Transcription of histone gene cluster by differential core-promoter factors (original) (raw)
Abstract
The 100 copies of tandemly arrayed Drosophila linker (H1) and core (H2A/B and H3/H4) histone gene cluster are coordinately regulated during the cell cycle. However, the molecular mechanisms that must allow differential transcription of linker versus core histones prevalent during development remain elusive. Here, we used fluorescence imaging, biochemistry, and genetics to show that TBP (TATA-box-binding protein)-related factor 2 (TRF2) selectively regulates the TATA-less Histone H1 gene promoter, while TBP/TFIID targets core histone transcription. Importantly, TRF2-depleted polytene chromosomes display severe chromosomal structural defects. This selective usage of TRF2 and TBP provides a novel mechanism to differentially direct transcription within the histone cluster. Moreover, genome-wide chromatin immunoprecipitation (ChIP)-on-chip analyses coupled with RNA interference (RNAi)-mediated functional studies revealed that TRF2 targets several classes of TATA-less promoters of >1000 genes including those driving transcription of essential chromatin organization and protein synthesis genes. Our studies establish that TRF2 promoter recognition complexes play a significantly more central role in governing metazoan transcription than previously appreciated.
[Keywords: TATA-box-binding protein (TBP), TBP-related factor 2 (TRF2), core-promoter recognition, histone gene cluster, ChIP-on-chip, chromatin organization]
Core promoters serve as the platform for the assembly of transcription initiation complexes critical for specifying accurate and regulated RNA synthesis. The eukaryotic cellular RNA polymerase II (Pol II) machinery has evolved to recognize multiple core-promoter elements such as the TATA box, Initiator, and DPE (Smale and Kadonaga 2003). Indeed, studies of metazoan core promoters revealed considerably greater _cis_-element diversification than previously expected. For example, TATA boxes, which were thought to be the most widely distributed prototypic core-promoter element recognized by the general transcription factor TBP (TATA-box-binding protein)/TFIID (consisting of TBP and TBP-associated factors, TAFs), are found in <20%∼30% of annotated promoters in Drosophila and human. Instead, the majority of core promoters fall into various distinct TATA-less categories (Ohler et al. 2002; Gershenzon and Ioshikhes 2005; Jin et al. 2006). Consistent with diversified core-promoter structures, recent studies identified a family of TBP-related factors (TRFs) (Crowley et al. 1993; Dantonel et al. 1999; Maldonado 1999; Ohbayashi et al. 1999; Rabenstein et al. 1999; Berk 2000; Tupler et al. 2001; Hochheimer and Tjian 2003), but their potential core-promoter recognition functions have remained elusive.
Metazoan cells have been found to use a diversified set of TBP-related molecules that display altered DNA-binding specificities (Hochheimer and Tjian 2003). In Drosophila, TRFs have been implicated in promoter-selective transcription for both Pol II and Pol III gene promoters (Hansen et al. 1997; Holmes and Tjian 2000; Takada et al. 2000; Hochheimer et al. 2002; Isogai et al. 2007). However, a comprehensive analysis of TRFs in promoter-selective recognition of Pol II core promoters has not been performed. Our earlier studies found that a multisubunit TRF2-containing complex includes the transcription factor DREF and is involved in targeting a subset of promoters containing the DNA replication-related element (DRE) (Hochheimer et al. 2002). The PCNA gene promoter contains such a DRE (Hirose et al. 1993) and represents a novel tandem core-promoter class composed of two distinct transcriptional start sites, each of which appears to be subject to regulation either by the TRF2/DREF complex or TBP/TAFs. While TRF2 recruitment to the core promoter via DREF may account for a subset of TRF2-dependent promoters, TRF2 is also found in complexes lacking DREF. For example, TRF2 and DREF display only a limited set of overlapping sites in Drosophila Schneider cells visualized by immunofluorescence staining (Y. Isogai and R. Tjian, unpubl.), suggesting that TRF2 may be playing multiple roles—some in conjunction with DREF and others independent of DREF. We therefore surmised that there may be additional important TRF2 target promoters that remained uncharacterized.
In order to gain a more comprehensive map of potential TRF2-dependent promoters, we conducted a genome-wide analysis of TRF2 recognition sites both by polytene chromosome staining as well as chromatin immunoprecipitation (ChIP) coupled with high-density tiling microarray detection (ChIP-on-chip). These approaches have revealed several important target genes that illustrate how TRF2 is used as an alternative core-promoter recognition factor. First, we provide biochemical and genetic evidence that two distinct sets of core-promoter recognition factors are responsible for directing transcription of the nucleosome core histone genes (H2A/B and H3/H4) and the linker histone H1. Genome-wide ChIP-on-chip analysis revealed that TRF2 recognizes and binds in vivo to a large number of TATA-less core promoters. Importantly, a majority of these TATA-less promoters are selectively recognized by TRF2, but not by TBP. Moreover, with salivary gland-specific depletion of TRF2, we found that TRF2 participates in regulation of chromatin organization and cell growth, by controlling Histone H1 and ribosomal protein gene expression. Taken together, these data establish that TRF2 is responsible for differentially recognizing and regulating a subset of TATA-less promoters that have shed the requirement for TBP through the usage of novel core-promoter structures. Remarkably, even coordinately expressed gene clusters such as the histone complex have evolved mechanisms to be differentially regulated by alternative core-promoter recognition machinery.
Results
Linking histone gene cluster to TRF2
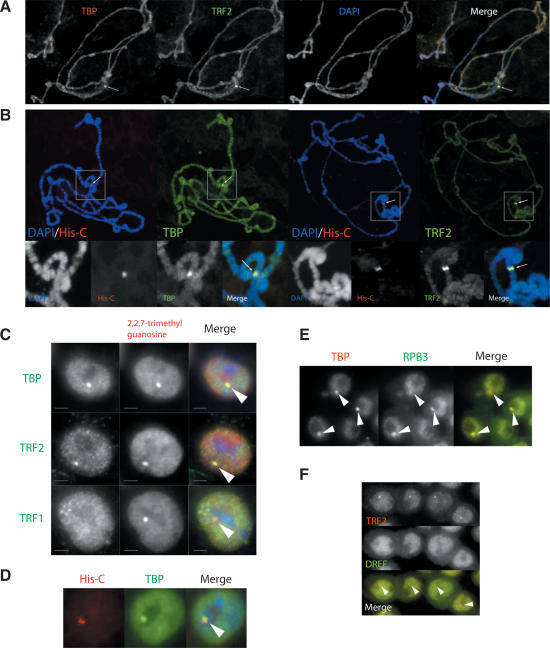
In an attempt to obtain an initial, relatively low-resolution picture of how TRF2 might be directly involved in promoter-selective transcription in vivo, we used polytene chromosome staining to examine both TBP and TRF2 occupancy on salivary gland DNA. We found that TRF2 localization is largely nonoverlapping relative to the more uniform localization of TBP, with one prominent exception at a locus proximal to the chromocenter that stained intensely with both TBP- and TRF2-specific antibodies (Fig. 1A). Interestingly, this genomic region was not stained by anti-DREF antibody, suggesting that DREF is not recruited to this locus (data not shown). Because of the particularly intense antibody staining as well as the cytological location, we reasoned that the observed signal may correspond to the histone genes, a gene cluster that is amplified to ∼100 copies per haploid genome (Lifton et al. 1978; Matsuo and Yamazaki 1989). Therefore, we used fluorescence in situ hybridization (FISH) to confirm the histone gene cluster locus and simultaneously determined if both TBP and TRF2 colocalize to the histone gene cluster by immunostaining. The unusually bright staining by anti-TBP and TRF2 antibodies indeed were found to overlap at the histone gene cluster DNA (Fig. 1B), establishing that both TBP and TRF2 are resident at this locus.
Figure 1.
Localization of TBP and TRF2 at the histone gene cluster and the HLB. (A) Polytene chromosomal staining of TRF2 (green) and TBP (red). DNA (blue) is stained by DAPI. Arrows mark the cytological locus stained intensely both by anti-TRF2 and anti-TBP antibodies. (B) Colocalization of TBP and TRF2 at the histone cluster. Polytene immunostaining combined with DNA FISH probing the histone gene cluster. (Left) TBP immunostaining (green) combined with histone gene cluster (His-C) DNA FISH (red). (Right) TRF2 immunostaining (green) combined with histone gene cluster DNA FISH (red). DNA (blue) was stained with DAPI. The magnified view for the histone cluster is displayed below. The arrows mark the site of the histone gene cluster indicated by the FISH signals. (C) Colocalization of TBP and TRF2 at the HLB. Drosophila S2 cells doubly stained with anti-TMG antibodies (red), a marker for the HLB, and anti-TBP-, TRF1-, or TRF2-specific antibodies (green). (D) Intranuclear clustering of TBP and the histone gene cluster. S2 cells stained with anti-TBP antibody (green) coupled with DNA FISH marking the histone gene cluster (red), indicated by the arrow. (E) Pol II localization at the HLB. Drosophila Kc cells are stained with anti-TBP (red) and anti-RPB3, a Pol II subunit (green), antibodies. Arrows indicate the HLB marked by the TBP staining. (F) TRF2 accumulates at the HLB independent of DREF. Drosophila S2 cells are stained with anti-DREF (green) and anti-TRF2 (red) antibodies.
