Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria (original) (raw)
Abstract
The TIM23 translocase mediates the ΔΨ- and ATP-dependent import of proteins into mitochondria. We identified Tim14 as a novel component of the TIM23 translocase. Tim14 is an integral protein of the inner membrane with a typical J-domain exposed to the matrix space. TIM14 genes are present in the genomes of virtually all eukaryotes. In yeast, Tim14 is essential for viability. Mitochondria from cells depleted of Tim14 are deficient in the import of proteins mediated by the TIM23 complex. In particular, import of proteins that require the action of mtHsp70 is affected. Tim14 interacts with Tim44 and mtHsp70 in an ATP-dependent manner. A mutation in the HPD motif of the J-domain of Tim14 is lethal. Thus, Tim14 is a constituent of the mitochondrial import motor. We propose a model in which Tim14 is required for the activation of mtHsp70 and enables this chaperone to act in a rapid and regulated manner in the Tim44-mediated trapping of unfolded preproteins entering the matrix.
Keywords: chaperone/import motor/mitochondria/protein translocation/TIM23 complex
Introduction
Transport of proteins from the cytosol across the outer membrane of mitochondria is facilitated by the TOM complex, and transport across and into the inner membrane by cooperation of the TOM complex with two distinct translocases, the TIM23 and TIM22 complexes (Paschen and Neupert, 2001; Endo and Kohda, 2002; Jensen and Dunn, 2002; Rehling et al., 2003). The TIM23 complex mediates import of the vast majority of proteins; virtually all matrix proteins and many inner membrane proteins use this translocase. The TIM23 complex has been studied in some detail, although deeper insight into its structure and function is lacking. It consists of a part that is tightly integrated into the membrane, and the import motor. The membrane part is made up of Tim23 and Tim17, which are believed to form the protein conduit (Dekker et al., 1997; Milisav et al., 2001; Truscott et al., 2001), and Tim50, which may have a role in the transfer of the preproteins from the TOM to the TIM23 complex (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). The mitochondrial import motor is attached to the membrane part at the matrix face of the inner membrane (Matouschek et al., 2000; Neupert and Brunner, 2002). So far, three constituents have been described, Tim44, mtHsp70 and Mge1. Tim44 is a peripheral membrane protein that is believed to interact with segments of preproteins appearing from the outlet of the protein conducting channel (Schneider et al., 1994). Tim44 recruits mtHsp70, a chaperone of the matrix space, to the entry site (Kronidou et al., 1994; Rassow et al., 1994; Schneider et al., 1994). MtHsp70 binds to unfolded chains as they enter the matrix, followed by its release from Tim44. Multiple cycles of binding and release of mtHsp70 to Tim44 and to the incoming polypeptide chain mediate the vectorial movement into the matrix. These reactions are driven by hydrolysis of ATP bound to mtHsp70. The molecular mechanism of this ATP-dependent cycling is still a matter of debate (Matouschek et al., 2000; Neupert and Brunner, 2002). It is unresolved how the high efficiency of protein translocation is achieved by the import motor.
The present study aimed to investigate whether the import motor of the TIM23 translocase is known in its entirety. We report the presence of a novel component, Tim14, an essential part of the TIM23 machinery. It is a member of the DnaJ protein family, consisting of a J-domain that faces the matrix space and a single transmembrane anchor in the inner membrane. Mitochondria isolated from cells depleted of Tim14 showed strong defects in the import of most precursors using the TIM23 translocase pathway. The only precursors of the TIM23 translocase that were not significantly affected were those which do not need a functional mitochondrial import motor. Tim14 interacts with Tim44 and mtHsp70 in an ATP-dependent manner and has a profound effect on the interaction of mtHsp70 with Tim44. A mutation in Tim14 that abolishes the function of the J-domain is lethal. Tim14 is apparently required for the efficient binding of mtHsp70 to the incoming polypeptide chain and for release from Tim44. We propose that the J-domain stimulates the ATPase activity of mtHsp70.
In conclusion, we have identified a central component of the mitochondrial import motor that had previously escaped detection. The existence of this new component has important implications on the mechanism of the mitochondrial protein import.
Results
Tim14, novel neighboring component of Tim44 in the mitochondria
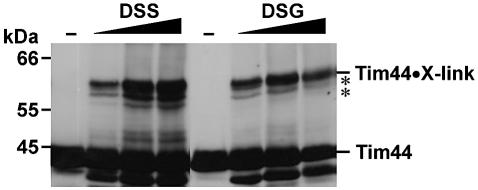
In an attempt to determine whether all components of the TIM23 translocase have been detected, we used a crosslinking approach to identify neighboring proteins of the known constituents of the complex. We had previously observed that several components can be crosslinked to Tim44 (Moro et al., 1999). Among the crosslinked adducts a prominent species of ∼60 kDa apparent molecular weight was found, pointing to a neighboring component of ∼15–20 kDa. In order to determine the significance of this observation and to identify the protein involved, we checked various crosslinkers. Using disuccinimidyl glutarate (DSG) and disuccinimidyl suberate (DSS), a major adduct of 60 kDa was observed besides several minor adducts (Figure 1). Such crosslinked adduct was also observed with DFDNB (data not shown). We conclude that an as yet unidentified component is present in mitochondria that interacts with Tim44 or is in close vicinity to Tim44.
Fig. 1. Tim44 can be crosslinked to a component of ∼15–20 kDa. Mitochondria isolated from wild-type yeast were incubated with the crosslinkers DSS and DSG. Samples were analyzed by SDS–PAGE and immunoblotting with antibodies against Tim44. The minor crosslinked adducts are labeled with asterisks.
To determine the identity of this protein we reasoned that if it was a component of the import machinery, it should be essential for viability as are all members of the TIM23 machinery. A second criterion was the approximate molecular weight, and a third was the presence of a mitochondrial import signal.
A gene that fulfilled these criteria corresponded to the reading frame YLR008c of Saccharomyces cerevisiae. This gene has the capacity to encode a protein with a molecular weight of 17.9 kDa, and is listed in various collections of putative genes that are essential for viability of S.cerevisiae (Winzeler et al., 1999). Thirdly, the gene appears to have a typical internal targeting signal, characterized by a positively charged segment located immediately after the transmembrane domain (Foelsch et al., 1996). We refer to the YLR008c gene product as Tim14.
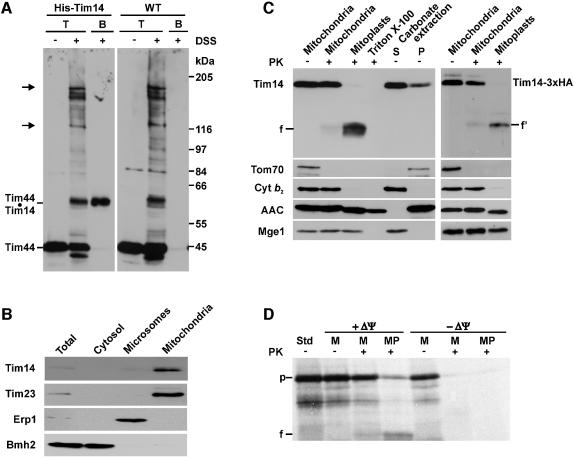
In order to verify that Tim14 is indeed the component that can be crosslinked to Tim44, we generated a yeast strain in which an octahistidinyl tag was present at the N-terminus of Tim14. Mitochondria isolated from this strain and from wild-type cells were subjected to crosslinking with DSS and subsequently analyzed for binding of Tim44-containing crosslinked adducts to the Ni-NTA–agarose beads. The overall crosslinking patterns of both types of mitochondria were virtually identical (Figure 2A). However, the Tim44-containing adduct of ∼60 kDa was specifically bound to the Ni-NTA–agarose beads only with mitochondria containing His-tagged Tim14, but not with wild-type mitochondria.
