Distinctive patterns of microRNA expression in primary muscular disorders (original) (raw)
Abstract
The primary muscle disorders are a diverse group of diseases caused by various defective structural proteins, abnormal signaling molecules, enzymes and proteins involved in posttranslational modifications, and other mechanisms. Although there is increasing clarification of the primary aberrant cellular processes responsible for these conditions, the decisive factors involved in the secondary pathogenic cascades are still mainly obscure. Given the emerging roles of microRNAs (miRNAs) in modulation of cellular phenotypes, we searched for miRNAs regulated during the degenerative process of muscle to gain insight into the specific regulation of genes that are disrupted in pathological muscle conditions. We describe 185 miRNAs that are up- or down-regulated in 10 major muscular disorders in humans [Duchenne muscular dystrophy (DMD), Becker muscular dystrophy, facioscapulohumeral muscular dystrophy, limb-girdle muscular dystrophies types 2A and 2B, Miyoshi myopathy, nemaline myopathy, polymyositis, dermatomyositis, and inclusion body myositis]. Although five miRNAs were found to be consistently regulated in almost all samples analyzed, pointing to possible involvement of a common regulatory mechanism, others were dysregulated only in one disease and not at all in the other disorders. Functional correlation between the predicted targets of these miRNAs and mRNA expression demonstrated tight posttranscriptional regulation at the mRNA level in DMD and Miyoshi myopathy. Together with direct mRNA–miRNA predicted interactions demonstrated in DMD, some of which are involved in known secondary response functions and others that are involved in muscle regeneration, these findings suggest an important role of miRNAs in specific physiological pathways underlying the disease pathology.
Keywords: skeletal muscle, muscular dystrophies, inflammatory myopathies
Primary muscle disorders involve different groups of diseases, including the muscular dystrophies, inflammatory myopathies, and congenital myopathies. The diseases are defined and classified in accordance with their clinical and pathological manifestations and the distribution of predominant muscle weakness.
The muscular dystrophies are the largest heterogeneous group of >30 different inherited disorders characterized by muscle wasting and weakness of variable distribution and severity, manifesting at any age from birth to middle years, and resulting in significant morbidity and disability (1). Whereas the most characterized forms involve mutations within genes encoding structural members of the dystrophin-associated glycoprotein complex of the muscle membrane cytoskeleton, other mutations interfere with mRNA processing, alter protein posttranslational modifications, or modify enzymatic activities.
Abnormalities of dystrophin are known as the most common cause of muscular dystrophy, accounting for both Duchenne muscular dystrophy (DMD), one of the most severe types with rapidly progressive skeletal muscle weakness, and the milder Becker muscular dystrophy (BMD) phenotype (2). The highly heterogeneous limb girdle muscular dystrophies (LGMDs) (3) is another major group of muscular dystrophies. Notably, mutated calpain-3 in patients with LGMD type 2A (LGMD2A) was the first enzyme, rather than structural protein, to be associated with muscular dystrophy (4). Mutations in dysferlin, a muscle membrane protein that plays a role in membrane repair, cause the LGMD type 2B (LGMD2B) and Miyoshi myopathy (MM) (5). Facioscapulohumeral muscular dystrophy (FSHD), a progressive muscle disease affecting mainly the muscles of the face and upper arms caused by deletions of a 3.3-kb repeat region located on 4q35.2 (6), is an additional common type of muscular dystrophy.
Among the group of congenital myopathies, nemaline myopathy (NM) is the most common nondystrophic congenital myopathy and is characterized by relatively nonprogressive proximal weakness of often, but not always, congenital onset and the presence of nemaline rod structures in the affected myofibers (7). Mutations in six different genes encoding the thin filament proteins and other skeletal muscle proteins account for the majority of disease cases.
Clinical and histopathologic overlap between the inherited muscular disorders, and the distinct idiopathic inflammatory myopathies is also being increasingly recognized (8). Polymyositis (PM), the most common of the inflammatory myopathies, is a T cell-mediated pathology in which a cellular immune response is a key feature in promoting muscle damage. Inclusion body myositis (IBM) is suspected to be a primary inflammatory myopathy, like dermatomyositis (DM) and PM, or a primary degenerative myopathic disorder, such as a dystrophy with secondary inflammation (9). The general distinction between immune-mediated and nonimmune-mediated muscle diseases becomes less defined as more is learned of the complex, underlying pathogenic mechanisms in both inflammatory myopathies and muscular dystrophies.