TBP and TRF2 colocalize at the histone locus in multiple cell types
To further probe TBP and TRF2 localization in other cell types, we examined TBP/TRF2 localization in the interphase nuclei of both S2 and Kc cells, which are derived from Drosophila embryos. Interestingly, while a majority of TBP and TRF2 proteins appeared distributed uniformly in a punctate pattern throughout the nucleoplasm of these interphase cells, both TBP and TRF2 can be clearly seen to colocalize at a specific nuclear spot in the vicinity of the nucleolus (Fig. 1C). This nuclear body appears to accumulate a specific subset of the transcription machinery including Pol II (Fig. 1E), but not TRF1 or DREF (Fig. 1C,F). Since the histone gene cluster has recently been physically linked to the histone locus body (HLB) (Liu et al. 2006), an antibody against 2,2,7-trimethylguanosine (TMG), which marks small nuclear RNAs that accumulate in the HLB (Liu et al. 2006), was used to confirm that the site of TBP and TRF2 accumulation corresponds to the HLB. Indeed, intense TMG accumulation is found coincident with the TBP/TRF2 localization (Fig. 1C). Using DNA FISH in combination with immunostaining of TBP, we found that this specific nuclear body is invariantly juxtaposed to the histone gene locus (Fig. 1D). Therefore, both TBP and TRF2 in Drosophila cells appear to be specifically targeted to the HLB, a nuclear body that is tightly linked to histone gene expression.
The Histone H1 promoter is selectively recognized by TRF2
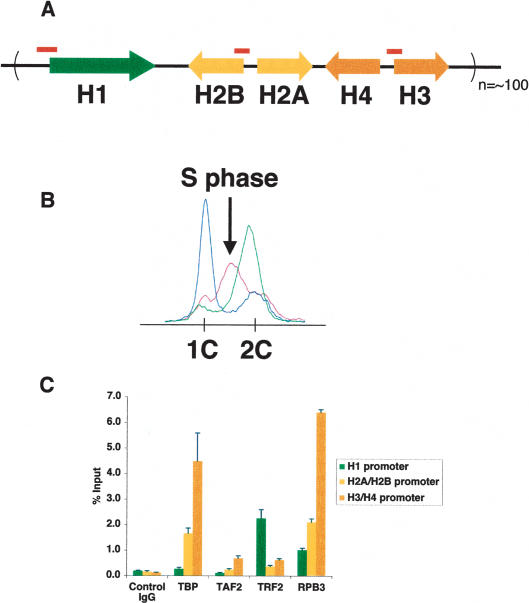
The apparent colocalization of TBP and TRF2 at the histone cluster at this relatively low resolution suggests that histone genes may represent another member of the tandem core-promoter class recognized by both TBP and TRF2. To test this hypothesis more directly, we examined the potential involvement of TBP and TRF2 at the histone locus at higher resolution by probing the individual core-promoter regions of H1, H2A/B, and H3/H4 genes (Fig. 2A). Using a ChIP assay with extracts derived from S-phase-synchronized Drosophila Kc cells (Fig. 2B), we found that TBP and TRF2 are recruited to the histone gene cluster in a segregated fashion. Remarkably, TBP selectively occupies H2A/B and H3/H4 core-promoter regions, but not H1, while TRF2 was found to occupy only the H1 promoter, and not H2A/B and H3/H4 (Fig. 2C). RNA Pol II is recruited to all five classes of histone gene promoters, consistent with the S-phase-specific transcription of the histone genes. As expected, TAF2, a subunit of the TFIID complex, does not occupy the Histone H1 gene promoter but is detectable at core histone promoters. TFIIA, which is known to bind directly to both TBP and TRF2 (Rabenstein et al. 1999), was also recruited to all five classes of histone promoters (data not shown). These striking findings suggest that even within a gene cluster, individual members may be regulated by distinct core promoter recognition machinery.
Figure 2.
Selective recruitment of TRF2/TBP in the histone gene cassette. (A) Schematics of the histone gene cassette. Positions of primer sets used to amplify the H1, H2A/B, and H3/H4 promoters are indicated by red bars. (B) Fluorescence-activated cell sorting (FACS) profile of synchronized Drosophila Kc cells. S-phase cells (purple) used for C are indicated by the arrow. G1 cells and G2 cells are indicated by blue and green, respectively. (C) Segregation of TBP and TRF2 recognition sites within the histone cluster. ChIP assays using S-phase-synchronized Kc cells. The graph represents the occupancy of TBP, TAF2, TRF2, and RPB3 at H1 (green), H2A/B (yellow), and H3/H4 (orange) core-promoter regions.
Since histone genes are generally expressed in an S-phase-dependent manner, we wondered whether the recruitment of TBP and TRF2 may correlate with different cell cycle phases. Therefore, we conducted ChIP experiments examining the status of TBP, TRF2, and Pol II occupancy at the H1 and H3/H4 promoters in G2 and S phases (Supplementary Fig. S2). Interestingly, while Pol II recruitment at the histone genes appears to be S-phase-dependent and a slight enrichment was observed for TBP at the H3/H4 promoters, TRF2 recruitment at the H1 promoter remained robust and invariant during all phases of the cell cycle. Drosophila cells can apparently use different strategies for transcriptional activation wherein recruitment of a core-promoter component such as TRF2 to the H1 promoter is not the rate-limiting step. This is in contrast to the more conventional situation observed at the heavy metal-inducible MtnA promoter, in which recruitment of TFIID provides a key step for activation (Marr et al. 2006).
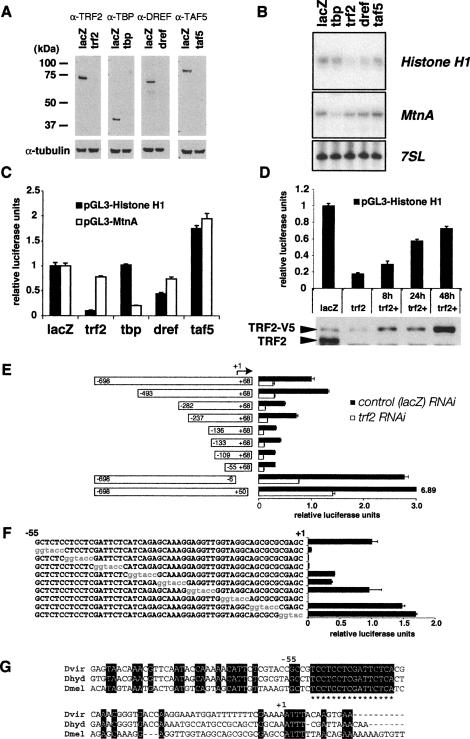
TRF2 is required for Histone H1 transcription
Since Histone H1 has apparently been singled out for recognition and binding by TRF2, we next probed whether TRF2, but not TBP, is actually responsible for potentiating the transcription of the Histone H1 gene. First, we used an efficient RNA interference (RNAi)-mediated knockdown assay in S2 cells to target TBP and TRF2 as well as other select subunits of TBP- or TRF2-containing complexes to determine whether the promoter activity of the Histone H1 gene requires TBP or TRF2 in Drosophila cells (Fig. 3A). After efficiently depleting TBP or a TAF (TAF5) by RNAi, Histone H1 steady-state mRNA levels remained largely unaffected (Fig. 3B). In contrast, RNAi depletion of TRF2 resulted in a significant and reproducible down-regulation of endogenous levels of Histone H1 mRNA as well as a Histone H1 reporter gene activity (approximately fivefold). Near complete depletion of DREF resulted in a more modest reduction (approximately twofold) of endogenous H1 mRNA (Fig. 3B) as well as H1 reporter activity (Fig. 3C). Since DREF does not appear to be recruited to the histone cluster and consensus DRE elements have not been found in the Histone H1 promoter, this observed effect may be indirect. Such an indirect down-regulation was expected since DREF depletion induces a cell cycle arrest by regulating a large number of DNA replication-related genes (Hyun et al. 2005). As an important control, we found that the integrity of the general Pol II transcriptional machinery remained largely unaffected upon TRF2 depletion. For example, heavy metal-inducible transcription from the TBP/TFIID-dependent Metallothionein A (MtnA) gene (Marr et al. 2006) remained robust even after near complete depletion of TRF2 (Fig. 3B). Assaying the effects of depleting TRF2 or TBP using luciferase reporter assays instead of measuring endogenous Histone H1 or MtnA confirmed our results (Fig. 3C). In addition, using a stable S2 cell line carrying an inducible V5-tagged Trf2 gene, we were able to efficiently (∼80%) rescue Histone H1 promoter activity by re-expressing exogenous TRF2 protein for 48 h (Fig. 3D). These data, taken together, suggest that Histone H1, unlike PCNA, is driven by a novel class of core promoters that are TRF2-dependent and TBP-independent.
