Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species (original) (raw)
Abstract
The resident prokaryotic microflora of the mammalian intestine influences diverse homeostatic functions of the gut, including regulation of cellular growth and immune responses; however, it is unknown how commensal prokaryotic organisms mechanistically influence eukaryotic signaling networks. We have shown that bacterial coculture with intestinal epithelial cells modulates ubiquitin-mediated degradation of important signaling intermediates, including β-catenin and the NF-κB inhibitor IκB-α. Ubiquitination of these proteins as well as others is catalyzed by the SCFβTrCP ubiquitin ligase, which itself requires regulated modification of the cullin-1 subunit by the ubiquitin-like protein NEDD8. Here we show that epithelia contacted by enteric commensal bacteria in vitro and in vivo rapidly generate reactive oxygen species (ROS). Bacterially induced ROS causes oxidative inactivation of the catalytic cysteine residue of Ubc12, the NEDD8-conjugating enzyme, resulting in complete but transient loss of cullin-1 neddylation and consequent effects on NF-κB and β-catenin signaling. Our results demonstrate that commensal bacteria directly modulate a critical control point of the ubiquitin–proteasome system, and suggest how enteric commensal bacterial flora influences the regulatory pathways of the mammalian intestinal epithelia.
Keywords: commensal, NEDD8, NF-κB, reactive oxygen species, Ubc12
Introduction
The mammalian intestinal epithelium is a remarkable tissue that coexists in intimate contact with up to 1014 prokaryotic organisms that comprise the normal flora (Backhed et al, 2005). The normal flora serves nutritional and metabolic functions, and can influence epithelial growth (Falk et al, 1998). This bacterial community is also necessary for normal immune function, although in certain genetic backgrounds it has been implicated as a causative agent in inflammatory bowel disease (Macdonald and Monteleone, 2005). The ability of commensal bacteria to modulate immune and other gut functions has prompted interest in exogenous administration of bacterial agents, ‘probiotics', as a therapeutic approach for inflammatory intestinal disorders (Weng and Walker, 2006). However, whereas host cell interactions with pathogenic enteric organisms are well studied, how a taxonomically complex and ecologically dynamic commensal microbial community can influence eukaryotic biology is largely unknown, although studies with gnotobiotic mice have established that the gut flora profoundly influences host intestinal cell gene regulation (Hooper et al, 2001).
We previously reported that multiple species of non-pathogenic bacteria could attenuate the key immunoregulatory NF-κB pathway via specific inhibition of IκB-α ubiquitination (Neish et al, 2000). In addition, we also observed that these bacterial–epithelial interactions inhibited ubiquitination of β-catenin, allowing nuclear accumulation and activation of Wnt signaling (Neish et al, 2000; Sun et al, 2004). Ubiquitin (Ub)-mediated degradation of IκB-α and β-catenin is mediated by a common Ub ligase, E3-SCFβ−TrCP, suggesting that commensal bacteria were exerting effects via a common mechanism of action. We subsequently showed that commensal bacteria could reduce neddylation of the Cul-1 subunit of E3-SCFβ−TrCP accounting for the blockade of the NF-κB pathway (Collier-Hyams et al, 2005). Neddylation denotes the covalent modification of the SCF (Skp1, Cdc53/Cullin, F box receptor) family of Ub ligases by the Ub-like protein NEDD8. The process is emerging as a central regulatory event in cellular processes that are controlled by protein degradation, including NF-κB and β-catenin (Pan et al, 2004; Petroski and Deshaies, 2005). Unlike covalent modification by the related Ub-like protein SUMO (‘sumoylation'), in which SUMO is conjugated to a large spectrum of mostly nuclear proteins, neddylation is apparently limited to substrates of the cullin family and thus acts as a regulatory modification specific to the SCF Ub ligases. In a variety of experimental models, stable mutations resulting in the loss of Cul-1 neddylation have profound functional consequences in cellular signaling pathways (Cope and Deshaies, 2003; Pan et al, 2004). Regulated physiological deneddylation has been reported in yeast's responses to UV (Groisman et al, 2003) and the perception of light in Arabidopsis seedlings (Wang et al, 2003), while our previous description of bacterially mediated loss of Cul-1 neddylation was the first reported example of this phenomenon in mammalian cells (Collier-Hyams et al, 2005).