Fig. 2. Tim14 is a component of the inner membrane of mitochondria. (A) Tim14 is the product of the YLR008c reading frame. Mitochondria isolated from a yeast strain in which a sequence encoding eight histidine residues was fused to the 5′ end of the YLR008c reading frame, and from wild-type cells, were subjected to crosslinking with DSS. An aliquot of each type of mitochondria was directly subjected to SDS–PAGE; the other aliquot was solubilized and incubated with Ni-NTA beads. Bound material was eluted and subjected to SDS–PAGE. Resolved proteins were blotted onto nitrocellulose membrane and immunodecorated with antibodies against Tim44. T, total mitochondria incubated in the absence or presence of DSS; B, material bound to Ni-NTA beads. Arrows indicate crosslinked adducts of Tim44 to mtHsp70. (B) Tim14 is located in mitochondria. Equal amounts of protein of subcellular fraction were subjected to SDS–PAGE and immunodecoration with antibodies against Tim14 and marker proteins of mitochondria (Tim23), microsomes (Erp1) and cytosol (Bmh2). (C) Tim14 is located in the inner membrane, exposing an N-terminal segment of ∼8 kDa into the intermembrane space and its C-terminus into the matrix. Left panel: isolated mitochondria, mitoplasts prepared by osmotic shock, Triton-solubilized mitochondria and the supernatant (S) and pellet (P) of carbonate extraction were incubated with or without proteinase K (PK; 100 µg/ml). Samples were subjected to SDS–PAGE and immunoblotting with antibodies against Tim14 and against various mitochondrial marker proteins. Cyt_b_2, cytochrome _b_2; AAC, ADP/ATP carrier; f, 13 kDa fragment of Tim14. Right panel: mitochondria containing a version of Tim14 carrying a 3HA-tag at its C terminus and derived mitoplasts were treated with proteinase K as indicated. Samples were analyzed by SDS–PAGE and immunoblotting with an antibody against the HA-tag and against the indicated mitochondrial marker proteins. f′, fragment. (D) Tim14 can be imported into isolated mitochondria. Reticulocyte lysate containing 35S-labeled Tim14 was incubated with mitochondria in the presence or absence of a membrane potential ΔΨ. Mitochondria were reisolated, aliquots were converted to mitoplasts and treated with proteinase K. Samples were subjected to SDS–PAGE and autoradiography. M, mitochondria; MP, mitoplasts; Std, 40% of input into import experiments; p, precursor of Tim14; f, 13 kDa fragment.
We confirmed by subcellular localization experiments that Tim14 is a mitochondrial protein. Antibodies against Tim14 recognized a single band of ∼21 kDa in yeast whole-cell extracts and in the mitochondrial fraction (Figure 2B).
The submitochondrial location of Tim14 was determined by treating mitochondria with proteinase K. With intact mitochondria Tim14 was not degraded, but when first mitoplasts were generated by hypoosmotic swelling, Tim14 was cleaved, yielding a fragment of ∼13 kDa (Figure 2C, left panel). When mitochondria were lyzed with Triton X-100 and then treated with proteinase K, the protein was degraded so that no fragment was detectable. When mitoplasts from a strain expressing Tim14 with a 3HA-tag at the C-terminus were treated with proteinase K, a fragment was observed that could be decorated with antibodies against the HA antigen, indicating that the C-terminal segment of Tim14 is facing the matrix (Figure 2C, right panel). Upon alkaline extraction of mitochondria the protein fractionated partly with the membranes, the larger proportion, however, was found in the supernatant. Apparently, Tim14 belongs to the class of proteins that span the inner membrane with one transmembrane segment but do not firmly interact with the membrane lipids.
The mitochondrial location of the Tim14 was also demonstrated by its ability to become imported into isolated mitochondria after synthesis in a cell-free system. Import was dependent on the mitochondrial membrane potential. When the mitochondria were converted to mitoplasts and treated with proteinase K, the imported protein was converted to an ∼13 kDa fragment (Figure 2D) This radioactive fragment co-migrated with the fragment generated from the endogenous Tim14 visualized by immunodecoration of the same blot.
We conclude that Tim14 is a protein that spans the inner membrane of mitochondria; an N-terminal segment is exposed to the intermembrane space, whereas the major part is located in the matrix space.
Tim14 is a J-domain protein essential for the viability of yeast cells
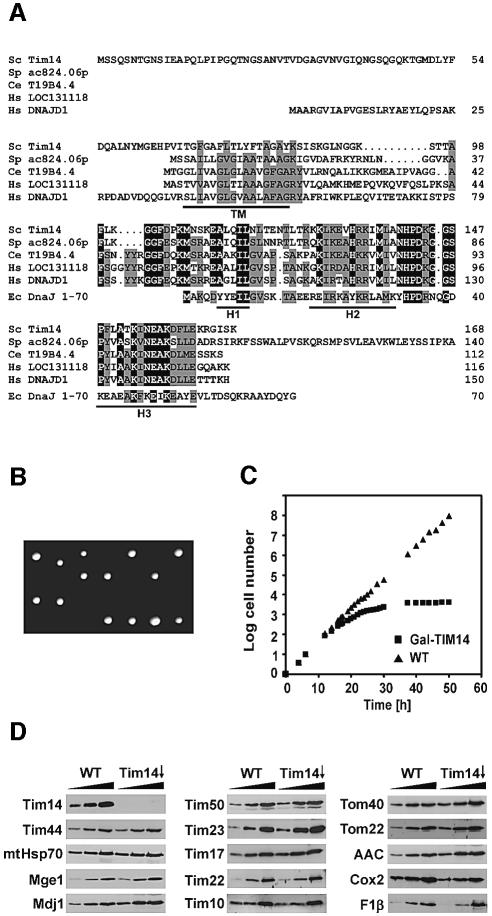
The amino acid sequence of Tim14 as deduced from the DNA sequence of reading frame YLR008c is shown in Figure 3A. It comprises an N-terminal hydrophilic segment of 65 amino acid residues, a predicted α-helical transmembrane domain of 18 residues and a hydrophilic C-terminal domain of 85 residues. This latter part contains a segment of ∼50 residues with strong similarity to J-domains known from DnaJ-like proteins (Bukau and Horwich, 1998; Kelley, 1998). Thus, the J-domain is located at the inner face of the mitochondrial inner membrane.