Figure 3.
TRF2 regulates Histone H1 promoter activity. (A) Western blot showing efficient RNAi-mediated knockdown of TRF2, TBP, TAF5, and DREF. α-Tubulin was used as a loading control. (B) Endogenous Histone H1 mRNA level is dependent on TRF2. Primer extension analysis of Histone H1, MtnA, and 7SL RNA transcripts. Ten micrograms of nuclear RNA isolated after RNAi-mediated depletion of indicated transcription factors were used to perform a primer extension reaction using specific primers for Histone H1, MtnA, and 7SL RNA. (C) Cloned Histone H1 promoter activity is dependent on TRF2. Luciferase reporter analysis using the Histone H1 promoter and the MtnA promoter. Luciferase reporter activity calibrated to pAct5C-hRenilla activity was plotted after dsRNA-mediated depletion of transcription factors indicated. (D) Rescue of Histone H1 activity by exogenous TRF2 expression. (Top) Luciferase reporter activity of pGL3-Histone H1 using S2 cells with inducible TRF2-V5 expression under the control of MtnA promoter. During 4 d of RNAi treatments, some cells received an extra dose of TRF2 protein under the control of MtnA promoter. The 2 d of dsRNA treatment were sufficient to cause a threefold reduction of pGL3 reporter activity (data not shown). (lacZ) Control RNAi; (trf2) trf2 RNAi; (trf2+) trf2 RNAi with re-expression of TRF2-V5 for the indicated number of hours. (Bottom) Western blot using anti-TRF2 antibody indicating the efficient RNAi of TRF2 and re-expression of TRF2-V5. (E) TRF2-dependent activity of the Histone H1 promoter is localized to the proximal promoter region. Truncations of −698 to +68 of the Histone H1 promoter fused to luciferase were generated and tested for activity in either lacZ control or trf2 RNAi backgrounds. The smallest fragment (−55 to +68) retained TRF2-dependent activity. (F) Identification of two proximal sequence boxes critical for the Histone H1 promoter activity by linker scanning mutagenesis. Luciferase activity from various Histone H1 promoter (−698 to +68) templates harboring mutations in the −55 to +68 region (shown in red) is plotted in the graph. (G) A promoter-proximal, TC-rich box critical for the Histone H1 activity is highly conserved among Drosophila species. ClustalW sequence alignments of the proximal Histone H1 promoter from Drosophila melanogaster (Dmel), Drosophila hydei (Dhyd), and Drosophila virilis (Dvir). The conserved sequences are marked by asterisks.
The TATA-less Histone H1 promoter is driven by TRF2
Using the Histone H1 promoter fused to the luciferase reporter, we next mapped a minimum fragment of the _cis_-regulatory region that retains TRF2-dependent transcription in S2 cells. Histone H1 promoter activity decreases gradually with progressive deletion of upstream promoter-proximal sequence. However, a core-promoter region 55 bp upstream of the transcriptional start site is sufficient to support TRF2-dependent promoter activity (Fig. 3E). Linker scanning mutagenesis experiments revealed at least two elements between −55 and +1 that are critical for Histone H1 promoter activity (Fig. 3F). One site, located from −55 to −40, is TC-rich and is highly conserved among different Drosophila species (Fig. 3G). Importantly, this core-promoter region of Histone H1 lacks a canonical TATA box found in core histone promoters. Taken together, these data suggest that selectivity of the Histone H1 gene is at least in part mediated by specific interactions between TRF2 and this TATA-less core promoter (see Fig. 6A, below). In addition, our attempts to test direct binding of recombinant TRF2 alone to this promoter-proximal sequence have thus far not been successful, suggesting that TRF2 likely requires additional partners to recognize and bind these _cis_-regulatory elements.
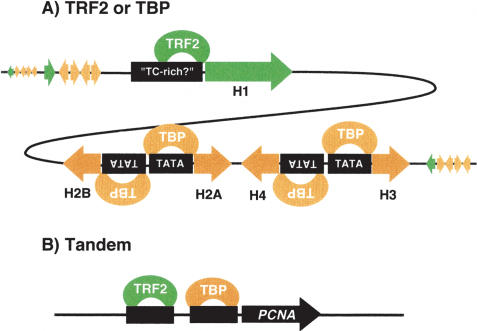
Figure 6.
Model of promoter-selective transcription in Drosophila mediated by TBP/TRF2. In Drosophila, at least three types of core-promoter architecture became apparent in this study. (A) A core promoter such as the TATA-less Histone H1 promoter is recognized and regulated uniquely by TRF2. In contrast, core-histone promoters harbor TATA boxes and are targeted uniquely by TBP. (B) Another type of core-promoter architecture is a tandem type, where both TRF2 and TBP/TFIID occupy proximal promoter regions, such as the ones seen in the PCNA promoter.
Genome-wide high-resolution survey of TRF2 target core promoters
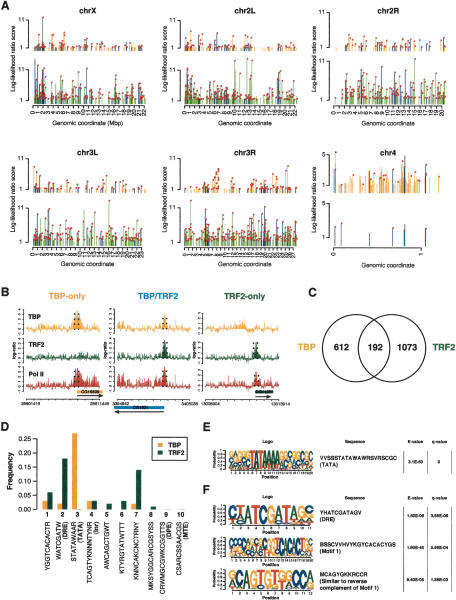
Differential regulation of linker and core histone genes in the histone gene cluster represents the first example of a bona fide target gene that appears to be selectively regulated by TRF2 but not by TBP. This finding suggests that TRF2 can be used to drive expression from two different types of core-promoter structures; i.e., tandem TRF2/TBP core promoters such as those represented by the promoter proximal regions of PCNA, as well as the TRF2-dependent but TBP-independent Histone H1 promoter. This led us to examine whether the mechanisms used to control Histone H1 expression might support a more versatile role of TRF2 in core-promoter recognition and transcriptional regulation than had previously been anticipated, especially given the mouse knockout studies that suggested a rather restricted role of TRF2 in spermiogenesis (Zhang et al. 2001; Martianov et al. 2002). To obtain a more global picture of core-promoter recognition mediated by TRF2, we screened for TRF2 as well as TBP and RNA Pol II-binding sites in S2 cells throughout the entire Drosophila genome. Using the ChIP-on-chip experimental platform we recently developed (Isogai et al. 2007), >1000 highly statistically significant binding sites for TRF2 were newly identified in addition to Histone H1 (Fig. 4A; Supplementary Table S8). A majority of the binding sites map to known 5′ ends of Pol II genes, and 60% of these TRF2 sites appear to also colocalize with RNA Pol II (Fig. 4A,B). This suggests that, unlike other classes of TRFs (i.e., TRF1 and TRF3), TRF2 may play a major role in RNA Pol II transcription. Genomic regions recognized by TRF2 as well as TBP appear to be distributed evenly throughout different chromosomes, except for chromosome 4. Interestingly, TRF2 was found to occupy only eight sites on chromosome 4, while TBP occupies 95 sites, most of which, however, are not occupied by Pol II in S2 cells. This observation is reminiscent of unusual chromatin and gene organization observed previously for chromosome 4 (Sun et al. 2000; Larsson et al. 2004), whose functional significance remains unclear.
Figure 4.
Genome-wide distribution of TRF2/TBP recognition sites. (A) Overview of TBP/TRF2/Pol II-binding sites in the Drosophila genome. The coordinates are in megabase pairs (Mb). TBP-binding sites are indicated in orange, TRF2-binding sites are indicated in green, and sites bound both by TBP and TRF2 are indicated in blue. Pol II (RPB3) binding for each peak is indicated by a circle (in magenta) at the tip of each peak. (B) Spatial structure of TRF2/TBP binding to core promoters. Three examples of the binding profiles of TRF2 and TBP on genomic tiling arrays. (Left) 5′ end of the CG15509 gene that is selectively bound by TBP and Pol II (RPB3). (Middle) 5′ end of CG1921 gene that is bound simultaneously by TBP, TRF2, and Pol II. (Right) 5′ end of CG11276 gene that is selectively bound by TRF2 and Pol II. The regions specified by broken lines are selected as binding regions by our statistical data analysis program TileHGMM. (C) Genome-wide survey of TBP- and TRF2-binding sites. Venn diagram showing the overlap between TBP and TRF2 occupied genomic regions. (D) Motif scanning analysis of core-promoter regions of TRF2- and TBP-bound promoters. Core-promoter sequences spanning −100 to approximately +50 relative to the annotated gene start sites of the top 100 promoters uniquely bound by either TBP or TRF2 (also occupied by Pol II) were probed for the presence of the 10 most commonly found core-promoter motifs (Ohler et al. 2002). We adopted the IUB nomenclature for the nucleotide sequences for degenerate base positions. (E) List of significant sequence motifs for TBP-bound core promoters by de novo motif finding. The _E_-value represents the significance of the motif among the TBP data set, whereas the _q_-value represents the discrimination from the TRF2 data set. (F) List of significant sequence motifs for TRF2-bound core promoters by de novo motif finding. The _E_-values represent the significance of the motif among the TRF2 data set, whereas the _q_-values represent the discrimination from the TBP data set.
