennecke et al. 2007). However, questions for which answers have yet to be elucidated include:
- What are the differences between piRNAs associated with Aub and Piwi in male and female gonads?
- Is Aub in testes physically associated with Su(Ste) piRNAs?
- If so, how is the Aub–piRNA complex involved in gene silencing at the molecular level?
Here we closely investigated the profiles of piRNAs associated with Piwi and Aub in ovaries and testes. Although cellular localizations of Piwi and Aub in ovaries differ from each other, they bind to a similar set of piRNAs. In contrast to this, we found that piRNAs associated with Aub in testes were rather unique. Of these piRNAs, the most abundant (∼46% of the total) were those corresponding to Su(Ste) antisense transcripts. Aub–piRNA complex immunopurified from testes was able to cleave target RNAs containing sequences of Su(Ste) and Stellate. The second largest class of piRNAs associated with Aub in testes contained only two kinds of sequences, both of which were mapped to tiny, repetitive regions on chromosome X. Interestingly, these two, termed AT-chX-1 and -2, showed strong complementarity to vasa (vas) mRNA, a germline-specific transcript involved in oocyte differentiation and cyst development (Lasko and Ashburner 1988; Styhler et al. 1998). Indeed, the protein levels of VAS in aub mutant testes were increased by about twofold compared to those in wild type (wt). The Aub–piRNA complex from testes even showed activity in cleaving target RNAs containing part vas mRNA. Immunofluorescence showed that Aub is localized in the cytoplasm, as is AGO2 (Findley et al. 2003); however, 21-nt siRNA incubated in ovary lysate was only loaded onto AGO2, not onto Aub. Taken together, we propose that both in ovaries and testes, Aub functions in cytoplasmic RNA silencing only through its association with 24–30-nt piRNAs. Our studies provide the first biochemical insights into gene silencing mechanisms mediated by Aub in accord with piRNAs in fly testes.
RESULTS
Expression of Aub in germline cells
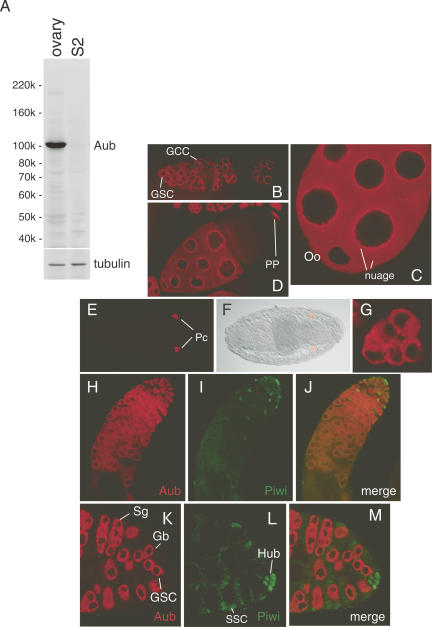
To biochemically elucidate the functional involvement of Aub in the presumptive RNA silencing process in fly gonads, we produced monoclonal antibodies against Aub. Western blotting on ovary and S2 cell lysates using the anti-Aub antibody revealed that Aub is strongly expressed in ovaries, but little in S2 cells (Fig. 1A). AGO1 and AGO2 expressed in S2 cells (Miyoshi et al. 2005) were not cross-reacted by anti-Aub (Fig. 1A). Western blotting on Piwi and AGO3 recombinant proteins produced in E. coli confirmed that the anti-Aub cross-reacts to neither of those (Supplemental Fig. 1A), indicating that the antibody is specific to Aub. In early embryos, Aub is strongly expressed, but its protein levels gradually diminish throughout development (Supplemental Fig. 1B).
FIGURE 1.
Aub expression. (A) Aub expression is detected in Drosophila ovaries, but not in S2 cells. Western blotting was performed on lysates of fly ovaries and S2 cells using an anti-Aub antibody produced in our laboratory. Anti-tubulin was utilized as a loading control. (B) Fluorescent image of a germarium region stained with anti-Aub, which clearly indicates that Aub is localized in the cytoplasm of germline stem cells (GSC) and germline cyst cells (GCC), but Aub appears not to be expressed in somatic cells in the germarium. (C) Fluorescent image of Aub in an egg chamber at stage 6. Aub is found both in oocytes (Oo) and nurse cells. In the latter, Aub accumulation at the peripheral region of the nuclei and in the cytoplasmic particles is observed (nuage). (D) Fluorescent image of Aub in an egg chamber at stage 10A. Around this stage, Aub accumulation to the posterior pole (PP) in oocytes is clearly found. (E) Fluorescent image of an embryo (stage 14) stained with anti-Aub. Pole cells are indicated as Pc. (F) The DIC image of the embryo shown in E and the immunostaining in E were merged. (G) This immunostaining image clearly shows that Aub is localized in the cytoplasm of pole cells in the interior of embryos. (H–M) Testes were double-stained with anti-Aub and anti-Piwi antibodies (Saito et al. 2006). Fluorescent images of (H,K) Aub and (I,L) Piwi. Merged images are shown in J and M, which clearly indicate that the expressions of Aub and Piwi are mutually exclusive in testes. Aub expression is restricted to GSC, gonialblast cells (Gb), and spermatogonia cells (Sg), while Piwi is only expressed in hub (Hub) and supporting somatic cells (SSC). All images shown in this figure represent a single confocal section.
To examine the expression patterns of endogenous Aub in ovaries and embryos, immunofluorescent staining was carried out. In the germarium, Aub was found in the cytoplasm of germline stem cells (GSC) and germline cyst cells (GCC) (Fig. 1B). There are strong accumulations of Aub at the cytoplasmic side of perinuclear regions, called the nuage, a germline-specific body conserved in species (Eddy 1975). In egg chambers at stages 6 and 10A, Aub was clearly detected in the cytoplasm of both nurse cells and oocytes (Fig. 1C,D). Nuage staining was observed only in nurse cells, not in oocytes at stage 6 (Fig. 1C), which is consistent with the notion that nuage is known to be lost from oocytes (Mahowald and Strassheim 1970). Clear, sharp accumulation of Aub was observed at the posterior of oocytes at stage 10A (Fig. 1D). Aub signals were under the detection limit in somatic cells such as terminal filament cells, cap cells, and follicle cells (Fig. 1B,C), where strong Piwi expression has been observed (Saito et al. 2006). These staining patterns rather resembled those previously reported with GFP-tagged Aub (GFP-Aub) overexpressed in fly ovaries (Harris and Macdonald 2001) and with polyclonal antibodies against Aub (Brennecke et al. 2007). In embryos, endogenous Aub was clearly accumulated in the cytoplasm of pole cells, as expected (Fig. 1E–G).