Fig. 3. Tim14 is a J-domain protein essential for yeast cell viability. (A) Deduced amino acid sequences of Tim14 proteins and of the J-domain of E.coli DnaJ. Sc, S.cerevisiae; Sp, S.pombe; Ce, C.elegans; Hs, Homo sapiens; Ec, DnaJ of E.coli (residues 1–70). The predicted single transmembrane domain (TM) is underlined. Identical residues are shown in black, similar residues in grey. H1–H3, characteristic α-helical segments of the J-domain of DnaJ are indicated. (B) TIM14 is an essential gene in yeast. Tetrad analysis of diploid yeast cells carrying a deletion in the TIM14 gene. (C) A yeast strain in which Tim14 is down-regulated stops growing. Cells carrying a TIM14 gene under control of the GAL10 promoter were first grown on galactose and then transferred to glucose-containing medium (Tim14↓); as control a wild-type strain (WT) was grown in parallel. (D) Cells harboring Tim14 under galactose control and wild-type cells were grown for 21 h after shift to glucose containing medium. Mitochondria (12.5, 25 and 50 µg) were analyzed by SDS–PAGE and immunoblotting with antibodies. F1β, subunit β of ATP-synthase; Cox2, subunit 2 of cytochrome oxidase.
DnaJ is a co-chaperone of Escherichia coli, and homologs are present in practically all prokaryotes and eukaryotes (Kelley, 1998), where DnaJ relatives are found in diverse cellular subcompartments, including mitochondria. In addition to the full-length homologs, a number of proteins have been identified that contain only the N-terminal J-domain (Kelley, 1998), which is responsible for the stimulation of the ATPase activity of members of the DnaK/Hsp70 family (Liberek et al., 1991; Szabo et al., 1994; Wall et al., 1994; Misselwitz et al., 1998). Tim14 belongs to this latter family. All eukaryotic organisms have apparent Tim14 orthologs that contain three common elements: (i) a highly conserved C-terminally located J-domain; (ii) a transmembrane domain preceding the J-domain; and (iii) a putative mitochondrial targeting signal directly at the C-terminal side of the transmembrane domain.
Figure 3A presents an alignment of sequences of S.cerevisiae, Schizosaccharomyces pombe, Caenorhab ditis elegans and human. There are two closely related homologs with the above characteristics in humans. In S.cerevisiae a homolog, Mdj2, was found that has similar characteristics, but the J-domain appears to be less related to Tim14 than the apparent orthologs. Mdj2 is encoded by a non-essential gene, yet its function is unclear as yet (Westermann and Neupert, 1997).
We deleted the TIM14 gene in diploid cells. These were subjected to tetrad analysis. Two out of four spores did not yield viable cells (Figure 3B). This confirms that TIM14 is an essential gene.
In order to generate a tool to study the function of Tim14, we constructed a yeast strain in which the gene was under the control of the GAL10 promoter. In the presence of galactose the cells grew like wild-type cells; in the presence of glucose, the cells slowed down in their growth after ∼12 h and virtually stopped growing after ∼26 h (Figure 3C). Mitochondria were isolated from cells grown for 21 h in the presence of glucose and from isogenic wild-type cells as a control. The levels of a number of mitochondrial proteins were determined by immunoblotting. Tim14 was virtually absent in mitochondria from cells grown with glucose (Figure 3D). In contrast, components of the TIM23 translocase such as Tim17, Tim23, Tim44, Tim50, Mge1 and mtHsp70 were present at roughly control levels. Tim22, the central component of the TIM22 translocase, was also present at normal levels. Other mitochondrial components, such as subunit 2 of the cytochrome oxidase complex, Cox2, F1β of the ATP synthase, the ADP/ATP carrier (AAC) and Mdj1 were also present at roughly control levels in the depleted cells (Figure 3D). Likewise, the concentrations of proteins of the intermembrane space such as Tim10, as well as the outer membrane protein, Tom40, were not significantly affected by down- regulation of Tim14.
Tim14 is required for the import of preproteins
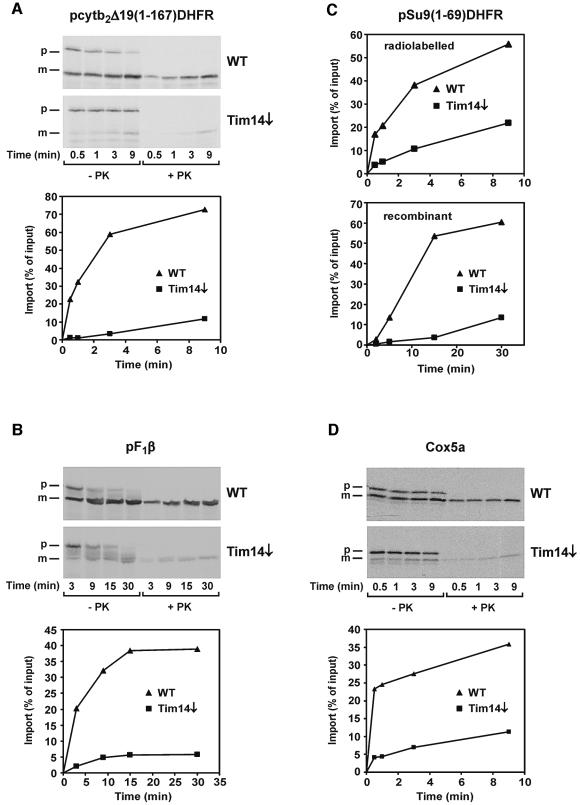
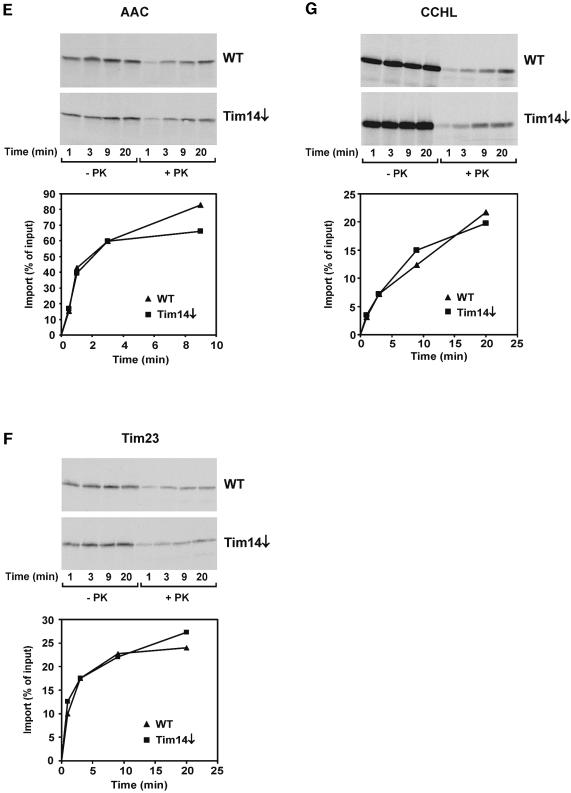
Most nuclear-encoded proteins of mitochondria which exert a function that is essential for the viability of yeast cells are involved in the topogenesis of mitochondrial proteins. We studied the import of a number of mitochondrial preproteins into isolated mitochondria from a strain depleted in Tim14. For comparison, mitochondria from wild-type cells were analyzed in parallel. Several preproteins with an N-terminal matrix-targeting signal (MTS) were checked. The import of all of these was strongly reduced in cells depleted of Tim14. Examples shown are the matrix-targeted precursor pcytb2Δ19(1–167)DHFR, a fusion protein consisting of the first 167 amino acid residues of cytochrome _b_2 and of DHFR (with an intact MTS but an inactivated sorting signal; Figure 4A), and pF1β (precursor of subunit β of F1dash;ATPase; Figure 4B). The import of pSu9(1–69)DHFR was also strongly reduced in the Tim14-depleted strain (Figure 4C, upper panel). In addition to the radiochemical amounts of this precursor, we also imported this precursor as recombinant protein, since this form can be added in amounts that saturate the import system. Again in this case, a drastic reduction of import was observed (Figure 4C, lower panel). Furthermore, a precursor of a mitochondrial inner membrane protein that uses the TIM23 complex, pCoxVa, was strongly affected in its import (Figure 4D). The import of the ADP/ATP carrier (Figure 4E) and Tim23 (Figure 4F), which use the TIM22 translocase, was not or only slightly reduced. In addition, proteins that use only the TOM complex but none of the TIM translocases, such as cytochrome c heme lyase (Figure 4G), were imported at control rates. We conclude that Tim14 is specifically required for import of proteins using the TIM23-mediated pathway.