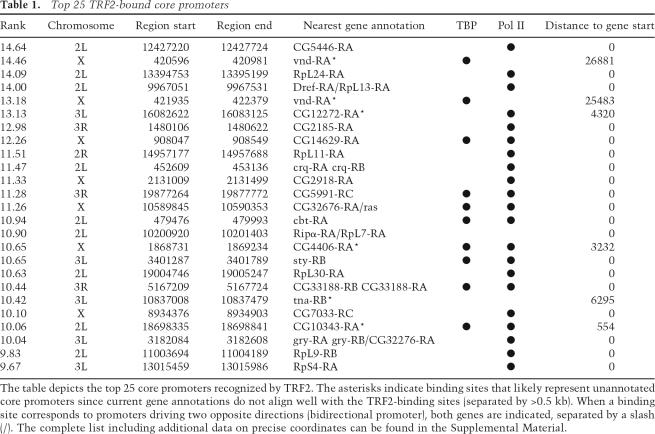
Consistent with our polytene chromosome staining, the majority (∼80%) of the TBP- and TRF2-binding sites mapped by high-resolution ChIP-on-chip were distinct and nonoverlapping, suggesting that TRF2 likely plays a critical and independent role in the recognition of a large number of genes that appear to be TBP-independent (Fig. 4A,C). However, ∼20% of the TRF2- and TBP-binding regions actually overlap (Fig. 4C). This finding suggests that the occurrence of tandem TBP and TRF2 core promoters such as was found for the PCNA promoter (Hochheimer et al. 2002) may also be quite widespread in the Drosophila genome. While genes linked to the core promoters targeted by TRF2 appeared to encode products of diverse cellular functions, it became apparent that TRF2 likely targets a cluster of ribosomal protein genes that appear to be coregulated (Table 1). Indeed, one of the most prominent targets of TRF2 belongs to a specific set of ribosomal protein genes (Supplementary Table S1). This finding suggests that TRF2 may be used to coordinate expression of related classes of functionally interdependent gene sets. Thus, our studies reveal that TRF2 targets a novel class of TBP-independent genes and suggest that this TRF may also be responsible for regulating distinct sets of coordinately controlled genes that are critical for proteins carrying out biochemically related events (i.e., protein synthesis).
Table 1.
Top 25 TRF2-bound core promoters
TRF2-bound promoters are associated with a distinct repertoire of core-promoter elements
In order to better understand the differential and selective core-promoter recognition by TRF2 revealed by our genome-wide survey, we first analyzed core promoters (−100 to +50) uniquely bound by TRF2 versus TBP by scanning the 10 most commonly found core-promoter sequence motifs reported for Drosophila (Ohler et al. 2002). This analysis revealed distinct core-promoter element preferences associated with TRF2- and TBP-bound promoters (Fig. 4D). As expected, TBP-bound promoters exhibit a strong enrichment for TATA boxes, with >85% of the first top 100 TBP target promoters containing variants of TATA boxes (data not shown). In contrast, TRF2-bound core promoters are virtually devoid of TATA boxes. Instead, TATA-less categories of core-promoter motifs appear preferentially associated with TRF2. Among the most enriched motifs is the DRE element (∼20%), which serves as a binding site for DREF, a subunit of one class of TRF2 complexes. Therefore, it seems evident that TBP- and TRF2-containing initiation complexes are differentially utilized for recognition of distinct core-promoter classes. Another unbiased analysis using de novo motif-finding platforms (Bailey and Elkan 1995; Isogai et al. 2007) confirmed that these distinct sequence elements uniquely linked to TBP versus TRF2 data sets are highly statistically significant. Again we see that TATA boxes are tightly linked to occupancy by TBP but not TRF2 (Fig. 4E). In contrast, DRE and Motif 1 are highly linked to occupancy by TRF2 over TBP (Fig. 4F). Thus, differential targeting of TRF2 to TATA-less classes of core promoters may, at least in part, rely on these discriminating sequence elements recognized by either TBP/TAF- or TRF2-containing initiation complexes.
In vivo requirements of TRF2 in fly embryogenesis and organ development
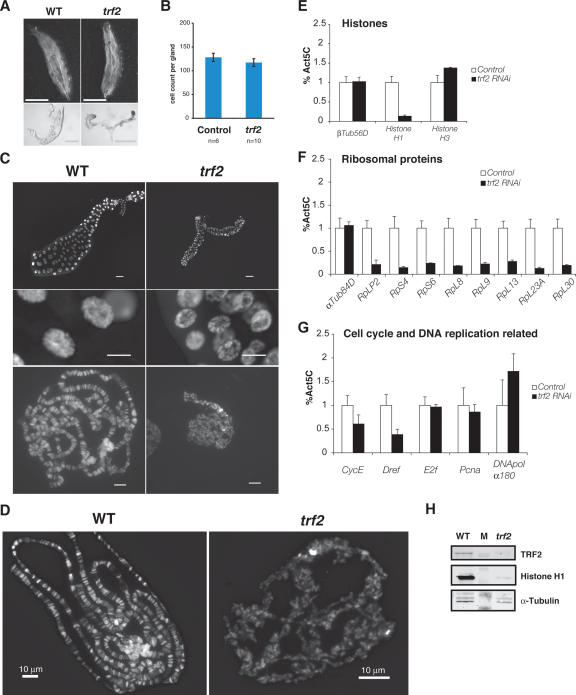
The identification of a large number of TRF2 recognition sites that appear TBP-independent prompted us next to examine whether these recognition sites represent bona fide functional targets of TRF2. Importantly, we asked whether these diverse TRF2-bound promoters drive specific pathways in vivo during fly development. Since our earlier studies in which TRF2 was depleted during embryogenesis resulted in lethality (Kopytova et al. 2006; M. Prestel and A. Hochheimer, unpubl.), we targeted depletion of TRF2 by RNAi in specific tissues, for example, by using a salivary gland-specific GAL4 driver. Salivary gland-specific expression of Trf2 double-stranded RNA (dsRNA) resulted in more than fivefold reduction of Trf2 mRNA, while TRF2 protein was reduced to near undetectable levels (Fig. 5H). Although these mutant Drosophila embryos developed to the third instar larval stage (L3), a majority of them failed to pupate or died during pupal stages. There appears to be no apparent size differences between mutant and wild-type larvae at L3, suggesting that a significant level of larval development remains unaffected by specific depletion of TRF2 in the salivary gland (Fig. 5A,C). Interestingly, however, salivary glands in the TRF2-depleted larvae display severe growth defects (Fig. 5A,C). Importantly, this defect appears to be cell growth-related rather than proliferation-deficient since cell numbers remained constant between mutant and wild-type glands (∼120 cells) (Fig. 5B) but with a dramatic reduction in cell and organ size. Most notably, nuclei in the TRF2-depleted glands appeared underdeveloped, consistent with a failure to successfully polytenize chromosomes (Fig. 5C). These specific developmental defects attributable to TRF2 depletion support the notion that critical biological pathways impinging on the growth of certain organs may be selectively regulated by TRF2.
Figure 5.
TRF2 is required for cell growth and chromatin organization. (A) Salivary-specific depletion of TRF2 affects salivary gland growth. Shown is a comparison between control (WT) and mutant (trf2) L3 larvae (top) and dissected salivary glands (bottom). Bar, 0.5 mm. Note that only half of each pair is shown for the control salivary glands. (B) Cell counts of control and trf2 mutant salivary glands. (C) DNA stain of control (WT) and mutant (trf2) salivary gland polytene chromosome. Bars: top, 0.1 mm; middle, 25 μm; bottom, 10 μm. Note that only half of a pair is shown for the control salivary glands. (D) DNA staining of control (WT) and mutant (trf2) polytene chromosome spreads. (E_–_G) RT–PCR analysis of mRNAs in TRF2-depleted salivary glands. Error bars represent the standard deviation from triplicates. (H) Western blot probing TRF2, Histone H1, and α-Tubulin in wild-type and TRF2-depleted salivary glands. M lane represents a marker lane.