Testis staining with anti-Aub revealed Aub in GSC, gonialblast, and spermatogonia cells (Fig. 1H,K). However, Aub expression was undetected in somatic cells surrounding gonialblast and spermatogonia cells, and in hub (Fig. 1H,K), where the strong expression of Piwi was observed (Fig. 1I,L; Cox et al. 2000; Saito et al. 2006). Hub is a tiny cluster of post-mitotic somatic cells localized at the apical tip of the testis and functions in the maintenance of GSC identity and influences its behavior (Ohlstein et al. 2004; Snee and Macdonald 2004). Double-staining of testes with anti-Aub and anti-Piwi confirmed that Aub signals were only detected in germ cells, whereas Piwi signals were restricted to somatic cells in testes (Fig. 1J,M). Namely, expression of these two proteins was mutually exclusive in respect to both cellular localization and cell type.
Comparison of piRNAs associated with Aub and Piwi in fly ovaries
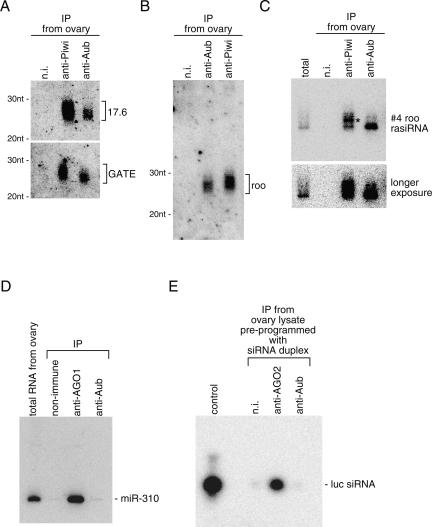
We previously identified small RNAs associated with Aub immunoprecipitated from ovary lysate with the anti-Aub antibody (Gunawardane et al. 2007). Silver staining patterns of protein and RNA components in the immunoprecipitate are shown in Supplemental Figure 2. Western blotting analyses on the immunoprecipitate using anti-AGO3 and anti-Piwi antibodies confirmed that the Aub complex does not contain either protein (Supplemental Fig. 2C). To gain insight into the piRNAs associated with Aub and Piwi in ovaries, we reexamined the compositions of those clones, which are listed in Supplemental Tables 1, 2, and 3. Northern blotting analyses demonstrated that piRNAs associated with Aub seemed also to be associated with Piwi (Fig. 2A–C). A size difference was clearly observed between piRNAs with Piwi and Aub (Fig. 2A–C). This agrees with previous data showing that 32P-end-labeled piRNAs associated with Piwi in fly ovaries contained molecules migrating slightly slower than those associated with Aub on denaturing acrylamide gels (Brennecke et al. 2007; Gunawardane et al. 2007). piRNAs associated with Aub and Piwi most likely contain an equal amount of monophosphate at the 5′-ends (data not shown) and are methylated at their 3′-ends (Saito et al. 2007). Thus, piRNAs with Aub might simply be shorter than those associated with Piwi.
FIGURE 2.
Detection of Aub association with piRNAs. (A) Northern blotting with probes for 17.6 and GATE retrotransposons shows that piRNAs derived from the transposable elements are involved in Aub and Piwi complexes immunopurified from ovary lysate. It should be noted that the Piwi complex contains piRNAs that are apparently a little longer than those within the Aub complex. (B) The same set of samples used in A was probed with a roo retrotransposon sequence. As in A, the fraction of piRNAs with Piwi seems to contain longer molecules compared with those with Aub. (C) An oligo recognizing #4 roo piRNA, originally found to exist in fly ovaries (Vagin et al. 2006), was used for Northern blotting analysis. The anti-Piwi lane clearly contains bands migrating slower than those observed in the total and anti-Aub lanes (an asterisk shows one such prominent band). (D) microRNAs are not loaded onto Aub in ovaries. RNA molecules within the AGO1 and Aub complexes immunopurified from ovary lysate were probed with a DNA oligo to detect miR-310. The band corresponding to miR-310 is only found in the anti-AGO1 lane. It should be noted that no miRNAs were obtained through the experiment identifying the small RNAs within Aub complex in ovaries, indicating that it is most likely that no miRNAs are loaded onto Aub in ovaries. (E) luc siRNA duplex was first added to ovary lysate. After incubation, AGO2 and Aub were immunopurified from the reaction mixture using specific antibodies. RNA molecules were isolated from the complexes and run on a denaturing acrylamide gel. In the anti-AGO2 lane, the luc guide siRNA appears, but not in the anti-Aub and (n.i.) non-immune lanes, indicating that luc guide siRNA is specifically loaded onto AGO2 in the ovary lysate.
We previously reported that miRNAs were not present in Piwi complex immunopurified from ovaries and that siRNAs pre-programmed in ovary lysate were not loaded on Piwi (Saito et al. 2006). We speculated that neither siRNAs nor miRNAs were loaded onto Aub, as was the case for Piwi. To examine this possibility, small RNAs isolated from Aub complex in ovaries were probed with an oligo that specifically recognizes miR-310, one of the miRNAs expressed in fly ovaries. A positive band was detected in just the AGO1 lane (Fig. 2D). Another oligo specifically recognizing miR-186 showed the same result (data not shown). This strongly suggested that Aub does not function together with miRNAs. Indeed, only a trace of miRNAs was obtained (nine of a total 925 Drosophila small RNAs; 0.97%) as Aub-associating small RNAs (Supplemental Table 1). We then assessed if the siRNA duplex of 21 nt in length, a triggering molecule of RNAi mediated by AGO2, was loaded onto Aub as well as AGO2 upon addition to ovary lysate. luc guide siRNA was efficiently loaded onto AGO2 but not onto Aub (Fig. 2E). These data suggested that the 21 nt siRNA loading machinery composed with Dicer-2 and R2D2 (Liu et al. 2003) was unable to load the small RNA onto both Piwi (Saito et al. 2006) and Aub (this study) proteins.