Fig. 4. Mitochondria deficient in Tim14 are defective in the import of preproteins using the TIM23 complex. A yeast strain in which TIM14 was under control of the Gal10 promoter was grown in the presence of glucose for 21 h to deplete Tim14. Mitochondria from this strain and from wild-type were incubated with radiolabeled preproteins, mitochondria were reisolated, treated with proteinase K and subjected to SDS–PAGE, autoradiography and densitometry. (A) pcytb2Δ19(1–167)DHFR, a precursor consisting of the first 167 amino acid residues of cytochrome _b_2 with a deleted sorting signal fused to dihydrofolate reductase; (B) pF1β, precursor to the β-subunit of ATP synthase; (C) pSu9(1–69)DHFR, matrix targeting signal of subunit 9 of ATP synthase from Neurospora crassa (residues 1–69) fused to DHFR (upper panel, precursor synthesized in reticulocyte lysate; lower panel, precursor expressed in E.coli); (D) pCox5a, precursor of subunit 5 of cytochrome oxidase; (E) AAC, ADP/ATP carrier; (F) Tim23; (G) CCHL, precursor of cytochrome c heme lyase. With those preproteins which become proteolytically processed by the matrix processing peptidase upon import, the processed forms in PK treated mitochondria were quantified. p, precursor from; m, processed form.
Tim14 is part of the TIM23 complex
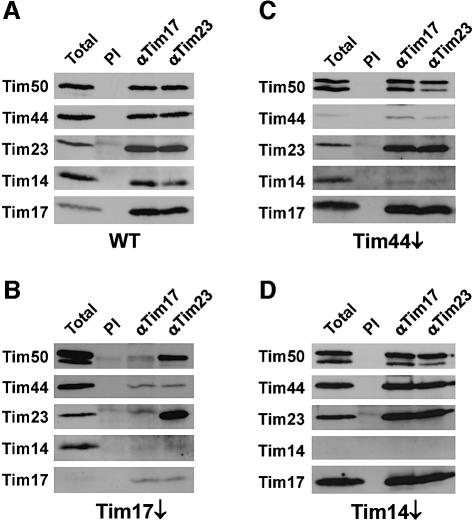
We examined whether Tim14 is present in the TIM23 complex. Antibodies against Tim17 and Tim23 were employed to test for interactions between the known components of the TIM23 complex and Tim14 by co-immunoprecipitation (Figure 5). With mitochondria from wild-type cells, antibodies against Tim17 and Tim23 efficiently co-precipitated Tim14, as well as Tim50 and Tim44 (Figure 5A). In order to gain deeper insight into the structural organization of the TIM23 translocase, we carried out co-immunoprecipitation experiments using mitochondria selectively depleted in one of its components. With mitochondria from Tim17-depleted cells, in which Tim23 was still present, practically neither Tim44 nor Tim14 were co-precipitated using antibodies against either Tim17 or Tim23 (Figure 5B). A similar result was obtained with mitochondria depleted in Tim23 (data not shown). Interestingly, Tim50 and Tim23 form a partial complex in the absence of Tim17. With mitochondria from Tim44-depleted cells, antibodies against Tim17 and against Tim23 co-precipitated the Tim17·Tim23·Tim50 subcomplex, but only minor amounts of Tim14 were co-precipitated, together with residual amounts of Tim44 (Figure 5C). Finally, in case of Tim14-depleted mitochondria, the antibodies against Tim17 and Tim23 pulled down the Tim17·Tim23·Tim50 subcomplex together with Tim44, with a similar efficiency to wild-type mitochondria (Figure 5D).
Fig. 5. Tim14 is part of the TIM23 complex. Mitochondria from (A) wild-type yeast cells, (B) Tim17-depleted cells, (C) Tim44- depleted cells and (D) Tim14-depleted cells were solubilized in digitonin containing EDTA and subjected to immunoprecipitation using antibodies against Tim17 and Tim23 prebound to protein A–Sepharose beads. Antibodies against Tim17 and Tim23 were used because these were able to efficiently recognize their antigens in lysate from digitonin solubilized mitochondria. As a control, antibodies from preimmune serum (PI) were used. The beads were harvested by centrifugation, washed and bound proteins were eluted with Laemmli buffer. Samples were analyzed by SDS–PAGE and immunoblotting with antibodies against the indicated TIM23 components.
These experiments demonstrate that Tim14 is indeed a component of the TIM23 translocase. They also suggest that Tim14 is recruited to the Tim17·Tim23 subcomplex by Tim44. Tim14 seems not only to be in the neighborhood of Tim44, as demonstrated by the crosslinking experiments, but to physically interact with components of the TIM23 machinery.
Tim14 is a central constituent of the mitochondrial import motor
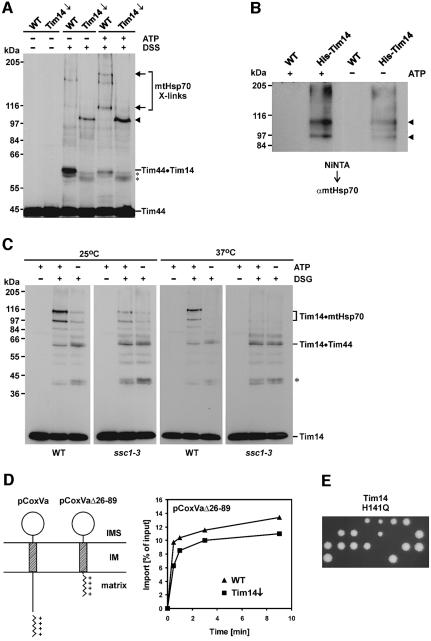
The interaction of Tim14 with Tim44 might suggest a role of Tim14 as part of the import motor. Such a function would imply dynamic interactions with other components of the motor in the course of its ATP-driven cycles. We performed crosslinking experiments with isolated mitochondria whose levels of matrix ATP were manipulated. Crosslinking between Tim14 and Tim44 was relatively weak when ATP was high in the matrix, but was strong when ATP was low (Figure 6A). On the other hand, crosslinked adducts, previously identified as being adducts of Tim44 with mtHsp70 (Schneider et al., 1994), were strong when ATP was high and weak when the ATP level was low. This ATP dependence is in agreement with previous co-immunprecipitation results (Schneider et al., 1994). Furthermore, in the strain where Tim14 was down-regulated, the crosslinking pattern of Tim44 was quite different. The crosslinked adducts of Tim44 with mtHsp70 were strongly reduced. Instead of the Tim44·Tim14 adduct, a few as yet unidentified crosslink bands of slightly lower size became prominent, and a new strong adduct of ∼100 kDa appeared. This might represent a dimer of Tim44; however, this remains to be proven. The changes of the crosslinking pattern in the mitochondria depleted of Tim14 most likely reflect conformational changes of the components of the mitochondrial import motor and not a complete disassembly, as the interaction of Tim44 with mtHsp70 was observed by co-immunoprecipitation from mitochondria depleted of Tim14 (data not shown). In any case, the presence or absence of Tim14 has a strong influence on the environment of Tim44.