Currently, although the number of genes identified increases every year, adding to our understanding and revealing the overall complexity of the pathogenesis of the various muscular disorders, and despite the well documented histological pathology of dystrophic tissue, the underlying molecular pathways remain poorly understood, and the decisive secondary factors responsible for the variability in the clinical phenotypes are still mainly unknown. Gene expression profiling of human and mouse normal and diseased skeletal muscle has generated more detailed insight in the molecular process underlying the different conditions (10–14). However, although each of these studies has identified a number of genes in various functional categories that are differentially expressed in the disease states, the substantial underlying disease mechanisms remain to be elucidated.
MicroRNAs (miRNAs) are a class of small, endogenous noncoding RNA molecules that posttranscriptionally regulate gene expression. Several hundred mammalian miRNAs have been identified, many of which are tissue-specific and/or temporally regulated in their expression (15). The function of only a small fraction of these has been described in detail and point to their involvement in a variety of developmental and physiological processes (16, 17).
Not surprisingly, miRNAs have been shown to play an important role in the regulation of muscle development. miRNA-1 and miRNA-133 are expressed in cardiac and skeletal muscle and are transcriptionally regulated by the myogenic differentiation factors MyoD, Mef2, and SRF (18–21). In Drosophila, deletion of the single miRNA-1 gene results in a defect in muscle differentiation or maintenance (20, 21). In contrast, overexpression of miRNA-1 in mouse cardiac progenitors has a negative effect on proliferation, where it targets the transcription factor Hand2, involved in myocyte expansion (21). Similar to the heart, miRNA-1 overexpression in cultured skeletal myoblasts promotes skeletal muscle differentiation, as does the related but skeletal muscle-specific miR-206 (18, 22) that has also been shown to mediate MyoD-dependent inhibition of follistatin-like-1 and Utrophin genes in myoblasts (23).
In light of their involvement in modulating cellular phenotypes, we hypothesized that miRNAs might be involved in the regulation of the pathological pathways leading to muscle dysfunction, and they might be different among the different pathways that lead to myofiber degeneration. In the present study, we describe a comprehensive miRNA expression profile in muscle tissues from a broad spectrum of primary muscle disorders, aiming to identify new or modifying elements involved in the regulatory networks of muscle and interpret these results within the framework of previous mRNA expression analysis that allowed the examination of the molecular pathophysiological pathways of dystrophic muscle. Further analyses of the overall differentially expressed miRNAs were applied to select potential target genes and unravel biological signaling pathways, potentially targeted by these miRNAs.
Results
Overview.
To identify miRNAs that might be involved in the secondary pathological pathways in various muscle diseases and discriminate between the miRNAs possibly involved in the underlying pathways, either specific to a given disease or shared among disease types, we have carried out a comparative miRNA expression profiling across a panel of 10 different groups of muscle disorders (DMD, BMD, FSHD, LGMD2A, LGMD2B, MM, NM, IBM, DM, and PM) with different clinical and pathological characteristics [detailed in supporting information (SI) Fig. 4 and SI Table 2) and unaffected human skeletal muscle.
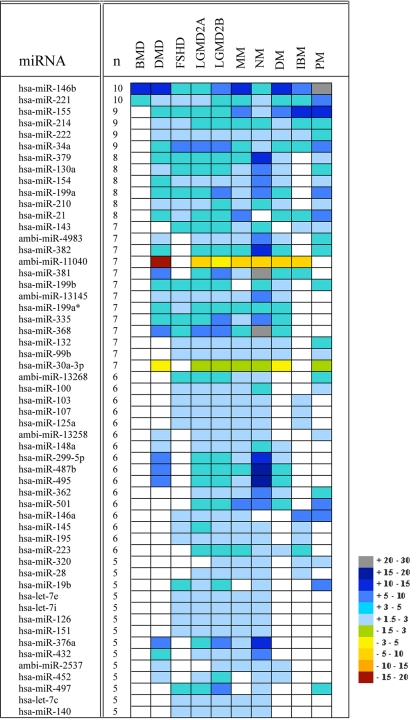
miRNA expression microarrays containing 428 human miRNAs from the miRBase database and novel human miRNA (Ambi-miRs) were used in this study. In total, a subset of 185 human miRNAs, corresponding to 43% of human miRNA probes present on the array, were found to be differentially expressed at a significant level (P < 0.05, false discovery rate < 0.05) in at least one of the 10 muscle conditions compared with the control panel (SI Table 3). Interestingly, of the differentially expressed miRNAs, most were up-regulated in the different diseases (39 in DMD, 62 in FSHD, 88 in LGMD2A, 87 in LGMD2B, 69 in MM, 140 in NM, 20 in IBM, 37 in PM, and 35 in DM) as compared with normal muscle tissue. Whereas a total of 151 different miRNA genes were found to be consistently up-regulated relative to the control, only 28 miRNAs were down-regulated among the various conditions. Overlying this broad commonality is the up-regulation of specific miRNAs and the specific down-regulation of others that allows us to assign a distinctive signature to each of the 10 conditions (Fig. 1 and SI Table 4).