Su(Ste) piRNAs are the most abundant associated with Aub in testes
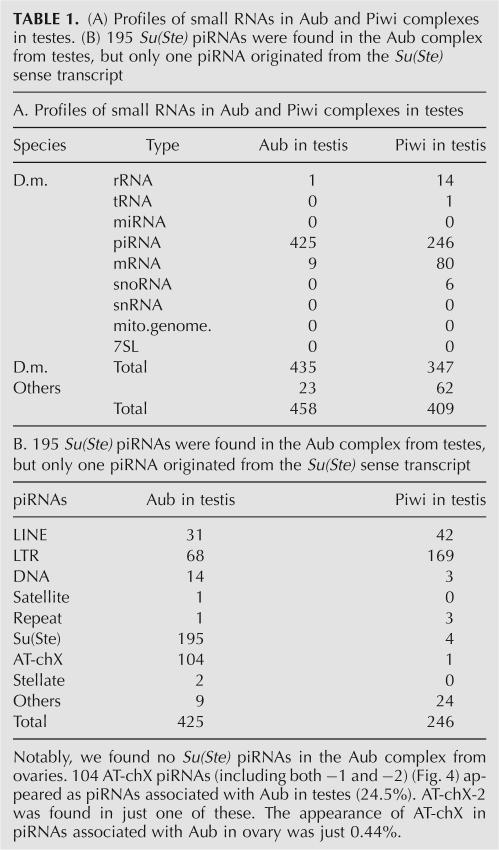
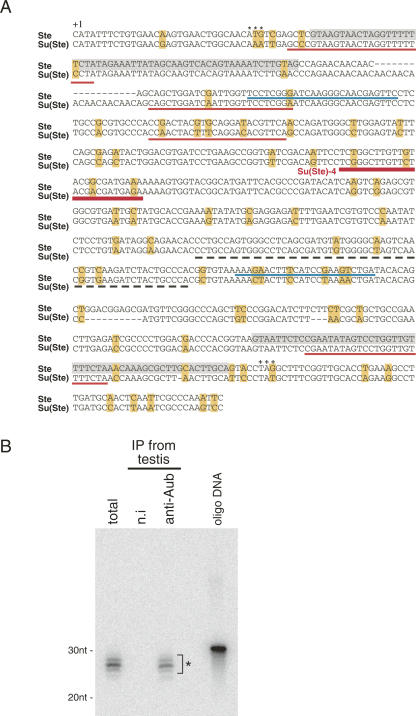
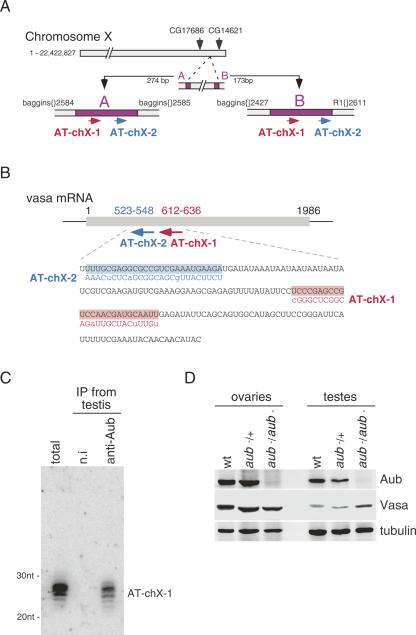
Immunoprecipitation of Aub from testes was also performed using the anti-Aub antibody. As in ovaries, small RNAs 24–30 nt long were observed (Supplemental Fig. 3). A small RNA library was constructed, and 458 clones were sequenced (Table 1). The list of piRNAs obtained is shown in Supplemental Table 4. Although piRNAs associated with Aub in testes also contain those derived from LINE and LTR-type transposons (Table 1) and show a strong preference for U at their 5′-ends (∼87%) (Supplemental Fig. 4), the most abundant class of piRNAs identified was made up of those derived from Su(Ste) antisense transcripts (195 of 425 piRNAs; ∼46%) (Fig. 3A; Table 1; Supplemental Table 4). Consistent with these findings, previously genetic studies have shown that both aub and Su(Ste) mutations cause an accumulation of Stellate mRNA in testes (Bozzetti et al. 1995, Aravin et al. 2004; Vagin et al. 2006). Su(Ste) piRNAs in a sense orientation were found only once in this screening. This bias was apparently not a consequence of any loading bias to Aub because it has been reported that even in total RNAs, Su(Ste) piRNAs in sense are hardly detected in testes (Vagin et al. 2006). piRNAs generated from Stellate sense transcript were also obtained in this experiment (two of 425) (Fig. 3A; Table 1; Supplemental Table 4). Notably, Su(Ste) piRNAs seemed not evenly produced from the precursor molecules (Fig. 3A). Particularly, one Su(Ste) piRNA, termed Su(Ste)-4, was so abundant that it appeared 120 times [including closely related ones such as Su(Ste)-4a shown in Supplemental Table 4] in all the clones sequenced. The association of Su(Ste)-4 with Aub in testes was further confirmed by Northern blotting analysis using a DNA oligo specific for the small RNA (Fig. 3B).
TABLE 1.
(A) Profiles of small RNAs in Aub and Piwi complexes in testes. (B) 195 Su(Ste) piRNAs were found in the Aub complex from testes, but only one piRNA originated from the Su(Ste) sense transcript
FIGURE 3.
Analyses of Stellate and Su(Ste) piRNAs associated with Aub in testes. (A) The nucleotide sequence of Stellate pre-mRNA (accession number: X15899) (Balakireva et al. 1992) was aligned with that of Su(Ste) (accession number: Z11734) (Balakireva et al. 1992). (+1) The transcription start site, (***) the translation start site, and (+++) the stop codon of the Stellate transcript. (Boxes in yellow) Nucleotides unmatched between Su(Ste) and Stellate sequences. (Shadowed areas with gray boxes) The intron sequences. Stellate and Su(Ste) piRNAs identified as being in the Aub complex are underlined in blue and red, respectively. (A thick red underline) Su(Ste)-4 piRNA, the most abundantly found through the experiment. (A black dot line) A particular region in Su(Ste) from which various piRNAs (29 of 425 piRNAs) were originated (Supplemental Table 4). Note that all the Su(Ste) piRNAs obtained in this experiment contained sequences of Su(Ste) antisense transcript. Stellate piRNAs were from the sense mRNA. (B) The Northern blot confirms that the Aub complex immunopurified from testes contains Su(Ste)-4 piRNA (*).