Fig. 6. Tim14 is a constituent of the mitochondrial import motor. (A) Tim14 interacts with Tim44 in an ATP-dependent manner. Mitochondria from wild-type and Tim14 depleted cells were subjected to crosslinking with DSS in the presence of low and high matrix ATP. Samples were analyzed by SDS–PAGE and immunoblotting with antibodies against Tim44. Crosslinked adducts to unidentified components close in size to the Tim44·Tim14 crosslink are indicated by asterisks. The arrowhead indicates a Tim44-containing adduct, which is seen only in mitochondria depleted of Tim14. Arrows indicate crosslinked adducts of Tim44 to mtHsp70 as demonstrated by immunodecoration with antibodies against mtHsp70 (not shown here). (B) Tim14 can be crosslinked to mtHsp70. Mitochondria from a strain harboring a Tim14 with an N-terminal His-tag and from wild-type were treated with DSG in the presence or absence of high levels of matrix ATP. Samples were solubilized and incubated with Ni-NTA beads. Bound material was eluted and subjected to SDS–PAGE and immunoblotting with antibodies against mtHsp70. Arrowheads indicate the Tim14–mtHsp70 crosslinked adducts. (C) Crosslinking of Tim14 to mtHsp70 is deficient in the ssc1-3 mutant at non-permissive temperature. Mitochondria were isolated from wild-type cells and from ssc1-3 cells. They were pre-incubated either at 25 or at 37°C for 10 min under conditions that either keep matrix ATP level high or lead to depletion of matrix ATP, and were treated with DSG. Samples were analyzed by immunoblotting with antibodies against Tim14. The asterisk indicates an unidentified crosslinked adduct of Tim14. (D) A precursor that is imported independently of the import motor is not inhibited in its import in mitochondria depleted of Tim14. The precursor pCoxVa(Δ26–89) was synthesized in reticulocyte lysate and imported into mitochondria from wild-type cells and from Tim14-depleted cells. Left panel: schematic representation of the topologies of precursor proteins pCoxVa and pCoxVa(Δ26–89). Right panel: analysis of the import kinetics was as in Figure 4. (E) A Tim14 variant with a mutation in the characteristic HPD motif of the J-domain does not support growth of yeast cells. The conserved histidine residue of the HPD motif of Tim14 (residue 141) was changed to a glutamine residue by in vitro mutagenesis, and the plasmid obtained was transformed into a diploid yeast strain that had one chromosomal allele of Tim14 deleted. Diploid cells were sporulated and individual spores were analyzed for growth on YPD medium. All viable spores obtained were not growing on minimal medium lacking histidine, indicating that these spores carried the chromosomal wild-type allele of Tim14.
Does Tim14 also interact with mtHsp70? Two kinds of experiments were performed. First, mitochondria were isolated from a strain carrying a His-tagged Tim14 and from wild-type cells. They were subjected to crosslinking with DSG in the presence of high and of low matrix ATP. They were then lyzed and the lysates were incubated with Ni-NTA beads. Bound material was eluted with imidazole-containing buffer; proteins were resolved by SDS–PAGE and analyzed by immunoblotting with antibodies against mtHsp70. Two crosslinked adducts of ∼93 and 110 kDa apparent molecular weight were observed. They were strong when matrix ATP was high and weak when matrix ATP was low (Figure 6B).
Secondly, mitochondria from wild-type and from the ssc1-3 strain, which carries a mtHsp70 with a temperature-sensitive mutation in the ATPase domain (Gambill et al., 1993), were incubated either at 25°C (to not induce inactivation of mtHsp70) or at 37°C (to inactivate mtHsp70). The level of matrix ATP was kept either high or was lowered by treatment of the mitochondria with apyrase and oligomycin, and then mitochondria were incubated with or without DSG. Mitochondrial proteins were resolved by SDS–PAGE and immunodecorated with antibodies against Tim14. In wild-type mitochondria the characteristic crosslinks between mtHsp70 and Tim14 were present when ATP was high, but not when it was low (Figure 6C). This was true also with wild-type mitochondria pretreated at 37°C, although the levels of crosslinked species were somewhat reduced by the exposure to a temperature of 37°C. With the ssc1-3 mitochondria, ATP-dependent crosslinking of Tim14 to mtHsp70 was seen upon preincubation of the mitochondria at 25°C (Figure 6C). In contrast, after exposure of the ssc1-3 mitochondria to 37°C, Tim14 could be no longer crosslinked to mtHsp70 (Figure 6C). Remarkably, in this mutant the interaction of Tim14 with Tim44 was still present, but its dependence on matrix ATP was lost.
These results show that Tim14 can interact with mtHsp70, or at least is in close proximity to mtHsp70, depending on the nucleotide bound to mtHsp70. In any case the interaction of these two components appears to be weak. The association of Tim14 with the TIM23 complex including mtHsp70 was observed when mitochondria were lyzed with the mild detergent digitonin, but not in the presence of Triton X-100, in which the interaction of mtHsp70 with Tim44 was preserved (Rassow et al., 1994; Schneider et al., 1994; Horst et al., 1997; data not shown). This weak interaction is not surprising in view of the fact that J-domains interact with DnaK/Hsp70 proteins in a transient, ATP-dependent manner.
As part of the mitochondrial import motor, Tim14 is predicted to be in close proximity to incoming polypeptide chains. We imported radiolabeled preprotein p_b_2Δ19(1–167)DHFRK5 (Schneider et al., 1994) into isolated mitochondria in the presence of methotrexate to arrest it as a translocation intermediate spanning both the TOM and the TIM23 complexes. This arrested precursor protein was crosslinked to Tim14, indicating that Tim14 is indeed in the close vicinity of a translocating chain (see Supplementary data, available at The EMBO Journal Online).