Fig. 1.
miRNAs common to various muscular disorders. The list includes 55 commonly dysregulated miRNAs in five or more types of muscular disorders. A color scale represents the relative intensity of the expression signal by means of fold change compared with the control group, with gray indicating high expression and red low expression. For a complete list see SI Table 4.
In addition, a set of six miRNAs (30b, 92, 361, 423, 29a, and 29b), was found to be expressed in an inconsistent pattern in few of the conditions (DMD, NM, FSHD, and LGMD2B) such that in one disease the miRNA is down-regulated, whereas in others it is up-regulated, and vice versa (SI Table 4). This finding might point out the differences in the pathology and genes involved in the different regulated networks.
Among the 185 differentially expressed miRNAs, the expression profile in human tissues has been previously established for 145 (see Materials and Methods). Of these, 60% (87/145) are known to be expressed in adult muscle (and in other tissues), whereas the expression of the other 58 miRNAs, mostly up-regulated, was not previously detected in adult muscle to our knowledge. Moreover, almost a fifth of these nonmuscle miRNAs (11/58 miRNAs) were detected in cells of the immune system, including lymphocytes and macrophages (SI Table 4). These findings are consistent with the persistent inflammatory response observed in many dystrophic skeletal muscles that leads to an altered extracellular environment, including an increased presence of inflammatory cells and elevated levels of various inflammatory cytokines.
Direct Quantification of miRNA Gene Expression for Validation of Microarray Results.
The Trilogy technology (24) for quantification of miRNA expression was selected for validating miRNA microarray data. Ten different miRNAs showing distinct expression patterns in the 10 different diseases (miR-21, miR-22, miR-29c, miR-30a-3p, miR-146b, miR-221, miR-368, miR-379, Ambi-miR-693, and Ambi-miR-11040) and two miRNAs with no significant variation (miR-10b and miR-100), were quantified in two independent replicates. Thirty-nine of the RNA samples previously profiled on the arrays were analyzed on the Trilogy platform with an average of three different samples analyzed for each miRNA in any given disease. The expression of miR-146b, miR-379, miR-221, and miR-368 was below the limits of detection of this assay. The relative variations of miRNA expression levels for miR-21, miR-22, miR-29c, miR-30a-3p (except for LGMD2A), and Ambi-miR 11040 were in concordance with the normalized array data, thus validating our array results (SI Fig. 5). Ambi-miR-693, however, which was found on the arrays as down-regulated compared with muscle biopsies from unaffected individuals in all of the examined diseases (LGMD2B, MM, and NM), was found here as being up-regulated. Although we have not determined the exact cause for this discrepancy, it should be noted that there is no up-front enrichment for small RNAs in the Direct assay (unlike our microarray assays), thus we cannot exclude the possibility that precursors are also being quantified.
Distinctive Patterns of miRNA Expression Are Associated with Different Types of Primary Muscle Disorders.
Five miRNAs (miR-146b, miR-221, miR-155, miR-214, and miR-222) (Fig. 1 and SI Table 4) were found to be consistently dysregulated in almost all samples analyzed in the study (with an exception for BMD in which the last three miRNAs were also dysregulated but with a fold change <1.5 and therefore are not included in SI Table 4), across the various diseases. This finding might suggest that these miRNAs are involved in a common underlying regulatory pathway among all diseases. By contrast, other miRNAs were dysregulated only in one given disease and not in any of the others: miR-486, miR-485-5p, miR-331, miR-30e-5p, miR-30d, miR-30a-5p, miR-26a, miR-22, miR-193b, miR-101, miR-95, Ambi-miR-7075, and Ambi-miR-13156, all in muscle biopsies taken from Duchenne patients; miR-517* in FSHD; Ambi-miR-10617 in LGMD2A; miR-301 in LGMD2B; and miR-302c* in MM. The finding of two different miRNAs uniquely dysregulated each in one of the dysferlinopathies and not in the other might point to the involvement of a different secondary regulatory mechanism in the two different phenotypes despite their being allelic diseases. In NM a much larger set of 36 different miRNAs was uniquely dysregulated.
Among the set of miRNAs dysregulated in the various dystrophies (DMD, BMD, and FSHD), 49 in DMD and 38 in FSHD are also dysregulated in various other nondystrophic muscle diseases. Nonetheless, narrow subsets of diseases with shared miRNA profiles were identified. These include: miR-29a in DMD and FSHD; miR-30c in DMD and MM; miR-30b, miR-92, miR-29c, miR-423, miR-361, miR-299-3p, and miR-181d in DMD and NM (Fig. 1 and SI Table 4). We also noted miRNAs with a shared profile across FSHD and the following diseases: miR-16 in FSHD and LGMD2A; miR-279 in FSHD and LGMD2B; and miR-99a, miR-93, miR-455, miR-20b, miR-18a, miR-17-5p, miR-152, miR-106a, and miR-106b, all in FSHD, LGMD2A, LGMD2B, and NM.