A second large class of piRNAs associated with Aub in testes was made up of those derived from two tiny regions (A, 274 nt; and B, 173 nt) on chromosome X (Fig. 4A). The A and B regions are both located between retrotransposons (Fig. 4A). Interestingly, one of these piRNAs, termed AT-chX-1, whose sequence appeared 103 times in this screening (∼24% of the total piRNAs), showed strong complementarity to vas mRNA (Fig. 4B), a germline-specific transcript involved in oogenesis (Lasko and Ashburner 1988; Styhler et al. 1998). Northern blotting using an oligo specific for AT-chX-1 verified its association with Aub in testes (Fig. 4C). AT-chX-2, another piRNA found in this subgroup, appeared only once in this experiment but also showed high complementarity to vas mRNA (Fig. 4B). Western blotting revealed that the protein levels of VAS in aub −/aub − mutant fly testes were increased by about twofold compared with those of wild-type (wt) and aub −/+ (Fig. 4D; Supplemental Fig. 5A). Mislocalization or ectopic expression of VAS was not observed in aub mutant testes (data not shown). We also found that VAS expression was not increased in piwi mutant testes (Supplemental Fig. 5B). Although it has been shown that the phosphorylation of VAS in ovaries is altered in mutants defective in piRNA production and/or function (Klattenhoff et al. 2007), we found no change in the electrophoretic mobility of VAS in aub mutant and wt testes (Fig. 4D; Supplemental Fig. 5A; data not shown), suggesting that the observed twofold changes in VAS abundance do not reflect an effect on its phosphorylation. In contrast, in ovaries, where little AT-chX-1 and -2 was found in the Aub complex (three of 689), we did not observe such an alteration of VAS levels (Fig. 4D) as reported (Wilson et al. 1996). Taken together, it is suggested that vas expression in testes might possibly be down-regulated by the Aub complex containing AT-chX-1 and -2 through RNA silencing mechanisms (see below).
FIGURE 4.
AT-chX-1 shows strong complementarity to vas mRNA. (A) The length of Chromosome X is 22,422,827 nt (accession number: AE014298). The A and B regions (shown in purple) where AT-chX-1 and -2 are coded span from 21,765,976 to 21,766,249 (274 nt) and from 21,773,609 to 21,773,781 (173 nt), respectively. The distance of the A and B regions is 7360 nt. AT-chX-1 in both regions corresponds to the sequences from 21,766,034 to 21,766,058 and from 21,773,667 to 21,773,691, respectively. AT-chX-2 in the A and B regions is from 21,766,122 to 21,766,147 and 21,773,775 to 21,773,780, respectively. The A and B regions are flanked by baggins{}2584 and baggins{}2585, and baggins{}2427 and R1{}2611, respectively. CG17686 and CG14621 are protein-coding genes located close to the A and B regions. (B) Schematic drawing of the vas mRNA. Nucleotide sequences of a partial vas mRNA (black) (173 nt; a region showing strong complementarity to the B region in A [∼81% identity] is shown) (accession number: NM165103), AT-chX-1 (red), and AT-chX-2 (blue) are indicated. Lowercase letters show nucleotides mismatched between vas mRNA and AT-chX-1 or AT-chX-2. Along with AT-chX-1, AT-chX-2 is also found in the Aub-associated small RNA identification experiment, but appeared just once. In contrast, AT-chX-1 appeared 103 times in 425 piRNA clones (∼24%). (C) Association of AT-chX-1 with Aub was confirmed by Northern blotting. The probe used was a DNA oligo containing a sequence complementary to AT-chX-1. (D) The protein levels of VAS are increased in the aub mutant fly testes. Ovary and testis lysates of wild-type (wt), aub − _/_+ and aub − /aub − were each probed with anti-Aub, anti-VAS, and anti-tubulin. In testes, VAS protein levels in aub − /aub − are about twofold compared with those in aub −/+ and wt, whereas they are not changed in the ovary lysates with or without Aub. This figure shows representative data from three individual experiments. Anti-tubulin was used for normalization. (wt) yellow white; (aub −/+) aubHN2/+; (aub − /aub −) aubHN2/aubQC42.
Piwi and piRNA association in testes
We also carried out a parallel examination of small RNAs residing within the Piwi complex in testes (Table 1; Supplemental Fig. 3; Supplemental Table 5). We detected only a few Su(Ste) piRNAs (four of 246 piRNAs) and no AT-chX-1 and -2 as Piwi-associating small RNAs (Table 1; Supplemental Table 5), which suggested that Piwi hardly contributes to the repression of Stellate and vas gene expression. It is, however, worth noting that the loss of Piwi in testes increases the production of Su(Ste) piRNAs (Vagin et al. 2006). The majority of the small RNAs associated with Piwi in testes were piRNAs derived from transposable elements and other repetitive sequences in the genome, as observed in ovaries (Saito et al. 2006). Interestingly, pieces of protein-coding sense mRNAs were conspicuous in this screening (80 of 347 small RNAs present in the Piwi complex in testes) (Table 1; Supplemental Table 5) as has been reported in the small RNA profiling study done for MIWI, a Piwi homolog expressed in mouse (Girard et al. 2006). However, the significance of the association of Piwi/MIWI in testes with small RNAs originating from sense mRNAs remains unclear.