Although the rapid generation of reactive oxygen species (ROS) is a cardinal feature of the cellular response of phagocytes to pathogenic and commensal bacteria, evidence is accumulating that ROS are also physiologically elicited in other cell types, including intestinal epithelia, in response to microbial signals (Lambeth, 2004; Terada, 2006). Drosophila requires commensal microbe-induced hydrogen peroxide (H2O2) to maintain gut epithelial homeostasis (Ha et al, 2005a, 2005b), and plants also utilize induced ROS in modulating signal transduction pathways in response to bacterial pathogens and symbionts (Kotchoni and Gachomo, 2006). In mammalian cells, ROS have been shown to serve as critical second messengers in multiple signal transduction pathways in response to proinflammatory cytokines and growth factors (Lambeth, 2004). Intriguingly, it was recently reported that the enzymes involved in sumoylation, which are paralogous to the neddylation enzymes, are negatively regulated by H2O2 (Bossis and Melchior, 2006), suggesting that neddylation may be regulated in a similar manner. We thus sought to characterize if ROS generation was also a feature of the mammalian intestinal epithelia in response to commensal strains, and further if ROS could influence SCF-regulated pathways.
Results
Bacterial contact results in rapid transient generation of ROS in epithelia in vitro and in vivo
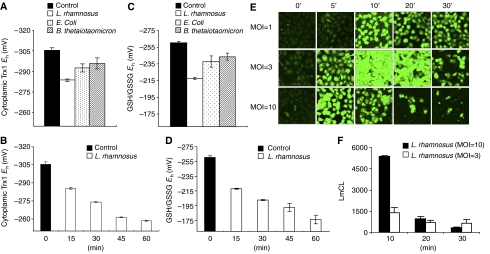
To determine if cells respond to bacterial contact with ROS production, we incubated cultured epithelial cells with representative commensal bacteria Lactobacillus rhamnosus, Bacteroides thetaiotaomicron, and Escherichia coli that we had previously shown had inflammatory inhibitory properties (Collier-Hyams et al, 2005), and assayed the two major antioxidant systems involved in peroxide elimination, thioredoxin-1 (Trx1) and glutathione (GSH) (Jones, 2006). As shown in Figure 1A and C, bacteria resulted in the oxidation of both Trx1 and GSH, with Lactobacillus inducing the greatest degree of change. Rapid oxidation of the Trx1 and GSH pools by Lactobacillus occurred progressively over the 1 h of coculture (Figure 1B and D). These data indicated that the cells were responding to a de novo oxidant stimulus. To detect the appearance of ROS, we employed the ROS indicators dichlorofluorescein diacetate (DCF) and dihydroethidium (DHE) that principally measure H2O2 and superoxide anion respectively. Again we observed rapid, transient and dose-dependent activation of ROS production in bacterially colonized epithelial cells (Figure 1E; Supplementary Figure S1), which was abrogated by pretreatment with the antioxidant _N_-acetylcysteine (NAC) and the NADPH oxidase inhibitor diphenyliodonium (DPI) (Supplementary Figure S2). Essentially identical data were obtained with several epithelial cell types (HeLa, Caco2, and T-84) (data not shown). In all cases, the response was greatest with Lactobacillus, whereas it was observed to a lesser extent with B. thetaiotaomicron and E. coli (data not shown). A rapid increase in the chemiluminescence of luminol, an ROS probe, was also observed in epithelial cells treated with Lactobacillus (Figure 1F), further corroborating that ROS was being induced in these cells.
Figure 1.