The LGMDs analyzed in this study (LGMD2A, LGMD2B, and MM) present a tighter intragroup correlation of miRNAs compared with the other dystrophies. Expression changes for 52 miRNAs were shared by all three diseases, and other dysregulated miRNAs were expressed in only one of the diseases or in the two dysferlin phenotypes. In addition, a set of miRNAs was detected as specifically differentially expressed in each of the LGMDs in concordance with NM (Fig. 1 and SI Table 4).
The most extensive dysregulation of miRNAs was observed in NM with >150 different miRNAs being dysregulated, and of these, 36 being dysregulated solely in this phenotype (SI Table 4). These results could be explained by the high genetic heterogeneity of the underlying disease cause, in which six different genes probably reflecting six different disease mechanisms are known to be involved in the disorder (7).
More than 60% (116) of the overall differentially expressed miRNAs in this study were implicated in at least one of the inflammatory myopathies studied, but not exclusively dysregulated in any of them. Excluding those miRNAs that were common among the three inflammatory myopathies but were also shared by most of the other diseases in the study (miR-146b, miR-221, miR-155, miR-214, and miR-222), not much overlap in the different dysregulated miRNAs was observed although >20 different miRNAs were found to be dysregulated in each of the conditions (Fig. 1 and SI Table 4). These results provide an opportunity to distinguish between those inflammatory-related miRNAs and classify the other 69 dysregulated miRNAs as being involved in other pathologic processes taking place in the different affected myofibers and could give rise to the heterogeneous phenotypes.
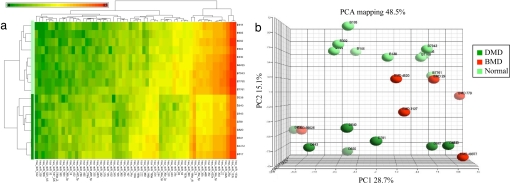
Hierarchical clustering (Fig. 2 and SI Fig. 6) and principal component analysis (PCA) of miRNAs selected by ANOVA clearly segregate and separate muscle biopsies taken from the different muscular disorders from the normal control muscle groups based on their miRNA expression profile.
Fig. 2.
Unsupervised hierarchical clustering and PCA of miRNA expression differentiate DMD from normal muscle. Sixty miRNAs with significantly different expression between DMD (n = 8, D) and normal individuals (n = 9, B) were identified by ANOVA. (a) Hierarchical clustering of 17 samples and 60 genes. Each row represents an individual, and each column represents an miRNA gene. A color code represents the relative intensity of the expression signal, with red indicating high expression and green indicating low expression. (b) PCA of ANOVA-selected miRNAs. In this plot, the first principle components (PC1) axis accounted for 28.7% of the variance in the data set and is a result of noise, possibly introduced by different muscle types and genders. The second principle component (PC2) accounts for 15.1% of the variance and segregates DMD from normal individuals. BMD samples (n = 6) are found as intermediate between DMD and normal muscle, with a distribution consistent with their phenotypic characteristics. The profiles from more severely affected patients (BMD49377 and BMD89026) are found with those of DMD patients, whereas the mildly affected BMD patients (BMD29 and BMD4620) are close to normal muscle.
Functional Correlation Between mRNA and Predicted miRNA Targets in Muscular Disorders.
To gain insight into the function of miRNAs during the disease process, we analyzed the functional correlation between miRNAs and mRNA expression to identify differentially expressed miRNA–mRNA modules and assess the extent of miRNA effects on mRNA expression. First, we inspected the functional correlation between dysregulated mRNAs and targets of dysregulated miRNAs, which allows capturing of indirect target effects, where miRNAs bind regulatory proteins, such as transcription factors, and the latter exert the main effect.
A total of 10 mRNA and 10 miRNA datasets were analyzed (SI Table 5). A meta miRNA predictor (MAMI) enabling maximal accuracy and tunable sensitivity and specificity in predictions was applied to predict targets of differentially expressed miRNAs (A.E., C. Freifield, A. T. Kho, I.E., M. Galdzicki, K. Naxerova, M. F. Ramoni, L.M.K., and I.S.K., unpublished work).