Immunoprecipitated Aub–piRNA complexes from ovaries and testes show Slicer activity
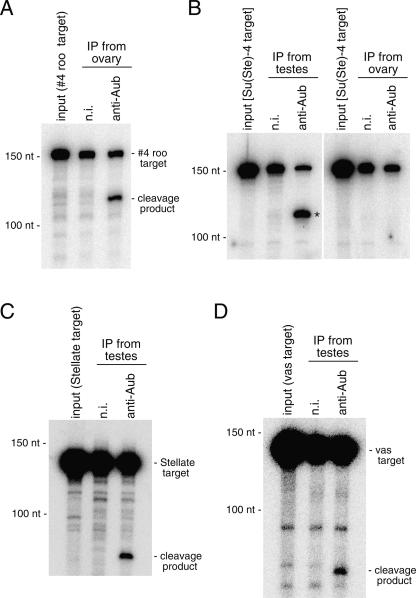
Previously, we demonstrated that Drosophila AGO1 and AGO2 exhibit Slicer activity in vitro (Miyoshi et al. 2005). Moreover, we recently showed that recombinant Piwi, Aub, and AGO3 produced in E. coli also exhibit Slicer activity (Saito et al. 2006; Gunawardane et al. 2007). We then assessed if the Aub–piRNA complexes immunopurified from ovaries and testes could, indeed, show the target RNA cleavage activity in vitro. A target RNA containing a sequence perfectly complementary to #4 roo piRNA, one of the piRNAs residing in the Aub complex in ovaries (Fig. 2C), was produced and used in the assays. As shown in Figure 5A, the #4 roo target RNA was efficiently cleaved with Aub complex immunopurified from ovaries, indicating that endogenous Aub residing in fly gonads also has Slicer activity. Similar experiments were carried out using Aub complex immunopurified from testes. An RNA target containing a sequence completely matching Su(Ste)-4 piRNA [Su(Ste)-4 target] or part of the Stellate sense mRNA (Stellate target), which should be recognized by the Aub–Su(Ste)-4 complex, was efficiently cleaved, as expected (Fig. 5B,C). These results are consistent with data demonstrating that mutations in aub cause an increase in both Stellate mRNA and sense Su(Ste) RNA (Schmidt et al. 1999; Aravin et al. 2001; Vagin et al. 2006). Under the same conditions, Aub complex obtained from ovaries, where Su(Ste) transcripts are presumably unexpressed, did not cleave the Su(Ste)-4 target (Fig. 5B). In fact, Su(Ste)-4 piRNA did not appear at all in the experiment identifying small RNAs in Aub complex in ovaries (Supplemental Table 1). Although the activity was relatively weak, a target RNA containing a sequence of vas mRNA showing complementarity to AT-chX-1 was also cleaved by Aub complex immunopurified from testes (Fig. 5D). These results imply that vas is a candidate for in vivo target genes for Aub–piRNA complexes in testes.
FIGURE 5.
The Aub-piRNA complexes show Slicer activity. (A) In vitro target RNA cleavage assay using a target containing a sequence complementary to #4 roo piRNA (Vagin et al. 2006) (#4 roo target). #4 roo target was radiolabeled with 32P at the 5′-end. The Aub complex immunopurified from ovaries was incubated with #4 roo target, and after the reaction RNAs were prepared and analyzed on a gel. The Aub complex shows its ability to cleave the target, whereas the control (n.i.) shows no such activity. (B) In vitro target RNA cleavage assay was carried out using the Aub complex immunopurified from testes and ovaries. An RNA target, Su(Ste)-4 target, which has a sequence completely complementary to Su(Ste)-4 piRNA was produced and used. An expected cleavage product (*) is observed in the Aub complex from testes, but not in that from ovaries, indicating that the Aub complex from ovaries does not have the ability to cleave the particular target RNA. (C) An RNA target that has a partial sequence of Stellate mRNA was used in the cleavage assay. The Aub complex used was immunopurified from testes. (D) An RNA target that has a partial sequence of vas mRNA was also cleaved with the Aub complex from testes.
DISCUSSION
The present study together with our previous findings (Saito et al. 2006; Gunawardane et al. 2007) clearly demonstrated that both Piwi and Aub, the Drosophila Argonaute members specifically expressed in gonads, are associated with a variety of piRNAs in both ovaries and testes.
Although Piwi and Aub both associate with piRNAs, we also see clear differences between them in terms of which cells in fly gonads are expressed and where in the cells they are localized. By detecting endogenous Piwi and Aub using the specific antibodies we produced, it was found that all germ cells including GSC in ovaries simultaneously express both proteins, but their cellular localization is clearly distinct from each other; Piwi is found just in the nucleus (Saito et al. 2006) and Aub in the cytoplasm (Fig. 1). In somatic cells such as cap cells, however, only Piwi seems to be expressed (Fig. 1; Saito et al. 2006). In testes, we found that Piwi is only expressed in somatic cells, but not in GSC and gonialblast/spermatogonia (Saito et al. 2006), where we found Aub is specifically expressed (Fig. 1). This suggests that at least in testes, Piwi and Aub function in different RNA silencing pathways. Even in ovaries, it has been shown that the loss of Piwi or Aub causes specific defects; for example, piwi1 ovarioles were mostly devoid of germline cells (Lin and Spradling 1997), Aub was shown to be required in the germline for the production of functional oocytes (Harris and Macdonald 2001), and aub mutations caused de-repression of retrotransposons (Vagin et al. 2004, 2006). Thus it is clear that Aub does not sufficiently compensate for the loss of Piwi function and vice versa. This is even though the two proteins are coexpressed in the same germ cells and many piRNAs are shared by both proteins.