Aberrant organization and structure in TRF2-deficient polytene chromosomes
Since the Histone H1 gene was identified as a direct TRF2 target gene in Drosophila cultured cells, we wondered if some of the defects in the mutant salivary gland may derive in part from a lack of the linker histone H1. We therefore visualized chromatin structure and integrity using polytene chromosomes, where interphase chromatin is amplified 100- to ∼1000-fold through endoreduplication (Andrew et al. 2000). While polytene chromosomes of wild-type salivary glands consist of uniformly thick bundles of endoreplicated chromatin bearing distinct banding patterns due to unique regions of condensed and decondensed chromatin, the chromosomes from TRF2-depleted salivary glands exhibited very few discernible banding patterns and are interspersed with numerous thin chromatin fibers that appeared improperly folded (Fig. 5D). To determine if this severe disorganization of TRF2-depleted polytene chromosomes may partially be due to alterations in chromatin structure, Histone H1 mRNA levels in total RNA derived from control and mutant salivary glands were analyzed using RT–PCR. Indeed, Histone H1 mRNA levels are significantly down-regulated, while Histone H3 mRNA levels remain intact (Fig. 5E). Importantly, H1 protein levels in TRF2-depleted salivary glands are also severely down-regulated if not completely abolished (Fig. 5H). Taken together, our data provide both in vivo and in vitro evidence that a differential regulation of Histone H1 versus the other core histone genes during fly development is dependent on the selective usage of an alternative core-promoter recognition machinery that harbors TRF2 rather than TBP/TFIID. These studies also provide in vivo evidence that H1 does indeed play a critical role in the maintenance of chromatin integrity as had been postulated and shown in vitro (Thoma et al. 1979).
TRF2 is required for expression of a cluster of ribosomal protein genes
The targeted depletion of TRF2 protein in salivary gland cells enabled us to examine whether a variety of TATA-less classes of core promoters bound by TRF2 in S2 cells are indeed dependent on TRF2 activity. Especially, we were intrigued by the finding that TRF2-depleted salivary glands are defective in cell growth because our previous ChIP-on-chip data suggested that one of the major classes of TRF2 target genes encodes ribosomal proteins (Table 1; Supplementary Table S5), which appear to be driven by TATA-less core promoters (Supplementary Fig. S6). In Drosophila, mutations in ribosomal protein genes lead to proliferation and growth defects (Lambertsson 1998). To examine if TRF2 is responsible for driving TATA-less ribosomal protein gene promoters, we conducted RT–PCR analysis of a dozen ribosomal protein genes expressed in salivary glands. Indeed, TRF2-deficient salivary glands display dramatically impaired levels of ribosomal protein mRNAs (Fig. 5F). In contrast, when we tested mRNA levels of several cell cycle and DNA replication-related genes including CycE, Dref, E2f, Pcna, and DNAPolα180, they appear largely unaffected (CycE, E2f, Pcna, and DNAPolα180) or exhibit only a modest down-regulation (Dref) in the TRF2-depleted cells (Fig. 5G). Therefore, the cell growth defects exhibited by salivary gland-specific depletion of TRF2 are likely linked to down-regulated protein synthesis caused by reduced levels of ribosomal proteins. Taken together, our in vivo analysis of TRF2 function in salivary gland cells suggests that TRF2 may be critically involved in several fundamental cellular functions including chromatin organization as well as protein synthesis, while underreplicated polytene chromosome may represent indirect effects.
Discussion
Regulation of histone genes via differential core-promoter targeting by TRF2 and TBP
In Drosophila, the five histone genes are found in a cluster that is tandemly amplified ∼100 times (Lifton et al. 1978; Matsuo and Yamazaki 1989). Despite the need to coordinate histone gene expression during the cell cycle, the ratio of linker and core histones can vary dramatically within each cell, among different tissues, and during embryonic development (Holmgren et al. 1985; Ruddell and Jacobs-Lorena 1985; Ner and Travers 1994). This observation suggested that Histone H1 gene expression may be differentially regulated relative to the patterns of core histone gene transcription. Here, our genome-wide survey of TRF2 target sites uncovered the finding that the histone gene cluster contains both TBP and TRF2 recognition sites. Most strikingly, these two core-promoter recognition factors are segregated within the histone cluster with TBP targeted to the core histone (H2A/B, H3, and H4) promoters, while TRF2 selectively directs transcription of the linker histone H1. This finding reveals a novel mechanism in which Histone H1 gene expression may be differentially regulated relative to the patterns of core histone gene transcription (Fig. 6A).
The finding that a TRF2-containing preinitiation complex is responsible for Histone H1 expression while the prototypic TBP/TFIID complex directs transcription of the core histones suggests that the expression of the linker histone H1 and core histones must be uncoupled under certain circumstances, possibly in a developmental-specific and cell type-specific manner. The analysis of TRF2-depleted salivary gland polytene chromosomes suggests that this is indeed the case. Remarkably, the polytene chromosomes in TRF2-deficient cells exhibited severe defects in chromosome organization and structure reminiscent of the failure to form 30-nm fibers in H1-depleted chromatin (Thoma et al. 1979; Fan et al. 2005; Maresca et al. 2005). Given that the Drosophila genome encodes only one H1 subtype compared with five to six in mammals (Fan et al. 2005), it is interesting that the H1 knockdown via TRF2 depletion resulted in a severely altered chromatin structure, which represents another in vivo evidence that histone H1 is indeed linked to organization of chromatin structure. Importantly, these TRF2-depleted cells appear to specifically down-regulate Histone H1 mRNA while leaving core histone transcripts intact. These findings suggest that TRF2 must serve as a key component of the transcriptional initiation complex evolved to differentially control linker histone versus core histone expression.
Targeting specific transcriptional complexes to distinct nuclear compartments
Transcription of nonpolyadenylated histone genes appears to be associated with a specific nuclear body (the HLB) through a physical coupling between the HLB and the histone gene cluster locus (Liu et al. 2006). The HLB is loaded with RNA synthesis and processing machinery, possibly serving as a “factory” for histone mRNA production. Thus, in order to rapidly produce histone transcripts during embryogenesis, Drosophila appears to have adapted an elegant strategy that involves tandemly amplified gene cassettes sequestered within a distinct nuclear address (the HLB). Interestingly, it appears that only specific subsets of transcription factors are deposited in the HLB. For example, among the three TBP paralogs in Drosophila (TBP, TRF1, and TRF2), only TRF2 and TBP that are used for linker and core histone transcription, in addition to Pol II, are “preloaded” within the HLB, perhaps to facilitate rapid as well as differential linker versus core histone transcript production. Therefore, the histone gene cluster presents an important paradigm wherein a distinct nuclear body loaded with specific transcriptional as well as post-transcriptional machinery becomes dedicated to the purpose of coordinately and differentially regulating five essential genes.
TRF2, a key recognition factor for TATA-less core promoters
Our high-resolution genome mapping of TRF2 recognition sites using the ChIP-on-chip platform has revealed >1000 novel binding sites, with 80% distinct from and 20% overlapping with TBP-binding sites. These results suggest that the TRF2-dependent and TBP-independent Histone H1 promoter is not an exception. Indeed, the H1 case may represent a more general case for how TRF2 can serve as an alternative core-promoter recognition factor at many Pol II genes. A comprehensive and detailed sequence motif analysis of the Drosophila genome revealed that TRF2-bound promoters significantly lack TATA boxes, while the TATA box is tightly correlated with TBP-binding sites. Instead, TRF2 appears to selectively recognize promoters containing other distinct core-promoter elements such as Motif 1, DRE, and Motif 7 (Ohler et al. 2002). In addition, functional analysis of transcripts derived from TRF2-depleted salivary glands confirmed that TRF2 activity is indeed required for directing these TRF2 target promoters (Fig. 5; Supplementary Fig. S3). Thus, our genome-wide analysis significantly strengthens the emerging picture that TRF2 likely evolved to recognize and regulate a large class of TATA-less core promoters.
One question concerning TRF2 function in promoter recognition is whether TRF2, like TBP, can directly recognize and bind to a distinct core-promoter element. TRF2 is likely to possess very different DNA-binding specificities from TBP since the amino acid residues critical for TATA-box recognition have been altered in TRF2 (Dantonel et al. 1999; Ohbayashi et al. 1999; Rabenstein et al. 1999). However, all our attempts to experimentally identify a direct TRF2-binding sequence have thus far failed. Similarly, our most recent computational efforts using TRF2 ChIP-on-chip data sets failed to identify any strong consensus core-promoter motifs comparable with the prototypic TATA box with its approximately −30-bp location relative to the start of transcription. Instead, we identified motifs such as the DRE and other uncharacterized elements with no set common position relative to the transcriptional start site. These findings are, however, consistent with our previous studies (Hochheimer et al. 2002) in which TRF2 failed to bind the core promoter by itself. Instead, it appears that TRF2 recruitment to at least a subset of core promoters relies on specific interactions between TRF2 and various other sequence-specific DNA-binding proteins, such as DREF. However, unlike previous studies, our genome-wide survey of TRF2- and TBP-binding sites in Drosophila revealed a considerably more comprehensive picture of how TRF2 may be used as an alternative core-promoter recognition factor. Importantly, mixing and matching various enhancer-binding factors (i.e., sequence-specific DNA-binding factors) and alternative core-promoter recognition factors (i.e., TFIID vs. TRF2) appears to be a powerful and perhaps common strategy for metazoan organisms to diversify transcriptional outputs.