A strong functional correlation was detected only in DMD and MM, suggesting that these two diseases have a tight posttranscriptional regulation at the mRNA level. In DMD, the correlation between the functions of down-regulated mRNAs and those of targets of up-regulated miRNAs reached significance with P < 0.0157. The functional correlation between up-regulated mRNA and targets of down-regulated miRNAs had a P < 0.023. In MM, the correlations' significances were P < 2.01E-06 and P < 0.03413 for functions of down-regulated mRNAs with targets of up-regulated miRNAs and up-regulated mRNAs with targets of down-regulated miRNAs, respectively.
miRNA–mRNA modules shared by DMD and MM include extracellular matrix (ECM) processes and cytoskeletal organization. Ion channel activity module was down-regulated in MM, and a strong down-regulation of a transcriptional activity module was observed in DMD. SI Fig. 7 shows an example of the miRNA–mRNA ECM module in DMD, where the overexpressed ECM protein-coding genes are regulated by both direct interaction with down-regulated miRNAs and through their mRNA targets. The overall network structure reveals tight posttranscriptional regulation whose alteration might contribute to secondary pathological processes in the dystrophic muscle in DMD, by either direct miRNA targeting or through secondary proteins.
Inference of miRNA Functions in Dystrophic Muscle Pathology.
Further insights into the biological pathways potentially regulated by miRNAs in the dystrophic process were obtained by direct comparison between the genes previously found as dysregulated in DMD (11) and the predicted target genes for the 62 differentially expressed miRNAs found in DMD. Fifty-seven mRNA–miRNA interactions were identified, representing 28 genes as targeted by at least one miRNA and dysregulated in DMD. About 42% of these genes were predicted to be targeted by multiple miRNAs (Table 1).
Table 1.
Direct miRNA–mRNA predicted targeting in DMD
| miRNA | Target | miRNA fold change | Target fold change |
|---|---|---|---|
| hsa-miR-30c | VIM | −2 | 2 |
| hsa-miR-148a | UCP3 | 2 | −3 |
| hsa-miR-130a | UBE2D1 | 2 | −3 |
| hsa-miR-101 | TUBB2A | −2 | 3 |
| hsa-miR-29c | TRO | −6 | 4 |
| hsa-miR-26a | SRPX | −2 | 3 |
| hsa-miR-101 | SPARC | −2 | 2 |
| hsa-miR-30c | RUNX1 | −2 | 6 |
| hsa-miR-197 | PRMT2 | −1 | 3 |
| hsa-miR-29c | PENK | −6 | 8 |
| hsa-miR-199a | PDE4D | 3 | −4 |
| hsa-miR-29a | PXDN | −2 | 2 |
| hsa-miR-214 | LMOD1 | 2 | −3 |
| hsa-miR-29c | HOM-TES-103 | −6 | 2 |
| hsa-miR-22 | HSPG2 | −3 | 2 |
| hsa-miR-22 | GPNMB | −3 | 2 |
| hsa-miR-210 | GPD1L | 2 | −2 |
| hsa-miR-21 | FAM50B | 3 | −1 |
| hsa-miR-197 | EEF1A1 | −1 | 2 |
| hsa-miR-26a | EPB41L3 | −2 | 4 |
| hsa-miR-101 | CFH | −2 | 4 |
| hsa-miR-29c | COL3A1 | −6 | 6 |
| hsa-miR-26a | COL1A2 | −2 | 5 |
| hsa-miR-29c | COL1A2 | −6 | 5 |
| hsa-miR-22 | CLIC4 | −3 | 2 |
| hsa-miR-193b | CLIC1 | −2 | 2 |
| hsa-miR-30c | CD99 | −2 | 2 |
| hsa-miR-30a-3p | ANXA1 | −4 | 2 |
| hsa-miR-30c | ACTN1 | −2 | 2 |
Earlier expression studies have demonstrated that significantly more mRNAs are overexpressed in dystrophic muscle than underexpressed compared with unaffected muscle (11), most likely because of an increase in protein turnover caused by the degenerative and regenerative nature of the disease. In the present analysis, muscle structure and regeneration and ECM genes were among these predicted miRNA targets. Interactions like proenkephalin–miR-29c; collagen, type I, alpha2–miR-29c; trophinin–miR-29c; RUNX1–miR-30a-5p, and PDE4D–miR-199a demonstrated high reciprocal fold change of the relevant miRNA and mRNA. At the mRNA level, dystrophin and several other structurally related proteins, which are substantially underexpressed in DMD muscle, were not predicted to correlate with any of these miRNAs.
Dysregulated miRNAs in Muscular Disorders Are Significantly Associated with Diverse Signaling Pathways.
With the understanding that identification of mRNA targets is predictive, we analyzed the functional enrichment of predicted targets of differentially expressed miRNAs in each of the muscle phenotypes in an attempt to uncover the functional meaning among these dysregulated miRNAs.