Although many studies have suggested connections between Aub and the silencing of the Stellate gene, including the disappearance of piRNAs originating from Su(Ste) transcripts in aub mutants (Aravin et al. 2001; Vagin et al. 2006), this study is the first to show that Aub in testes is physically associated with Su(Ste) piRNAs and that the Aub–piRNA complex from testes is able to cleave Stellate mRNA. The Aub–piRNA complex was also able to cleave a target RNA, showing complete complementarity to one of the Su(Ste) piRNAs, Su(Ste)-4, demonstrating that the Aub–piRNA complex can act in RNA silencing both on the locus piRNA is derived from and loci that show similarity to the piRNA. We found that two piRNAs, AT-chX-1 and -2, originating from tiny repetitive regions located between retrotransposons on chromosome X, showed considerable complementarity to vas mRNA. Cleavage on the target RNA sequence in the RNAi pathway has been shown to occur between the +10 and +11 positions of the guide siRNA (the 5′-end of the guide siRNA is assigned as +1) (Elbashir et al. 2001; Schwarz et al. 2004). Base-pairings between siRNA and the target RNA at the +10 and +11 positions is required for the cleavage. If these roles were also applicable to piRNAs, we speculated that AT-chX-1 (and -2) would possibly guide cleavage of vas mRNA. Indeed, the vas target was cleaved by the Aub complex purified from testes (Fig. 5D), although the cleavage efficiency was relatively low compared with that for Stellate mRNA. In agreement with this observation, the levels of vas mRNA were moderately increased by loss of Aub expression, whereas those of Stellate mRNA were severely diminished under the same conditions (Supplemental Fig. 5C). The Aub–piRNA complex may also repress the translation of vas mRNAs, just as Argonaute–miRNA complexes repress their mRNAs in various circumstances (Ambros 2004; Du and Zamore 2005; Kloosterman and Plasterk 2006). It has been reported that spindle-E (spnE) mutations cause piRNAs not to be accumulated in gonads, as in the case of aub mutations (Savitsky et al. 2006; Vagin et al. 2006). It should be noted that similar effects on derepression of vas gene expression were also observed in testes of spnE mutants (data not shown), in which the amounts of AT-chX-1 are markedly reduced (data not shown). As well, we found that vas overexpression (EGFP-VAS transgenic flies) (Sano et al. 2002) results in oversized apexes in testes, where spermatogonial cells and spermatocytes are located (Supplemental Fig. 5D). Interestingly, aub mutants also showed similarly oversized apexes in the testes. These results indicate a connection between such phenotypes and increased levels of VAS proteins. At present, we do not know what are the biological consequences of vas gene silencing in testis, but our data support a model in which vas gene expression is regulated in male germ cells by Aub in concert with the piRNAs.
Notably, >70% of Aub-associated piRNAs in testes were either Su(Ste) piRNAs or AT-chXs. In contrast, Piwi-associated piRNAs in testes are mostly derived from these piRNAs. At present, we do not know where this strong bias comes from. Piwi and AGO2 are not required for Stellate silencing (Vagin et al. 2006). This study showed that siRNAs and miRNAs were not loaded onto Aub (Fig. 2D,E). These findings support the view that a distinct pathway exists for loading of piRNAs onto Aub in testes. This strong bias could simply be due to a lack of Piwi expression in primary spermatocytes where Su(Ste) is expressed (Aravin et al. 2004). Given the model of piRNA biogenesis (Brennecke et al. 2007; Gunawardane et al. 2007), identification of AGO3-associated piRNAs in testes will shed light on the mechanism of the preferential association of Su(Ste) piRNAs and AT-chXs with Aub.
In this study, we fortuitously encountered a type of piRNA that originated from non-annotated, intergenic regions and was likely involved in triggering the silencing of protein-coding mRNA targets in collaboration with Aub. More comprehensive biochemical characterization of piRNAs associated with Aub, Piwi, and/or AGO3 in Drosophila should shed light on the biological processes involved in the production of the gametes necessary for development of new individuals and thus species perpetuation.
MATERIALS AND METHODS
Drosophila strains
The y w strain was used as a wild type. The strains bearing aub mutations, aubHN2 and aubQC42, were provided by P. Zamore (University of Massachusetts). Females of aubHN2 and males of aubQC42 were crossed to yield aub heterozygote flies, aubQC42/aubHN2.
Western blot analysis
Two hundred amino acids at the N terminus of Aub fused with GST were used as the antigen to immunize mice. Anti-Aub monoclonal antibodies were produced essentially as described previously (Ishizuka et al. 2002). Western blotting was performed as described previously (Miyoshi et al. 2005). Ten micrograms of proteins from each sample were loaded on gels (Figs. 1A, 4D). Anti-tubulin was from the Developmental Studies Hybridoma Bank (1:1000 dilution). Anti-VAS was provided by A. Nakamura (Kobe-RIKEN CDB) and used at 1:2500 dilution.
Immunofluorescence
Testes and ovaries were dissected manually from adult flies in 1× PBS. Embryos were collected and dechorionated. Immunostaining was performed following standard procedures. Anti-Aub was purified from culture supernatants of hybridoma cells using Thiophilic-Superflow Resion (BD Biosciences) and directly labeled using a HiLyte Fluor 555 Labeling Kit-NH2 (Dojindo Molecular Technologies). Culture supernatants of anti-Piwi hybridoma cells were used without dilution. Alexa 488-conjugated anti-mouse IgG (Molecular Probe) was used as the secondary antibody to detect the anti-Piwi antibody. All images were collected using a Zeiss LSM510 laser scanning microscope.
Small RNA cloning and sequence analysis
Cloning of small RNAs associated with Aub in ovaries and testes and Piwi in testes was carried out essentially as described (Saito et al. 2006) with minor modifications. Immunoprecipitation was performed essentially as described previously (Miyoshi et al. 2005). Immunoprecipitation buffer contained 30 mM HEPES-KOH (pH 7.3), 150 mM KOAc, 2 mM MgOAc, 5 mM DTT, 0.1% NP-40, 2 μg/mL Pepstatin, 2 μg/mL Leupeptin, and 0.5% Aprotinin. About 300 ovaries or about 1000 testes were used per immunoprecipitation. After immunoprecipitation, total RNAs were isolated from the immunopurified complexes with phenol:chloroform and precipitated with ethanol. RNAs were dephosphorylated with CIP (NEB) and labeled with [γ-32P]ATP with T4 polynucleotide kinase (TaKaRa) for visualization. For cloning of small RNAs isolated from Aub complex in ovaries, we used adapters and primers (i.e., RT and PCR primers) as described by Saito et al. (2006). Adapters (MI-5′ Linker and MI-3′ Linker) and primers used for small RNAs associated with Aub and Piwi in testes were included in the DynaExpress miRNA Cloning Kit (BioDynamics Laboratory). The sequences of each oligo were as follows:
- MI-3′ Linker: 5′-pCTGTAACTCGGGTCAATddC-3′ (DNA);
- MI-5′ Linker: 5′-AUCGUCUCGGGAUGAAA-3′(RNA);
- 3′ RT primer: 5′-ATTGACCCGAGTTACAG-3′ (DNA);
- 5′ Primer: 5′-ATCGTCTCGGGATGAAA-3′ (DNA).