Widespread occurrence of tandem core promoters in Drosophila
Our genome-wide ChIP-on-chip analysis also provides strong evidence that metazoan organisms make much more use of tandem core promoters containing both TFIID and TRF2 recognition sites than might have been anticipated (Fig. 6B). Whether or not this type of dual core-promoter structure represents a case of redundant pathways or is subject to selective and differential regulation of downstream targets remains unclear. Interestingly, two previously characterized TRF2 targets (PCNA and DNApolα180) appear unaffected when TRF2 is depleted in salivary glands, possibly due to the ability of such dual core promoters to use alternative transcription complexes. Thus, the possibility that TRF2 may be used in lieu of TBP/TFIID to diversify transcriptional outputs in response to specific signals cannot be ruled out. It would be of interest for future studies to determine how these two distinct core-promoter recognition factors TBP/TRF2 operating at dual tandem promoters may be coordinated. Are these core-promoter recognition complexes at tandem core promoters recruited by common or distinct activator proteins? Since salivary gland depletion of TRF2 protein resulted in developmental defects, TRF2 may be necessary to selectively up-regulate genes required for specific developmental pathways.
Evolutionary conservation of TRF2 functions in metazoans
The identification of direct TRF2 target genes in the present study has revealed a striking link between TRF2 and specific biological processes such as chromatin organization and protein synthesis. Since TRF2 is conserved among many metazoan organisms, its role in various model organisms has been of considerable interest. Several studies found that inactivating TRF2 in nematode, fly, fish, and frog all resulted in lethality due to a block in embryogenesis (Dantonel et al. 2000; Kaltenbach et al. 2000; Veenstra et al. 2000; Muller et al. 2001; Kopytova et al. 2006). In contrast, germ cell-specific functions of TRF2 have also been reported for Drosophila and mice (Martianov et al. 2001; Zhang et al. 2001; Kopytova et al. 2006). In particular, while TRF2-null mice appear to display a modest non-Mendelian ratio of inheritance, the major defect manifests as a lack of spermiogenesis (Martianov et al. 2001; Zhang et al. 2001). Although these studies revealed that TRF2 provides nonredundant functions during development, these genetic studies were unable to link TRF2 to selective core-promoter recognition functions in vivo. For instance, direct TRF2 target genes responsible for these previously observed phenotypes have not been identified or characterized. The identification of histone H1 and ribosomal proteins as key gene products misregulated in TRF2-depleted Drosophila organs not only provides candidate TRF2 target genes responsible for the chromatin defects observed in TRF2-depleted Drosophila germ cells (Kopytova et al. 2006), but also underscores the potential role of TRF2 in other organisms. For example, TRF2-null mice display a major defect in chromocenter formation in spermatids (Martianov et al. 2002). This suggests that, consistent with TRF2-mediated H1 regulation in Drosophila somatic cells, TRF2 may also target genes that are essential for chromatin structure in mammalian gonads. However, the precise molecular targets and mechanisms of TRF2 action may differ. Indeed, a recent report points to the involvement of a human DREF homolog in regulating transcription from a TATA-box-containing histone H1 promoter in human cells (Ohshima et al. 2003). In contrast, in Drosophila, we found that the H1 gene is TATA-less and does not appear to be regulated by DREF.
In addition, our finding that Drosophila TRF2 directs the expression of a large number of gene products critical for essential cell function such as growth (i.e., ribosomal subunits and histones) would be consistent with the lethality associated with the loss of TRF2 in most organisms. These findings also suggest that in mammals TRF2 may play an important role regulating essential cell functions in tissues other than testis. The biological context of TRF2 usage as an alternative core-promoter recognition factor may well be more universal than we had anticipated.
Materials and methods
Antibodies
The rabbit polyclonal antibody against TRF2 was generated using GST fusion proteins of the C-terminal nonconserved region of Drosophila TRF2. Rabbit polyclonal serum against TBP was generated using his-tagged TBP. These sera were extensively purified using antigen affinity chromatography. RPB1- and RPB3-specific antibodies were similarly purified from anti-holo-Pol II antibodies (Skantar and Greenleaf 1995) using antigen affinity columns. Anti-DREF, anti-TAF5, and anti-TAF2 antibodies were described previously (Weinzierl et al. 1993; Hirose et al. 1996; Marr et al. 2006). Anti-TMG antibody was purchased from Oncogene.
Cell culture
Drosophila S2 and Kc cells were grown in M3-BPYE medium (Drosophila Resource Center). Synchronization of Kc cells was conducted as described (MacAlpine et al. 2004). Log-phase cultures (∼2 × 106 cells per milliliter) of Kc cells were first incubated with 0.2 nM ponasterone A for 24 h to obtain G2 cells. Cells were then rinsed with 1× PBS three times, resuspended in an equal volume of fresh M3-BPYE medium containing 1.5 mM hydroxyurea, and cultured for an additional 18 h to obtain G1/S cells. These Kc cells were rinsed again with 1× PBS three times, resuspended in fresh M3-BPYE, and cultured for an additional 2∼3 h to obtain S-phase cells. To confirm the synchrony of cell populations, fluorescence-activated cell sorting (FACS) sorting with propidium iodide was conducted as described (Darzynkiewicz and Juan 1997) using a staining buffer containing 1 mg/mL glucose, 20 μg/mL propidium iodide, and 1 mg/mL RNase A in 1× PBS. RNAi was conducted by treating cells with 10 μg of dsRNA per 0.4 × 106 cells per well seeded in a 24-well plate or 100 μg of dsRNA per 5 × 106 cells seeded in a 10-cm plate, and assays were conducted after the indicated number of days. For RNAi in S2 cells, the template for dsRNA synthesis was derived from amplifying the 5′ untranslated region (UTR) of the Trf2 gene from Trf2 cDNA clone (Rabenstein et al. 1999). For dsRNA against DREF, we used sequences spanning 1716∼2124 (relative to ATG), and dsRNA for TBP and TAF5 have been described (Marr et al. 2006). Reporter plasmids were transfected simultaneously using Effectene (Qiagen) for luciferase reporter assays as per the manufacturer’s instructions.
Measurement of Histone H1 promoter activity
Primer extension analysis was performed using nuclear RNAs isolated from Drosophila S2 cells after 5 d of dsRNA treatment. Histone H1 and 7SL RNA were detected in nuclear RNA, and MtnA transcripts were detected in total RNA from cells treated for 2 h with 0.5 mM CuSO4. Nuclear RNA was prepared using methods described previously (Herold et al. 2003). RNA was purified using TRI-reagent (Sigma). The pGL3-H1 promoter was constructed by inserting −698 to +68 of the Histone H1 promoter into pGL3(R2.1)-Basic vector (for Fig. 3C,D) or pGL3-Basic vector (for Fig. 3E,F) (Promega). Various H1 promoter constructs harboring deletions and mutations were generated using pGL3-H1 vector by exonuclease digestions (Henikoff 1984) and site-directed mutagenesis (Promega), respectively. Luciferase reporter assays with the Histone H1 promoter and the MtnA promoter in Figure 3C were conducted after 5 d of dsRNA treatment. The luciferase assays in Figure 3E were performed after 3.5 d of dsRNA treatment. As an internal standard, we used phRL-null plasmid (Promega) harboring the Act5C distal promoter (pAct5C-hRenilla). The rescue experiments shown in Figure 3D were conducted using S2 cells harboring a TRF2-V5 expression cassette from the MtnA promoter. The cells were first transfected with the pGL3-H1 reporter as well as pAct5C-hRenilla internal control plasmids and were treated with either control lacZ or trf2 (5′UTR) dsRNA. The re-expression of TRF2-V5 was induced by the addition of CuSO4 to a final concentration of 0.05 mM, for the indicated length of time prior to the harvest. The cells were harvested simultaneously on day 4, and luciferase activities were assayed. Each luciferase assay was conducted in triplicate, and standard deviation is indicated by error bars in the graphs.
Cytology
Polytene chromosomal staining and DNA FISH were conducted as described (Lavrov et al. 2004). Immunostaining and DNA FISH in S2 and Kc cells were described previously (Marr et al. 2006). For RPB3 immunostaining in Kc cells, cells were first treated with 2 mM thymidine overnight, which increased the S-phase population by 10%∼20%. FISH probes were generated using the pCR4 vector containing 5 kb spanning the histone cluster (Lifton et al. 1978) using methods described previously (Marr et al. 2006).
ChIP and ChIP-on-chip
The ChIP experiments in this study were conducted essentially as described (Isogai et al. 2007). A formaldehyde concentration of 1.0% was used for cross-linking S-phase-synchronized Kc cells, and 0.5% was used for asynchronous S2 cells for the ChIP-on-chip experiments. We conducted ChIP-on-chip experiments using affinity-purified TRF2, TBP, and Pol II antibodies. Pol II ChIP-on-chip experiments were conducted using RPB1 and RPB3 antibodies, and the merged data set was used for the subsequent analysis throughout this study, unless otherwise indicated. Detailed description of ChIP, ChIP-on-chip assays, and statistical analysis of the data with the TileHGMM platform (Keles 2007), and de novo sequence analysis using MEME (Bailey and Elkan 1995) and cosmo (Bembom et al. 2007), can be found elsewhere, as they have been described previously (Isogai et al. 2007).