In muscle biopsies from Duchenne patients, the 39 up-regulated miRNAs were identified to potentially target the 3′ UTRs of ≈5,000 genes (of which 807 are muscle-expressed genes) and >4,400 genes were identified to be targeted by the 23 miRNAs down-regulated in the disease (SI Table 6). Notably, the 11 miRNAs dysregulated exclusively in DMD were predicted to target >400 genes of diverse functions, by more than one prediction tool (SI Table 6). The detailed information for the other diseases is summarized in SI Table 6.
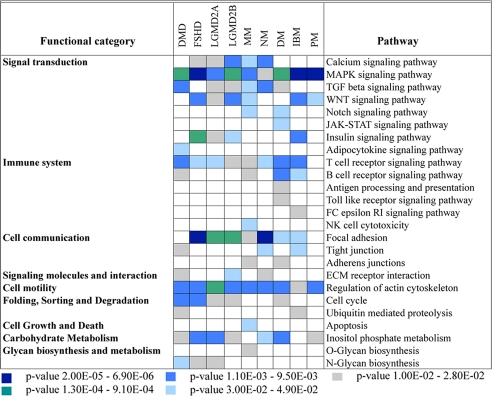
To analyze the role that these differentially expressed miRNAs play in the regulatory networks in muscular disorders, we have used the KEGG database and the DAVID bioinformatics resources (26) to identify significantly overrepresented biological pathways. Fig. 3 shows the overrepresented pathways identified in at least one muscle phenotype, using the overall predicted targets in any given disease.
Fig. 3.
Overrepresented miRNA regulatory pathways in primary muscular disorders. Fisher's exact test was used to identify significant enrichment for pathway annotations among predicted targets of the dysregulated miRNAs in the different diseases. Each column corresponds to a single disease, and each row corresponds to a KEGG pathway with an overrepresentation of miRNA targets. Pathways have been grouped in larger functional categories according to the KEGG annotation. Only pathways with at least one significant association are shown, and the confidence for enrichment of targets in a given pathway is shown by color-coding the P value ranges.
Genes that were commonly targeted by the dysregulated miRNAs in muscle specimens from DMD patients, for instance, were significantly clustered in 12 biochemical pathways with some, like TGF-β (P = 3.50E-03), being targeted by both up-regulated and down-regulated miRNAs, suggesting an extensive miRNA regulation of this pathway in DMD. The overall analysis highlighted 25 different pathways as being significantly targeted by the different miRNAs involved in the primary muscle diseases studied. Of those, six major signal transduction signaling pathways previously described (9, 27) as involved in various aspects of muscular disorders, such as TGF-β, calcium signaling pathway, Wnt, Notch, and MAPK signaling pathways, were found to be significantly targeted by the dysregulated miRNAs described in this study (Fig. 3).
Pathways related to the immune response were also significantly enriched in this data set in all diseases, with T cell receptor signaling pathway (P = 4.50E-03) being most abundant. Consistent with previous studies and more recent mRNA expression analysis, the immune-related pathways were highly enriched in two of the inflammatory myopathies, DM and IBM (and surprisingly not in PM patients, maybe because of previous steroids treatment and/or the amount of inflammatory cell infiltrates). Cellular pathways, including cell motility (P = 6.50e-04), cell communication (P = 2.40E-05), degradation (P = 6.60E-03), and others were also found to be extensively regulated by these miRNAs (Fig. 3).
Discussion
Significant progress has been made in the understanding of muscle dysfunction and the causative mechanism behind the major muscular dystrophies has been explored, but knowledge of the underlying regulatory network(s) is still incomplete. Compelling evidence has demonstrated the substantial regulatory role of miRNAs in muscle development and more recently in the etiology of cardiac failure (28). In light of these findings, we have examined miRNAs involved in major myopathological diseases in humans to gain insight into the specific regulation of genes that are disrupted in pathological muscle conditions.
A total of 185 miRNAs with statistically significant differential expression were identified in the 10 distinct forms of muscular dystrophies analyzed in the present work. Of those, a subgroup of 18 miRNAs was identified that correctly predict and distinguish the various diseases from the normal muscle tissue, with >90% accuracy in most groups (SI Table 7). In contrast to many previous studies, mostly in cancer, showing a global reduction of mature miRNA levels compared with normal tissues, an increase in abundance for many miRNAs was observed in the different muscular disorders. In this report, we provide evidence that miRNAs have a potential role in the pathophysiology of primary muscle diseases and present the complete suite of known miRNAs with altered expression in these diseases. These miRNA signatures provide the basis for a list of common target genes whose misregulation may contribute to the pathology of these disorders.
Secondary to the genetic defects, necrosis and inflammation play a crucial role in the pathogenesis of the different muscular dystrophies and myopathies, and expression profiling of various diseased muscles revealed distinct patterns of immune or immune modulatory pathways rather than nonspecific processes (11, 30). Although the immunopathology of these disorders is not fully understood, several miRNAs previously described as immune-related were found as commonly dysregulated among the various dystrophies. Together with the different patterns of dysregulated miRNAs unique to each of the different diseases, this pattern offers insights into the complexities of the inflammatory process taking place in the different affected muscle fibers.