First-strand cDNA synthesis was performed with Stratascript RT (Stratagene) or Reverse Transcriptase (BioDynamics Laboratory). KOD plus (TOYOBO) or Ex Taq polymerase (TaKaRa) was used as the polymerase. PCR products were cloned into the EcoRV site of the pBS SK+ vector (for small RNAs associated with Aub in ovaries) or pTAC-1 (for small RNAs associated with Aub or Piwi in testes) (BioDynamics Laboratory) and sequenced. RNA sequences between 5′ and 3′ adapters were analyzed by undertaking searches in annotated genomic databases (UCSC: http://genome.ucsc.edu/cgi-bin/hgBlat; NCBI: http://www.ncbi.nlm.nih.gov/blast/; and FlyBase: http://flybase.bio.indiana.edu/blast/) to determine whether the cloned RNAs map to the genome. We only used the best matches up to a maximum of two differences (mismatch, insertion, or deletion) for each small RNA sequence. For repeat annotation, we used results from the RepeatMasker program (UCSC BLAT), NCBI BLAST, or FlyBase BLAST.
Northern blot analysis
Total RNA either from fly ovaries/testes or immunoprecipitated complexes was isolated using ISOGEN (Nippon Gene). Five micrograms of total RNA were used in Figures 2, C and D, 3B, and 4C. Northern blotting was performed as reported previously (Saito et al. 2006). Probes used for roo piRNA and miR-310 were as described previously (Saito et al. 2006). Probes used for detecting #4 roo piRNA, Su(Ste)-4 and AT-chX-1 were as follows:
- For #4 roo piRNA: 5′-TCGACTCAGTGGCACAATAAAT-3′;
- for Su(Ste)-4: 5′-TCGGGCTTGTTCTACGACGATGAGA-3′;
- for AT-chX-1: 5′-GCCCGAGCCGTCTAACGATGAAACA-3′.
DNA fragments for detecting piRNAs that originated from 17.6 (accession number: XD1472) and GATE (accession number: AJ010298) retrotransposons were obtained by PCR on the Drosophila melanogaster genomic DNA and cloned into a pBS SK+ vector. The primers used are as follows:
- For 17.6: 5′-ATTCTTGTAAACAAATCTTA-3′ and 5′-CAGCCTCTCACAAATTCAAT-3′;
- for GATE: 5′-TCGAGCAGGGGCACTTCTCAGTC-3′ and 5′-ACTATGGCATGCGCGTCGCCA-3′.
PCR was again performed using primers for the T7 and T3 promoter sequences on each construct, and the PCR products were used as templates for in vitro transcription using MAXIscript T7 and T3 kits (Ambion) in the presence of 32P-UTP. RNAs transcribed were extracted with phenol:chloroform, precipitated with ethanol, and partially hydrolyzed as described previously (Saito et al. 2006).
siRNA loading assay
Three hundred ovaries were homogenized in a hypotonic buffer (30 mM HEPES-KOH at pH 7.4, 2 mM MgOAc, 5 mM DTT, and 1 mg/mL Pefablock SC) to prepare ovary lysate. luc siRNA duplex (Miyoshi et al. 2005) was incubated in the lysate for 1 h at 26°C. Of the luc siRNA duplex, only guide siRNA was labeled with [γ-32P]ATP for visualization. Aub and AGO2 complexes were then immunopurified from the mixture using the specific antibodies. Sodium chloride was added to the lysates to 800 mM just before immunoprecipitation was started. Reaction mixtures were rocked for 1 h at 4°C, and the beads were washed extensively with a washing buffer (30 mM HEPES-KOH at pH 7.4, 800 mM NaCl, 2 mM MgOAc, 5 mM DTT, 1 mg/mL Pefablock SC). RNAs were isolated from the immunoprecipitates, separated on a denaturing polyacrylamide gel, and visualized on BAS-2500 (Fuji).
In vitro target RNA cleavage assay
Target RNA cleavage assays using immunoprecipitates were performed in a reaction buffer containing 30 mM HEPES-KOH (pH 7.4), 100 mM KOAc, 2 mM MgOAc, 5 mM DTT, 10 mM creatine phosphate, 0.5 mM ATP, 30 μg/mL creatine kinase (Roche), 0.1 U/mL RNasin (Promega), and 0.5 μg of yeast RNA (Ambion). After reaction for 3 h at 26°C, RNAs were isolated from the whole reaction mixtures, run on gels, and visualized on the BAS 2500. To make a target harboring a sequence completely complementary to miR-310, a short DNA fragment produced by annealing a set of oligo DNAs (5′-AGCTTAAAGGCCGGGAAGTGTGCAATAA-3′ and 5′-AGCTTTATTGCACACTTCCCGGCCTTTA-3′) was inserted in a pBS SKII+ vector digested with HindIII (pBS310). To make a target harboring a sequence completely complementary to #4 roo piRNA (#4 roo piRNA target), a short DNA fragment produced by annealing a set of oligo DNAs (5′-GATCCATAGTCGACTCAGTGGCACAATAAATAAAGGC-3′ and 5′-GGCCGCCTTTATTTATTGTGCCACTGAGTCGACTATG-3′) was inserted in a pBS310 vector digested with BamHI and NotI. To make a target harboring a sequence completely complementary to Su(Ste)-4 piRNA [Su(Ste)-4 target], a short DNA fragment produced by annealing a set of oligo DNAs (5′- GATCTCGGGCTTGTTCTACGACGATGAGA-3′ and 5′- GGCCTCTCATCGTCGTAGAACAAGCCCGA-3′) was inserted in a pBS310 vector digested with BamHI and NotI. To make a target harboring a partial sequence of Stellate sense mRNA (Stellate target), a short DNA fragment produced by annealing a set of oligo DNAs (5′-AATTCTCTGGCTTGTTGTACGGCGATGAAAG-3′ and 5′-GATCCTTTCATCGCCGTACAACAAGCCAGAG-3′) was inserted into a pBS SK+ vector digested with BamHI and EcoRI. To make a target harboring a partial sequence of vas mRNA (vas target; as a target for AT-chX-1), a short DNA fragment produced by annealing a set of oligo DNAs (5′-TTCCTCCCGAGCCGTCCAACGATGCAATTGAGA-3′ and 5′-TCTCAATTGCATCGTTGGACGGCTCGGGAGGAA-3′) was inserted in a pBS SKII+ vector that was digested with BamHI and NotI and blunted. PCR was again performed using primers for the T7 and T3 promoter sequences, and the PCR products were then used as templates for in vitro transcription using a MEGAscript T7 kit (Ambion). The resultant RNAs were radiolabeled at the 5′-G cap by guanylyltransferase (Ambion) as previously described (Okamura et al. 2004) or with [γ-32P]ATP using T4 polynucleotide kinase and gel-purified. vasa target (Fig. 5D) was labeled with pCp at the 3′-end and gel-purified before use.