De novo sequence analysis
To perform de novo sequence analysis, we first extracted nonoverlapping data sets of TBP- and TRF2-binding sites that are co-occupied by Pol II and are within 500 bp of a known gene start site. Each data set was analyzed by MEME and cosmo, and the resulting significant motifs (_E_-value <1) were collected as a combined data set. We then scored the sequences in each data set by each motif in the combined motif data set using the PATSER program (Hertz and Stormo 1999). For each motif, we compared the mean of the two score distributions in TBP- and TRF2-bound data sets with a Wilcoxon rank sum test. After obtaining a _P_-value from this test for each motif, we adjusted the _P_-values for multiplicity, converting them to _q_-values (Storey 2002), and reported the discriminating motifs with a _q_-value <0.01.
Generation of trf2 RNAi flies
A DNA fragment comprising Trf2 coding sequence from 24 to 586 bp was amplified by PCR using Trf2 cDNA (Rabenstein et al. 1999) and XbaI-site-containing oligonucleotides GGTCAG CATTCTAGAGGCCAATTTGAACGGCGG and CGTGCT CATTCTAGACAATTGGCTGGTGAAGGT. The PCR product was cut with XbaI and cloned as inverted repeats in two consecutive steps into the plasmid pWIZ (Lee and Carthew 2003) using XbaI-compatible AvrII and NheI sites. pWIZ:Trf2 was used as a template to transcribe intron-spliced hairpin RNA under the control of Gal4/UAS to induce Trf2 gene silencing. pWIZ:Trf2 was introduced into the germline by _P_-element-mediated transformation (Spradling and Rubin 1982). Two independent transformant lines were established and mapped. We subsequently crossed homozygous pWIZ:Trf2 females to males carrying homozygous copies of the salivary gland driver GAL42314 (gift of J. Großhans). Females with a UAS-GFP construct served as a control.
Immunoblot using salivary gland extracts
Glands were dissected and boiled immediately in 2× Laemmli buffer, followed by benzonase nuclease treatment for 1 h at room temperature. The wild-type (WT) lane contains four pairs of salivary glands boiled in Laemmli buffer, and the trf2 lane contains six pairs of trf2 mutant salivary glands. The blot was probed with anti-α-tubulin (DM1A; Sigma), anti-TRF2, and anti-H1 (gift of J. Kadonaga), followed by secondary antibodies (IRDye 800CW Donkey anti-Rabbit IgG and IRDye 800CW Donkey anti-Mouse IgG; Biomol). The blot was imaged using a LI-COR Odyssey scanner.
Preparation of RNA from salivary glands of trf2 RNAi larvae
Thirty pairs of salivary glands from trf2 RNAi fly lines arrested at L3 or control larvae were dissected and lysed in Trizol reagent (GIBCO) and subsequently purified by RNeasy plus kit (Qiagen). Salivary glands from L3 larvae of the GAL4-driver line 2314 crossed to UAS-GFP flies served as a control. Five-hundred nanograms of total RNAs were used for reverse transcription using SuperScript III reverse transcriptase (Invitrogen) per the manufacturer’s instructions. These cDNAs were subjected to quantitative PCR using Opticon 2 (MJ Research). We used TBP-dependent Act5C transcripts as an internal standard for calibration. The primer sequences used for the amplification are available on request.
Acknowledgments
We are indebted to H. Mitlöhner for masterly dissection of larvae and preparation of salivary glands, and to Y. Berghöfer-Hochheimer for salivary gland polytene chromosome spreads and immunofluorescence microscopy. We are grateful to P. Becker and C. Dulac for their generous support. We also thank the following individuals who provided critical reagents and resources for this study: A. Greenleaf for anti-Pol II antibodies; J. Großhans for the salivary gland GAL4 driver fly line; F. Hirose for anti-DREF antibodies; J. Kadonaga for anti-H1 antibodies; M. Marr for the Escherichia coli expression vector for TBP; K. Wright for the template plasmid for TAF5 dsRNA synthesis; T. Laverty for instructions on polytene chromosome dissection; Z. Cande and B. Meyer for the microscopes; and M. Botchan, Y. Fong, J. Gall, J. Liu, R. Losick, and M. Marr for critical reading of the manuscript. R.T. is an investigator of the Howard Hughes Medical Institute and Director of the Li Ka-Shing Center for Biomedical and Health Sciences.
Footnotes
References
- Andrew D.J., Henderson K.D., Seshaiah P., Henderson K.D., Seshaiah P., Seshaiah P. Salivary gland development in Drosophila melanogaster. Mech. Dev. 2000;92:5–17. doi: 10.1016/s0925-4773(99)00321-4. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C., Elkan C. Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Mach. Learn. 1995;21:51–80. [Google Scholar]
- Bembom O., Keles S., van der Laan M.J., Keles S., van der Laan M.J., van der Laan M.J. Supervised detection of conserved motifs in DNA sequences with cosmo. Statistical applications in genetics and molecular biology. 2007 doi: 10.2202/1544-6115.1260. vol. 6, issue 1, article 8. http://www.bepress.com/sagmd/vol6/iss1/art8. [DOI] [PubMed] [Google Scholar]
- Berk A.J. TBP-like factors come into focus. Cell. 2000;103:5–8. doi: 10.1016/s0092-8674(00)00098-2. [DOI] [PubMed] [Google Scholar]
- Crowley T.E., Hoey T., Liu J.K., Jan Y.N., Jan L.Y., Tjian R., Hoey T., Liu J.K., Jan Y.N., Jan L.Y., Tjian R., Liu J.K., Jan Y.N., Jan L.Y., Tjian R., Jan Y.N., Jan L.Y., Tjian R., Jan L.Y., Tjian R., Tjian R. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature. 1993;361:557–561. doi: 10.1038/361557a0. [DOI] [PubMed] [Google Scholar]
- Dantonel J.C., Wurtz J.M., Poch O., Moras D., Tora L., Wurtz J.M., Poch O., Moras D., Tora L., Poch O., Moras D., Tora L., Moras D., Tora L., Tora L. The TBP-like factor: An alternative transcription factor in metazoa? Trends Biochem. Sci. 1999;24:335–339. doi: 10.1016/s0968-0004(99)01436-x. [DOI] [PubMed] [Google Scholar]
- Dantonel J.C., Quintin S., Lakatos L., Labouesse M., Tora L., Quintin S., Lakatos L., Labouesse M., Tora L., Lakatos L., Labouesse M., Tora L., Labouesse M., Tora L., Tora L. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol. Cell. 2000;6:715–722. doi: 10.1016/s1097-2765(00)00069-1. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z., Juan G., Juan G. DNA content measurement for DNA ploidy and cell cycle analysis. In: Robinson J.P., et al., editors. Current protocols in cytometry. Wiley; New York: 1997. pp. 7.5.1–7.5.24. [DOI] [PubMed] [Google Scholar]
- Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I., Stein A., Woodcock C.L., Skoultchi A.I., Woodcock C.L., Skoultchi A.I., Skoultchi A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Gershenzon N.I., Ioshikhes I.P., Ioshikhes I.P. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 2005;21:1295–1300. doi: 10.1093/bioinformatics/bti172. [DOI] [PubMed] [Google Scholar]
- Hansen S.K., Takada S., Jacobson R.H., Lis J.T., Tjian R., Takada S., Jacobson R.H., Lis J.T., Tjian R., Jacobson R.H., Lis J.T., Tjian R., Lis J.T., Tjian R., Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Herold A., Teixeira L., Izaurralde E., Teixeira L., Izaurralde E., Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G.Z., Stormo G.D., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Hirose F., Yamaguchi M., Handa H., Inomata Y., Matsukage A., Yamaguchi M., Handa H., Inomata Y., Matsukage A., Handa H., Inomata Y., Matsukage A., Inomata Y., Matsukage A., Matsukage A. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase α and proliferating cell nuclear antigen. J. Biol. Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- Hirose F., Yamaguchi M., Kuroda K., Omori A., Hachiya T., Ikeda M., Nishimoto Y., Matsukage A., Yamaguchi M., Kuroda K., Omori A., Hachiya T., Ikeda M., Nishimoto Y., Matsukage A., Kuroda K., Omori A., Hachiya T., Ikeda M., Nishimoto Y., Matsukage A., Omori A., Hachiya T., Ikeda M., Nishimoto Y., Matsukage A., Hachiya T., Ikeda M., Nishimoto Y., Matsukage A., Ikeda M., Nishimoto Y., Matsukage A., Nishimoto Y., Matsukage A., Matsukage A. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J. Biol. Chem. 1996;271:3930–3937. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- Hochheimer A., Tjian R., Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes & Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Hochheimer A., Zhou S., Zheng S., Holmes M.C., Tjian R., Zhou S., Zheng S., Holmes M.C., Tjian R., Zheng S., Holmes M.C., Tjian R., Holmes M.C., Tjian R., Tjian R. TRF2 associates with DREF and directs promoterselective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Holmes M.C., Tjian R., Tjian R. Promoter-selective properties of the TBP-related factor TRF1. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- Holmgren P., Johansson T., Lambertsson A., Rasmuson B., Johansson T., Lambertsson A., Rasmuson B., Lambertsson A., Rasmuson B., Rasmuson B. Content of histone H1 and histone phosphorylation in relation to the higher order structures of chromatin in Drosophila. Chromosoma. 1985;93:123–131. doi: 10.1007/BF00293159. [DOI] [PubMed] [Google Scholar]