In contrast to studies associating the overexpression of miR-155 with malignancy in humans (31), the present report describes the ubiquitous up-regulation of this immune-related miRNA (32, 33) in a completely different and unrelated context, raising intriguing questions about its functional role in the pathological process in muscle.
Despite the elucidation of several clinically relevant signal transduction pathways that can lead to disease progression, the means by which these pathways are coordinated with respect to the development and progression of muscle disease process remain obscure. Induction of miR-146 expression by activated NF-κB has been recently demonstrated by Taganov et al. (34), and its role in the immune system was also described. Evidence of perturbation of NF-κB signaling has been described in the process of modulating the immune response in several different dystrophies (35–37) and the inflammatory myopathies (38). It will be important to identify and analyze miR-146 downstream target genes and gain insights into the signaling pathways altered by the aberrant up-regulated expression of this miRNA in the different primary muscle diseases.
Currently, the major difficulty for functional studies of miRNAs is in determining their specific target genes at the transcriptional or translational level. Available prediction algorithms frequently predict hundreds of target genes for any single miRNA, and it is likely that this high number of genes contains a significant fraction of false positives. To restrict this high number and enrich for more reliable predicted targets, we have applied a meta predictor tool recently developed that integrates the leading prediction methods into an improved predictor. Beyond the predicted lists of targets the significant associations inferred between the sets of functional targets predicted for the overall miRNAs in each of the diseases and specific cellular pathways can be used to shape some initial hypotheses on how alteration of miRNA expression may be directly involved in different types of diseased muscles.
Furthermore, the functional correlation between the differentially expressed mRNAs and miRNAs as a module in DMD revealed a tight posttranscriptional regulation network at the mRNA level whose alteration might contribute to increased immune response, by either direct miRNA targeting or through secondary proteins. Together with the specific mRNA–miRNA predicted interactions, some of which are directly involved in compensatory secondary response functions like connective tissue infiltration, and others that are involved in muscle regeneration, these findings raise the opportunity for therapeutic intervention at the miRNA level in preventing specific physiological pathways underlying the disease. However, because it remains difficult to estimate the true false-positive rate of the overall target prediction, a better understanding of the biological significance of these miRNAs and the alterations found in the different muscle diseases would be ultimately achieved by the development of experimental models.
Conclusion.
Considerable advances have been made in understanding the mechanisms, both transcriptional and translational, that lead to altered gene expression under dystrophic conditions. Our results point to an additional dimension of regulation of muscle function mediated by miRNAs. An important aim for the future will be to experimentally assess the predicted targets of the miRNAs responsible for adverse skeletal muscle remodeling in the different diseases. The overall discovery of dysregulated miRNAs in the different diseases is expected not only to broaden our biological understanding of these diseases, but more importantly, to identify candidate miRNAs as potential targets for future clinical applications.
Materials and Methods
Patient Samples and RNA Isolation.
A total of 88 muscle specimens, representing 11 different human muscle conditions, were available for this study, all in compliance with the involved institutions approved protocols. RNAs were isolated with the mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer's instruction for total RNA isolation. More detailed information is provided in SI Text, SI Table 2, and SI Fig. 4.
miRNA Array Analyses.
RNA samples were processed by Asuragen Services (Austin, TX) according to the standard operating procedures of the company as described (39). For a detailed description see SI Text. Microarray data processing and analyses are described in detail in SI Text.
miRNA Quantification Assay.
Direct miRNA detection and quantification was performed by using the Direct miRNA assay (US Genomics) as described (24).
Assessment of miRNA and mRNA Expression in Normal Human Tissues.
Four independent data sets of miRNA expression across normal human tissues were used to assess miRNAs expression in adult skeletal muscle (25, 29, 39, 40). The assessment of mRNA expression in normal adult skeletal muscle is described in SI Text.
Functional Inference of miRNA and miRNA–mRNA Correlation Analysis.
Functional inference of miRNA and miRNA–mRNA correlation analysis are detailed in SI Text.
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. Tim Davison and Charles Johnson (Asuragen) for their expertise in the microarray application and excellent assistance; Dr. Marco Ramoni for data analysis; and Elicia Estrella, Kamila Naxerova, Michal Galdzicki, Joon Lee, and members of L.M.K.'s laboratory for helpful comments and suggestions. K.M.F. is supported by National Center for Research Resources Grant M01-RR00064 (to the University of Utah, Dr. Lorris Betz). M.M. and C.L. are supported by the Associazione Amici del Centro Dino Ferrari, Telethon Project GTF02008, and Eurobiobank Project QLTR-2001-02769. A.H.B. is supported by the Muscular Dystrophy Association, National Institutes of Health Grant R01-AR044345, and generous gifts from the Lee and Penny Anderson Family Foundation and the Joshua Frase Foundation. L.M.K. is an Investigator with the Howard Hughes Medical Institute.