Certain preproteins that use the TIM23 translocase are not dependent on the mitochondrial import motor driven by ATP-dependent binding of mtHsp70 to preproteins during import. These are precursors containing an N-terminal MTS that is immediately followed by a transmembrane segment. The MTS is translocated across the TIM23 complex to the matrix driven by the membrane potential ΔΨ, and the transmembrane segment is inserted into the inner membrane. Apparently, the transmembrane segment gets so close to the putative site in the TIM23 complex which mediates lateral exit that it can insert into the inner membrane without the necessity of being further ‘pulled in’ by mtHsp70. We used a derivative of pCoxVa, pCoxVa(Δ26–89), in which the segment between the MTS and the transmembrane anchor was shortened. Import of this precursor was shown to require Δψ, but not the function of mtHsp70 (Gartner et al., 1995). Import of this preprotein was virtually unimpeded in the strain down-regulated in Tim14 (Figure 6D), in contrast to what was observed with wild-type form of pCoxVa (see Figure 4D). This observation suggests that Tim14 is required for import by the TIM23 complex only when the mtHsp70-dependent import motor is involved.
Finally, we constructed a yeast expression plasmid encoding a mutant form of Tim14. The HPD motif in Tim14, a characteristic of all J-domain proteins, was mutated to QPD. This mutant form of DnaJ was reported to be unable to stimulate the ATPase activity of DnaK (Wall et al., 1994; Mayer et al., 1999). The expression plasmid encoding the QPD mutant of Tim14 was transformed into the diploid strain in which one chromosomal allele of TIM14 was deleted. The resulting strain was subjected to tetrad analysis. Two out of four spores were inviable with all tetrads analyzed, showing that Tim14 needs an intact HPD motif for its activity (Figure 6E). This again suggests strongly that Tim14 functions in the stimulation of mtHsp70.
Discussion
We have identified Tim14, a new component with an essential role in mitochondrial protein import. We named it Tim14 for the following reasons: assigning the affix 18 (according to the molecular weight in kilodaltons of the yeast protein) to this Tim was not possible, because Tim18 is already taken, likewise Tim17 (Pfanner et al., 1996; Kerscher et al., 2000; Koehler et al., 2000). Since all the other putative orthologs found in the data bases have lower molecular weights of ∼11–16 kDa, we use Tim14.
The discovery of Tim14 substantially changes and advances our knowledge of the structural organization and function of the import machinery. Tim14 is a constituent of the TIM23 translocase. It is anchored to the mitochondrial inner membrane such that it exposes a classical J-domain into the matrix. This J-domain is very similar to the J-domain of the bacterial co-chaperone DnaJ (Bukau and Horwich, 1998; Kelley, 1998). According to sequence similarity and structure prediction, Tim14 contains the typical helical segments, H1–H3, found to make up the tertiary fold of the J-domain of DnaJ (Szyperski et al., 1994; Kelley, 1998); also, the canonical HPD motif is present. Tim14 lacks, however, all other structural elements present in DnaJ.
The J-domain of DnaJ is known for its ability to stimulate the ATPase activity of DnaK (Liberek et al., 1991; Szabo et al., 1994; Wall et al., 1994; McCarty et al., 1995; Russell et al., 1999); this stimulation was shown to occur by the interaction of the J-domain with the ATPase domain of DnaK (Greene et al., 1998; Suh et al., 1998). We suggest that Tim14 has a major function in stimulating the ATPase activity of mtHsp70. Indeed, a yeast mutant in which the HPD motif was non-functional was not viable. Furthermore, as we report here, Tim14 can be crosslinked to mtHsp70 and Tim44 in an ATP-dependent manner. In summary, these findings strongly suggest a role of Tim14 in the protein import motor of the mitochondria. Thus, the J-domain protein Tim14 may exert a similar function in protein import as the J-domain of Mdj1p in protein folding in the matrix (Rowley et al., 1994).
We propose the following model for the mechanism of the import motor. The unfolded preprotein appearing from the outlet of the TIM23 import channel binds to Tim44. Tim44 recruits mtHsp70 in the ATP form, and the preprotein chain enters the peptide binding domain of mtHsp70. Tim14 stimulates the ATPase activity of mtHsp70. As a result, the peptide binding pocket closes and tightly binds the preprotein. The mtHsp70 dissociates from Tim44 and the preprotein bound to mtHsp70 can move into the matrix, but not in a retrograde fashion. The mitochondrial nucleotide exchange factor Mge1 mediates the release of bound ADP from mtHsp70 followed by release of the preprotein so that mtHsp70 can be engaged in the next cycle. The activation of mtHsp70 by Tim14 is an essential element in this proposed mechanism as in this way mtHsp70 can rapidly cycle on and off from Tim44. Net inward movement into the matrix would be the result.
The three components Tim14, Tim44 and mtHsp70 can be seen as parts of a molecular machine that, at the various stages of their action, change their positions relative to each other. ATP hydrolysis by mtHsp70 is driving these movements and thereby the translocation of the preproteins.
Tim44 appears to have an additional function in recruiting Tim14 and mtHsp70 either directly or indirectly to the Tim17·Tim23 subcomplex, which forms the protein import channel. Neither Tim23 nor Tim17 alone is sufficient to mediate this recruitment. In conclusion, Tim44 may be viewed as a scaffold that integrates the various partial reactions.
In the light of our results, the evolutionary conservation between the DnaK–DnaJ–GrpE folding machine and the mitochondrial import motor seems much more evident. Tim44 and Tim14 together seem to represent a DnaJ-type co-chaperone. In the bacterial system DnaJ contains the J-domain at its N-terminal part, which interacts with the ATPase domain of DnaK and stimulates its ATPase activity and a peptide binding segment in the more C-terminal part (Liberek et al., 1991; Szabo et al., 1994; Wall et al., 1994; McCarty et al., 1995; Banecki et al., 1996; Greene et al., 1998). In the mitochondrial system, Tim14 carries the J-domain, and Tim44 has the ability to bind translocated segments of the unfolded polypeptide chains (Schneider et al., 1994). There may be other functional aspects of these systems, which are conserved as well. The peptide binding domain of DnaK was reported to be able to interact with DnaJ (Suh et al., 1998, 1999). In mitochondria, the peptide binding domain of mtHsp70 was reported to interact with Tim44 (Moro et al., 2002), although a different view has been put forward (Krimmer et al., 2000).
Several questions arise from the mechanism proposed above. One is how the ATP/ADP cycle of mtHsp70 is controlled so that mtHsp70 is not in an idling state when no polypeptide substrate is presented by Tim44. One possibility among several is that the action of Tim14 is regulated by other, known or unknown, components of the TIM23 complex that sense the presence or absence of a substrate in the translocase. A further point that needs to be clarified is the sequence of events after binding of mtHsp70 to a segment of the incoming polypeptide chain. In analogy to the DnaJ·DnaK system (Liberek et al., 1991; Szabo et al., 1994; McCarty et al., 1995; Hartl, 1996; Bukau and Horwich, 1998), first ATP hydrolysis may occur, followed by dissociation of mtHsp70 from Tim44, as proposed above. Alternatively, the mtHsp70 in the ATP form could dissociate from Tim44, and ATP hydrolysis stimulated by Tim14 might occur afterwards. The association of Tim14 with the inner membrane, however, argues strongly for the conversion of mtHsp70·ATP to mtHsp70·ADP at the membrane as suggested previously (Schneider et al., 1994; Neupert and Brunner, 2002). This is in line with the fact that Hsp70 chaperones bind substrate tightly only in the ADP form. mtHsp70 must have a higher affinity for the substrate than the other parts of the TIM23 complex to be able to drive inward movement of the preprotein. However, ATP hydrolysis was also proposed to occur after release of mtHsp70 from the membrane (Voos and Rottgers, 2002). Lastly, the sequence of the events in the import motor are relevant to the problems as to how mtHsp70 facilitates inward movement of the substrate polypeptide. Release of the mtHsp70·ATP from Tim44 would exclude a powerstroke type function of mtHsp70 (Liu et al., 2003). In case of release of the ADP form, this reaction step would not allow a discrimination between different mechanisms proposed (Neupert and Brunner, 2002).