- Hyun J., Jasper H., Bohmann D., Jasper H., Bohmann D., Bohmann D. DREF is required for efficient growth and cell cycle progression in Drosophila imaginal discs. Mol. Cell. Biol. 2005;25:5590–5598. doi: 10.1128/MCB.25.13.5590-5598.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Takada S., Tjian R., Keles S., Takada S., Tjian R., Keles S., Tjian R., Keles S., Keles S. Novel TRF1/BRF target genes revealed by genome-wide analysis of Drosophila Pol III transcription. EMBO J. 2007;26:79–89. doi: 10.1038/sj.emboj.7601448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V.X., Singer G.A., Agosto-Perez F.J., Liyanarachchi S., Davuluri R.V., Singer G.A., Agosto-Perez F.J., Liyanarachchi S., Davuluri R.V., Agosto-Perez F.J., Liyanarachchi S., Davuluri R.V., Liyanarachchi S., Davuluri R.V., Davuluri R.V. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics. 2006;7:114. doi: 10.1186/1471-2105-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L., Horner M.A., Rothman J.H., Mango S.E., Horner M.A., Rothman J.H., Mango S.E., Rothman J.H., Mango S.E., Mango S.E. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol. Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Keles S. Mixture modeling for genome-wide localization of transcription factors. Biometrics. 2007;63:10–21. doi: 10.1111/j.1541-0420.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- Kopytova D.V., Krasnov A.N., Kopantceva M.R., Nabirochkina E.N., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Krasnov A.N., Kopantceva M.R., Nabirochkina E.N., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Kopantceva M.R., Nabirochkina E.N., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Nabirochkina E.N., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Yerokhin M.M., Simonova O.B., Simonova O.B., et al. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol. Cell. Biol. 2006;26:7492–7505. doi: 10.1128/MCB.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv. Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Larsson J., Svensson M.J., Stenberg P., Makitalo M., Svensson M.J., Stenberg P., Makitalo M., Stenberg P., Makitalo M., Makitalo M. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc. Natl. Acad. Sci. 2004;101:9728–9733. doi: 10.1073/pnas.0400978101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov S., Dejardin J., Cavalli G., Dejardin J., Cavalli G., Cavalli G. Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol. Biol. 2004;247:289–303. doi: 10.1385/1-59259-665-7:289. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Carthew R.W., Carthew R.W. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Lifton R.P., Goldberg M.L., Karp R.W., Hogness D.S., Goldberg M.L., Karp R.W., Hogness D.S., Karp R.W., Hogness D.S., Hogness D.S. The organization of the histone genes in Drosophila melanogaster: Functional and evolutionary implications. Cold Spring Harb. Symp. Quant. Biol. 1978;42:1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Liu J.L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J.G., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J.G., Buszczak M., Clatterbuck S., Goodman R., Gall J.G., Clatterbuck S., Goodman R., Gall J.G., Goodman R., Gall J.G., Gall J.G. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D.M., Rodriguez H.K., Bell S.P., Rodriguez H.K., Bell S.P., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E. Transcriptional functions of a new mammalian TATA-binding protein-related factor. J. Biol. Chem. 1999;274:12963–12966. doi: 10.1074/jbc.274.19.12963. [DOI] [PubMed] [Google Scholar]
- Maresca T.J., Freedman B.S., Heald R., Freedman B.S., Heald R., Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J. Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr M.T., Isogai Y., Wright K.J., Tjian R., Isogai Y., Wright K.J., Tjian R., Wright K.J., Tjian R., Tjian R. Coactivator cross-talk specifies transcriptional output. Genes & Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Fimia G.M., Dierich A., Parvinen M., Sassone-Corsi P., Davidson I., Fimia G.M., Dierich A., Parvinen M., Sassone-Corsi P., Davidson I., Dierich A., Parvinen M., Sassone-Corsi P., Davidson I., Parvinen M., Sassone-Corsi P., Davidson I., Sassone-Corsi P., Davidson I., Davidson I. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell. 2001;7:509–515. doi: 10.1016/s1097-2765(01)00198-8. [DOI] [PubMed] [Google Scholar]
- Martianov I., Brancorsini S., Gansmuller A., Parvinen M., Davidson I., Sassone-Corsi P., Brancorsini S., Gansmuller A., Parvinen M., Davidson I., Sassone-Corsi P., Gansmuller A., Parvinen M., Davidson I., Sassone-Corsi P., Parvinen M., Davidson I., Sassone-Corsi P., Davidson I., Sassone-Corsi P., Sassone-Corsi P. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: Requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development. 2002;129:945–955. doi: 10.1242/dev.129.4.945. [DOI] [PubMed] [Google Scholar]
- Matsuo Y., Yamazaki T., Yamazaki T. tRNA derived insertion element in histone gene repeating unit of Drosophila melanogaster. Nucleic Acids Res. 1989;17:225–238. doi: 10.1093/nar/17.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F., Lakatos L., Dantonel J., Strahle U., Tora L., Lakatos L., Dantonel J., Strahle U., Tora L., Dantonel J., Strahle U., Tora L., Strahle U., Tora L., Tora L. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol. 2001;11:282–287. doi: 10.1016/s0960-9822(01)00076-8. [DOI] [PubMed] [Google Scholar]
- Ner S.S., Travers A.A., Travers A.A. HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J. 1994;13:1817–1822. doi: 10.1002/j.1460-2075.1994.tb06450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T., Makino Y., Tamura T.A., Makino Y., Tamura T.A., Tamura T.A. Identification of a mouse TBP-like protein (TLP) distantly related to the Drosophila TBP-related factor. Nucleic Acids Res. 1999;27:750–755. doi: 10.1093/nar/27.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U., Liao G.C., Niemann H., Rubin G.M., Liao G.C., Niemann H., Rubin G.M., Niemann H., Rubin G.M., Rubin G.M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima N., Takahashi M., Hirose F., Takahashi M., Hirose F., Hirose F. Identification of a human homologue of the DREF transcription factor with a potential role in regulation of the histone H1 gene. J. Biol. Chem. 2003;278:22928–22938. doi: 10.1074/jbc.M303109200. [DOI] [PubMed] [Google Scholar]
- Rabenstein M.D., Zhou S., Lis J.T., Tjian R., Zhou S., Lis J.T., Tjian R., Lis J.T., Tjian R., Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddell A., Jacobs-Lorena M., Jacobs-Lorena M. Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc. Natl. Acad. Sci. 1985;82:3316–3319. doi: 10.1073/pnas.82.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skantar A.M., Greenleaf A.L., Greenleaf A.L. Identifying a transcription factor interaction site on RNA polymerase II. Gene Expr. 1995;5:49–69. [PMC free article] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Spradling A.C., Rubin G.M., Rubin G.M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Storey J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B Methodol. 2002;64:479–498. [Google Scholar]
- Sun F.L., Cuaycong M.H., Craig C.A., Wallrath L.L., Locke J., Elgin S.C., Cuaycong M.H., Craig C.A., Wallrath L.L., Locke J., Elgin S.C., Craig C.A., Wallrath L.L., Locke J., Elgin S.C., Wallrath L.L., Locke J., Elgin S.C., Locke J., Elgin S.C., Elgin S.C. The fourth chromosome of Drosophila melanogaster: Interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Lis J.T., Zhou S., Tjian R., Lis J.T., Zhou S., Tjian R., Zhou S., Tjian R., Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A., Koller T., Klug A., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupler R., Perini G., Green M.R., Perini G., Green M.R., Green M.R. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Veenstra G.J., Weeks D.L., Wolffe A.P., Weeks D.L., Wolffe A.P., Wolffe A.P. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312–2315. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- Weinzierl R.O., Dynlacht B.D., Tjian R., Dynlacht B.D., Tjian R., Tjian R. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature. 1993;362:511–517. doi: 10.1038/362511a0. [DOI] [PubMed] [Google Scholar]
- Zhang D., Penttila T.L., Morris P.L., Teichmann M., Roeder R.G., Penttila T.L., Morris P.L., Teichmann M., Roeder R.G., Morris P.L., Teichmann M., Roeder R.G., Teichmann M., Roeder R.G., Roeder R.G. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science. 2001;292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]