Abbreviations
miRNA
microRNA
DMD
Duchenne muscular dystrophy
BMD
Becker muscular dystrophy
FSHD
facioscapulohumeral muscular dystrophy
LGMD
limb-girdle muscular dystrophies
LGMD2A
LGMD type 2A
LGMD2B
LGMD type 2B
NM
nemaline myopathy
MM
Miyoshi myopathy
IBM
inclusion body myositis
DM
dermatomyositis
PM
polymyositis
PCA
principal component analysis
ECM
extracellular matrix.
Footnotes
The authors declare no conflict of interest.
References
- 1.Davies KE, Nowak KJ. Nat Rev. 2006;7:762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- 2.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 3.Laval SH, Bushby KM. Neuropathol Appl Neurobiol. 2004;30:91–105. doi: 10.1111/j.1365-2990.2004.00555.x. [DOI] [PubMed] [Google Scholar]
- 4.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 5.Bansal D, Campbell KP. Trends Cell Biol. 2004;14:206–213. doi: 10.1016/j.tcb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Tawil R, Van Der Maarel SM. Muscle Nerve. 2006;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal PB, Greenleaf RS, Tomczak KK, Lehtokari VL, Wallgren-Pettersson C, Wallefeld W, Laing NG, Darras BT, Maciver SK, Dormitzer PR, et al. Am J Hum Genet. 2007;80:162–167. doi: 10.1086/510402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman EP, Rao D, Pachman LM. Rheum Dis Clin North Am. 2002;28:743–757. doi: 10.1016/s0889-857x(02)00031-5. [DOI] [PubMed] [Google Scholar]
- 9.Dalakas MC. Nat Clin Pract Rheumatol. 2006;2:219–227. doi: 10.1038/ncprheum0140. [DOI] [PubMed] [Google Scholar]
- 10.Lennon NJ, Kho A, Bacskai BJ, Perlmutter SL, Hyman BT, Brown RH., Jr J Biol Chem. 2003;278:50466–50473. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 11.Haslett JN, Sanoudou D, Kho AT, Bennett RR, Greenberg SA, Kohane IS, Beggs AH, Kunkel LM. Proc Natl Acad Sci USA. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Hum Mol Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 13.Sanoudou D, Haslett JN, Kho AT, Guo S, Gazda HT, Greenberg SA, Lidov HG, Kohane IS, Kunkel LM, Beggs AH. Proc Natl Acad Sci USA. 2003;100:4666–4671. doi: 10.1073/pnas.0330960100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg SA, Sanoudou D, Haslett JN, Kohane IS, Kunkel LM, Beggs AH, Amato AA. Neurology. 2002;59:1170–1182. doi: 10.1212/wnl.59.8.1170. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez-Garcia I, Miska EA. Development (Cambridge, UK) 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 17.Kloosterman WP, Plasterk RH. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon C, Han Z, Olson EN, Srivastava D. Proc Natl Acad Sci USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokol NS, Ambros V. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Samal E, Srivastava D. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 22.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. Nat Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- 25.Liang Y, Ridzon D, Wong L, Chen C. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 27.Bassel-Duby R, Olson EN. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 28.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 29.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YW, Zhao P, Borup R, Hoffman EP. J Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taganov KD, Boldin MP, Chang KJ, Baltimore D. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baghdiguian S, Martin M, Richard I, Pons F, Astier C, Bourg N, Hay RT, Chemaly R, Halaby G, Loiselet J, et al. Nat Med. 1999;5:503–511. doi: 10.1038/8385. [DOI] [PubMed] [Google Scholar]
- 36.Macaione V, Aguennouz M, Rodolico C, Mazzeo A, Patti A, Cannistraci E, Colantone L, Di Giorgio RM, De Luca G, Vita G. Acta Neurol Scand. 2007;115:115–121. doi: 10.1111/j.1600-0404.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 37.Acharyya S, Villalta SA, Bakkar N, Bupha-Intr T, Janssen PM, Carathers M, Li ZW, Beg AA, Ghosh S, Sahenk Z, et al. J Clin Invest. 2007;117:889–901. doi: 10.1172/JCI30556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monici MC, Aguennouz M, Mazzeo A, Messina C, Vita G. Neurology. 2003;60:993–997. doi: 10.1212/01.wnl.0000049913.27181.51. [DOI] [PubMed] [Google Scholar]
- 39.Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E. RNA. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baskerville S, Bartel DP. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information