J-domain-containing proteins play important roles in the transport of proteins from the cytosol into the lumen of the endoplasmic reticulum (ER). Sec63 and Mtj1 are such components (Corsi and Schekman, 1997; Misselwitz et al., 1999; Chevalier et al., 2000; Dudek et al., 2002). Their J-domains stimulate the ATPase activity of the Hsp70-type chaperone, BiP, in the lumen of the ER (Corsi and Schekman, 1997; Misselwitz et al., 1998; Chevalier et al., 2000). It is remarkable that protein translocases as different in their composition and mechanisms as the Sec61 complex and the TIM23 complex have rather similar motors that drive the movement of proteins across membranes.
Recently, the mechanism of the mitochondrial import motor was investigated by an elegant in vitro assay, using the isolated components Tim44, mtHsp70 and Mge1 (Liu et al., 2003). The inclusion of Tim14 in such experiments will provide further insight into the details of the sequence of events and the kinetics of the numerous steps of the import cycle.
Materials and methods
Yeast strains, cell growth and plasmids
The wild-type strains D273-10b and YPH499 were used (Mokranjac et al., 2003). A heterozygous deletion strain of TIM14 was generated by replacing one allele of the TIM14 gene (from codon 1 to the stop codon) with the HIS3 gene in the diploid yeast strain YPH501 by homologous recombination. The Gal-TIM14 strain was constructed by replacing the 99 bp upstream of the TIM14 reading frame with a Gal10 promoter-containing cassette in YPH499. A His-Tim14 strain expressing Tim14 with a His8 tag at the N-terminus under control of the Gal10 promoter was constructed as described for the Gal-TIM14 strain; a cassette was used containing the GAL10 promoter followed by the coding sequence of the His8 tag. The nucleotides encoding the 3HA-tag were inserted after the coding sequence of Tim14 on the chromosome of YPH501 by homologous recombination according to published methods (Knop et al., 1999), generating the TIM14-3HA strain. For depletion of Tim14, Gal-Tim14 cultures were shifted from lactate medium containing 0.1% galactose to lactate medium containing 0.1% glucose for the indicated time periods. Control wild-type cultures were grown in the same way. Otherwise, cells were grown on lactate medium containing 0.1% galactose.
The Tim14 H141Q mutant was generated by site directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). As template for the PCR the plasmid pGEM4-Tim14 containing the nucleotides encoding the Tim14 protein was used. For identification of positive clones a silent mutation was introduced at the nucleotide position 412 generating a _Nhe_I restriction site. The resulting mutated nucleotide sequence of TIM14 was cloned into the yeast expression vector pVT102U generating the plasmid pVT102U-Tim14(H141Q). The diploid yeast strain TIM14/tim14::HIS3 was transformed with pVT102U-Tim14(H141Q) and subjected to sporulation and tetrad analysis.
Antibodies against Tim14
The nucleotide sequence encoding amino acid residues 50–168 of Tim14 was cloned into the pQE-30 vector (Qiagen). The protein was expressed in E.coli XL1-Blue cells and purified on a Ni-NTA–agarose column under denaturing conditions according to manufacturer’s instructions. Antibodies against the protein were raised in rabbits and affinity purified prior to use.
Chemical crosslinking
Intact mitochondria, resuspended in import buffer (without BSA), were subjected to chemical crosslinking with DSG or DSS. Crosslinker was added from a 100-fold stock in DMSO. After 30 min incubation on ice, excess crosslinker was quenched by addition of 0.1 M glycine pH 8.8. Mitochondria were reisolated and analyzed by SDS–PAGE and immunodecoration. Where indicated, mitochondrial ATP was depleted prior to crosslinking by addition of apyrase (10 U/ml) and oligomycin (10 µM) or kept high by addition of ATP (4 mM), NADH (5 mM), creatine phosphate (10 mM) and creatine kinase (0.1 mg/ml) for 10 min at 25°C. When crosslinking adducts were analyzed by binding to Ni-NTA–agarose beads, re-isolated mitochondria were solubilized with 1% SDS in 50 mM Na-phosphate pH 8.0, 100 mM NaCl, 10% glycerol, 10 mM imidazole, 1 mM PMSF for 15 min at 25°C. Samples were diluted 20-fold in the same buffer containing 0.2% Triton X-100 and, after a clarifying spin, added to 50 µl Ni-NTA–agarose beads. After 1 h agitation at 4°C, beads were washed and bound proteins eluted with Laemmli buffer containing 300 mM imidazole for 5 min at 95°C.
Miscellaneous
Subcellular and submitochondrial fractionations were performed as described previously (Rowley et al., 1994). Import of preproteins into isolated mitochondria was carried out according to published procedures (Mokranjac et al., 2003). For co-immunoprecipitation experiments mitochondria (1 mg/ml) were solubilized in 20 mM Tris pH 7.5, 80 mM KCl, 10% glycerol, 5 mM EDTA, 1 mM PMSF with 1% digitonin for 30 min on ice, and further processed as described previously (Mokranjac et al., 2003).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are very grateful to Professor Michael Brunner for stimulating discussions, to Ulrike Gärtner and Heiko Germeroth for excellent technical assistance, and to Carsten Bornhövd and Stephan Meier for providing subcellular yeast fractions and the CoxVa constructs. This work was supported by grants from the Deutsche Forschungsgemeinschaft, SFB 594 (B3, B4), the Bundesministerium für Bildung und Forschung (MITOP) and the Fonds der Chemischen Industrie.