Accession numbers
The accession numbers are: Aub-associated piRNAs in ovary, AB296387-AB297014; Aub-associated piRNAs in testis, AB297015-AB297190; and Piwi-associated piRNAs in testis, AB297191-AB297373.
SUPPLEMENTAL DATA
Supplemental materials are available upon request by sending an e-mail message containing the keyword “Aub-supplement” to siomim@genome.tokushima-u.ac.jp.
ACKNOWLEDGMENTS
We thank P. Zamore for providing the aub mutant stocks aubHN2 and aubQC42 and A. Nakamura for the anti-VAS antibody and technical advice. We also thank H. Kose, T. Suzuki, and V. Vagin for technical advice. We thank members of the Siomi laboratory for discussion and comments on the manuscript. K.S. is a postdoctoral fellow of the 21st Century COE Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). This study was supported in part by grants to M.C.S. and H.S. from MEXT and the New Energy and Industrial Technology Development Organization (NEDO).
Footnotes
REFERENCES
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Klenov, M.S., Vagin, V.V., Bantignies, F., Cavalli, G., Gvozdev, V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakireva, M.D., Shevelyov, Y.Y., Nurminsky, D.I., Livak, K.J., Gvozdev, V.A. Structural organization and diversification of Y-linked sequences comprising Su(Ste) genes in Drosophila melanogaster . Nucleic Acids Res. 1992;20:3731–3736. doi: 10.1093/nar/20.14.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzetti, M.P., Massari, S., Finelli, P., Meggio, F., Pinna, L.A., Boldyreff, B., Issinger, O.G., Palumbo, G., Ciriaco, C., Bonaccorsi, S., et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the β subunit of casein kinase 2. Proc. Natl. Acad. Sci. 1995;92:6067–6071. doi: 10.1073/pnas.92.13.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Aravin, A.A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell. 2007;128:1–15. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Cox, D.N., Chao, A., Baker, J., Chang, L., Qiao, D., Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.N., Chao, A., Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Du, T., Zamore, P.D. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Eddy, E.M. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;20:1993–1997. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley, S.D., Tamanaha, M., Clegg, N.J., Ruohola-Baker, H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Forstemann, K., Horwich, M.D., Wee, L., Tomari, Y., Zamore, P.D. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, A., Sachidanandam, R., Hannon, G.J., Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Gunawardane, L.S., Saito, K., Nishida, K.M., Miyoshi, K., Kawamura, Y., Nagami, T., Siomi, H., Siomi, M.C. A Slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila . Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris, A.N., Macdonald, P.M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., Siomi, M.C., Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova, A.I., Klenov, M.S., Gvozdev, V.A. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell, J.R., Yamaguchi, S., Carthew, R.W. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes & Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., Lee, Y.S., Carthew, R.W. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2006;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff, C., Bratu, D.P., McGinnis-Schultz, N., Koppetsch, B.S., Cook, H.A., Theurkauf, W.E. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kloosterman, W.P., Plasterk, R.H.A. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lasko, P.F., Ashburner, M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Leuschner, P.J., Ameres, S.L., Kueng, S., Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H., Spradling, A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Mahowald, A.P., Strassheim, J.M. Intercellular migration of centrioles in the germarium of Drosophila melanogaster. An electron microscopic study. J. Cell Biol. 1970;45:306–320. doi: 10.1083/jcb.45.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga, C., Tomari, Y., Shin, C., Bartel, D.P., Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H., Siomi, M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein, B., Kai, T., Decotto, E., Spradling, A. The stem cell niche: Theme and variations. Curr. Opin. Cell Biol. 2004;16:693–699. doi: 10.1016/j.ceb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Ishizuka, A., Siomi, H., Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.S., Barford, D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem. Sci. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Rand, T.A., Peterson, S., Du, F., Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Saito, K., Nishida, K.M., Mori, T., Kawamura, Y., Miyoshi, K., Nagami, T., Siomi, H., Siomi, M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Sakaguchi, Y., Suzuki, T., Suzuki, T., Siomi, H., Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes & Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, H., Nakamura, A., Kobayashi, S. Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster . Mech. Dev. 2002;112:129–139. doi: 10.1016/s0925-4773(01)00654-2. [DOI] [PubMed] [Google Scholar]
- Sarot, E., Payen-Groschene, G., Bucheton, A., Pelisson, A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky, M., Kwon, D., Georgiev, P., Kalmykova, A., Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes & Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., Palumbo, G., Bozzetti, M.P., Tritto, P., Pimpinelli, S., Schafer, U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster . Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Tomari, Y., Zamore, P.D. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Snee, M.J., Macdonald, P.M. Live imaging of nuage and polar granules: Evidence against a precursor–product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 2004;117:2109–2120. doi: 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- Styhler, S., Nakamura, A., Swan, A., Suter, B., Lasko, P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Szakmary, A., Cox, D.N., Wang, Z., Lin, H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Zamore, P.D. Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., Zamore, P.D. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Du, T., Zamore, P.D. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, V.V., Klenov, M.S., Kalmykova, A.I., Stolyarenko, A.D., Kotelnikov, R.N., Gvozdev, V.A. The RNA interference proteins and Vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster . RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- Vagin, V.V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Williams, R.W., Rubin, G.M. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Connell, J.E., Macdonald, P.M. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D. RNA silencing: Genomic defence with a slice of pi. Nature. 2007;19:864–865. doi: 10.1038/446864a. [DOI] [PubMed] [Google Scholar]
- Zaratiegui, M., Irvine, D.V., Martienssen, R.A. Noncoding RNAs and gene silencing. Cell. 2007;23:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]