References
- Banecki B., Liberek,K., Wall,D., Wawrzynow,A., Georgopoulos,C., Bertoli,E., Tanfani,F. and Zylicz,M. (1996) Structure–function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem., 271, 14840–14848. [DOI] [PubMed] [Google Scholar]
- Bukau B. and Horwich,A.L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell, 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Chevalier M., Rhee,H., Elguindi,E.C. and Blond,S.Y. (2000) Interaction of murine BiP/GRP78 with the DnaJ homologue MTJ1. J. Biol. Chem., 275, 19620–19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi A.K. and Schekman,R. (1997) The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol., 137, 1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P.J., Martin,F., Maarse,A.C., Bomer,U., Muller,H., Guiard,B., Meijer,M., Rassow,J. and Pfanner,N. (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70–Tim44. EMBO J., 16, 5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J. et al. (2002) A novel type of co-chaperone mediates transmembrane recruitment of DnaK-like chaperones to ribosomes. EMBO J., 21, 2958–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T. and Kohda,D. (2002) Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta, 1592, 3–14. [DOI] [PubMed] [Google Scholar]
- Foelsch H., Guiard,B., Neupert,W. and Stuart,R.A. (1996) Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J., 15, 479–487. [PMC free article] [PubMed] [Google Scholar]
- Gambill B.D., Voos,W., Kang,P.J., Miao,B., Langer,T., Craig,E.A. and Pfanner,N. (1993) A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol., 123, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner F., Voos,W., Querol,A., Miller,B.R., Craig,E.A., Cumsky,M.G. and Pfanner,N. (1995) Mitochondrial import of subunit Va of cytochrome c oxidase characterized with yeast mutants. J. Biol. Chem., 270, 3788–3795. [DOI] [PubMed] [Google Scholar]
- Geissler A. et al. (2002) The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell, 111, 507–518. [DOI] [PubMed] [Google Scholar]
- Greene M.K., Maskos,K. and Landry,S.J. (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl Acad. Sci. USA, 95, 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U. (1996) Molecular chaperones in cellular protein folding. Nature, 381, 571–579. [DOI] [PubMed] [Google Scholar]
- Horst M., Oppliger,W., Rospert,S., Schonfeld,H.J., Schatz,G. and Azem,A. (1997) Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J., 16, 1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. and Dunn,C. (2002) Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim. Biophys. Acta, 1592, 25–34. [DOI] [PubMed] [Google Scholar]
- Kelley W.L. (1998) The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci., 23, 222–227. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Sepuri,N.B. and Jensen,R.E. (2000) Tim18p is a new component of the Tim54p–Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell, 11, 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Murphy,M.P., Bally,N.A., Leuenberger,D., Oppliger,W., Dolfini,L., Junne,T., Schatz,G. and Or,E. (2000) Tim18p, a new subunit of the TIM22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol. Cell. Biol., 20, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimmer T., Rassow,J., Kunau,W.H., Voos,W. and Pfanner,N. (2000) Mitochondrial protein import motor: the ATPase domain of matrix Hsp70 is crucial for binding to Tim44, while the peptide binding domain and the carboxy-terminal segment play a stimulatory role. Mol. Cell. Biol., 20, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronidou N.G., Oppliger,W., Bolliger,L., Hannavy,K., Glick,B.S., Schatz,G. and Horst,M. (1994) Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc. Natl Acad. Sci. USA, 91, 12818–12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K., Marszalek,J., Ang,D., Georgopoulos,C. and Zylicz,M. (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl Acad. Sci. USA, 88, 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., D’Silva,P., Walter,W., Marszalek,J. and Craig,E.A. (2003) Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science, 300, 139–141. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Pfanner,N. and Voos,W. (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO rep., 1, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P., Laufen,T., Paal,K., McCarty,J.S. and Bukau,B. (1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol., 289, 1131–1144. [DOI] [PubMed] [Google Scholar]
- McCarty J.S., Buchberger,A., Reinstein and Bukau,B. (1995) The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol., 249, 126–137. [DOI] [PubMed] [Google Scholar]
- Milisav I., Moro,F., Neupert,W. and Brunner,M. (2001) Modular structure of the TIM23 preprotein translocase of mitochondria. J. Biol. Chem., 276, 25856–25861. [DOI] [PubMed] [Google Scholar]
- Misselwitz B., Staeck,O. and Rapoport,T.A. (1998) J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell, 2, 593–603. [DOI] [PubMed] [Google Scholar]
- Misselwitz B., Staeck,O., Matlack,K.E. and Rapoport,T.A. (1999) Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J. Biol. Chem., 274, 20110–20115. [DOI] [PubMed] [Google Scholar]
- Mokranjac D., Paschen,S.A., Kozany,C., Prokisch,H., Hoppins,S.C., Nargang,F.E., Neupert,W. and Hell,K. (2003) Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J., 22, 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro F., Sirrenberg,C., Schneider,H.C., Neupert,W. and Brunner,M. (1999) The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J., 18, 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro F., Okamoto,K., Donzeau,M., Neupert,W. and Brunner,M. (2002) Mitochondrial protein import: molecular basis of the ATP-dependent interaction of MtHsp70 with Tim44. J. Biol. Chem., 277, 6874–6880. [DOI] [PubMed] [Google Scholar]
- Neupert W. and Brunner,M. (2002) The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol., 3, 555–565. [DOI] [PubMed] [Google Scholar]
- Paschen S.A. and Neupert,W. (2001) Protein import into mitochondria. IUBMB Life, 52, 101–112. [DOI] [PubMed] [Google Scholar]
- Pfanner N. et al. (1996) Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem. Sci., 21, 51–52. [PubMed] [Google Scholar]
- Rassow J., Maarse,A.C., Krainer,E., Kubrich,M., Muller,H., Meijer,M., Craig,E.A. and Pfanner,N. (1994) Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol., 127, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P., Pfanner,N. and Meisinger,C. (2003) Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J. Mol. Biol., 326, 639–657. [DOI] [PubMed] [Google Scholar]
- Rowley N., Prip-Buus,C., Westermann,B., Brown,C., Schwarz,E., Barrell,B. and Neupert,W. (1994) Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell, 77, 249–259. [DOI] [PubMed] [Google Scholar]
- Russell R., Wali Karzai,A., Mehl,A.F. and McMacken,R. (1999) DnaJ dramatically stimulates ATP hydrolysis by DnaK: insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry, 38, 4165–4176. [DOI] [PubMed] [Google Scholar]
- Schneider H.C., Berthold,J., Bauer,M.F., Dietmeier,K., Guiard,B., Brunner,M. and Neupert,W. (1994) Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature, 371, 768–774. [DOI] [PubMed] [Google Scholar]
- Suh W.C., Burkholder,W.F., Lu,C.Z., Zhao,X., Gottesman,M.E. and Gross,C.A. (1998) Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl Acad. Sci. USA, 95, 15223–15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh W.C., Lu,C.Z. and Gross,C.A. (1999) Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem., 274, 30534–30539. [DOI] [PubMed] [Google Scholar]
- Szabo A., Langer,T., Schroder,H., Flanagan,J., Bukau,B. and Hartl,F.U. (1994) The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ and GrpE. Proc. Natl Acad. Sci. USA, 91, 10345–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski T., Pellecchia,M., Wall,D., Georgopoulos,C. and Wuthrich,K. (1994) NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc. Natl Acad. Sci. USA, 91, 11343–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott K.N., Kovermann,P., Geissler,A., Merlin,A., Meijer,M., Driessen,A.J., Rassow,J., Pfanner,N. and Wagner,R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol., 8, 1074–1082. [DOI] [PubMed] [Google Scholar]
- Voos W. and Rottgers,K. (2002) Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta, 1592, 51–62. [DOI] [PubMed] [Google Scholar]
- Wall D., Zylicz,M. and Georgopoulos,C. (1994) The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem., 269, 5446–5451. [PubMed] [Google Scholar]
- Westermann B. and Neupert,W. (1997) Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. J. Mol. Biol., 272, 477–483. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Esaki,M., Kanamori,T., Tamura,Y., Nishikawa,S. and Endo,T. (2002) Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell, 111, 519–528. [DOI] [PubMed] [Google Scholar]