Commensal bacteria induce generation of epithelial ROS and cause oxidative stress. (A, B) Commensal bacteria cause oxidation of Trx1 pools. Caco2 cells were colonized with different commensal bacterial strains (L. rhamnosus (open bars), B. thetaiotaomicron (bars with diagonals), E. coli (dotted bars)) for 15 min as shown in panel A or with Lactobacillus over a time course as shown in panel B. Cells treated with PBS served as controls (filled bars). The graphs show the _E_h for Trx1, and data are represented as means±s.e. All values denote a statistically significant difference from control (P<0.05). (C, D) Redox analysis of GSH/GSSG following colonization of IEC-6 cells with different commensal bacterial strains (L. rhamnosus (open bars), B. thetaiotaomicron ( bars with diagonals), E. coli (dotted bars)) for 15 min as shown in panel C or with Lactobacillus over a period of time as shown in panel D. Cells treated with PBS served as control (open bars). Graphs show the _E_h for GSH/GSSG. Data are represented as means±s.e. All values denote a statistically significant difference from control (P<0.05). (E) Bacteria stimulate rapid epithelial ROS generation. IEC-6 cells were colonized with Lactobacillus, washed, and incubated with 5 μM of CM-H2DCF-DA. Fluorescence was measured by confocal laser scanning microscopy (Zeiss). (F) Kinetics of bacterially induced ROS generation in epithelial cells. ROS production was measured in trypsinized HeLa cells incubated with Lactobacillus by luminol assay. Peak relative luminescence is shown. Data represent the mean of three independent assays and are expressed in units of luminol chemiluminescence (LmCL).
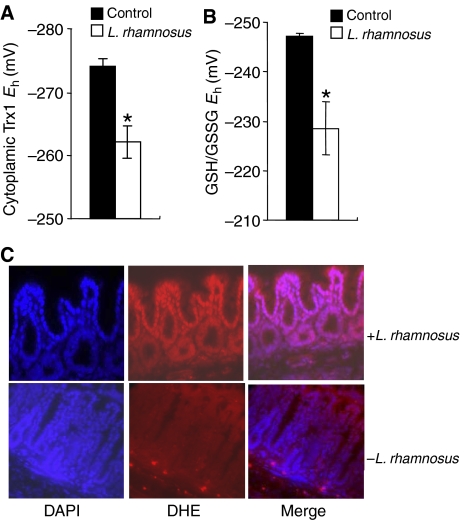
In vivo, Lactobacillus and to a lesser extent B. thetaiotaomicron (data not shown) also caused oxidative stress in the mucosa of murine small bowel 30 min post-oral inoculation of bacteria, as measured by the oxidation of Trx1 and GSH pools in tissue lysates prepared from proximal jejunum (Figure 2A and B). Additionally, ROS generation (superoxide anion) was also observed specifically in murine colon epithelia 30 min after rectal administration of Lactobacillus, by instillation of DHE into the colon (Figure 2C). Treatment with Lactobacillus for 30 min showed increased nuclear staining of the oxidized form of DHE (red), predominantly in the epithelia, indicating increased production of ROS in comparison with that of untreated control (Figure 2C). Based on these in vivo and in vitro assays, we concluded that commensal intestinal bacteria induce rapid generation of ROS within minutes of contact with epithelial surfaces.
Figure 2.
In vivo changes in intracellular redox in mouse intestinal epithelial cells contacted by commensal bacteria. (A) _Lactobacillus_-induced Trx1 oxidation in epithelial cells as analyzed by redox Western analysis. The graph shows the _E_h values for Trx1 in proximal jejunum 30 min after treatment with a bolus of bacteria (open bars) as compared with untreated controls (filled bars). Data are represented as means±s.e. Asterisks denote a statistically significant difference from control (P<0.05). (B) _Lactobacillus_-induced changes in the redox pools of GSH/GSSG. The graph shows the _E_h values for GSH/GSSG in the jejunum 30 min after treatment with a bolus of bacteria (open bars) as compared with untreated controls (filled bars). Data are represented as means±s.e. The asterisks denote a statistically significant difference from control (P<0.05). (C) In situ detection of superoxide in mouse colon treated with Lactobacillus. After 30 min of infusion of Lactobacillus, mouse colon was stained with DHE and imaged as described. Under identical tissue processing and imaging conditions, nuclear fluorescence in mouse colon treated with bacteria (top panels) is markedly increased compared with colon from untreated control mouse (bottom panels).
Bacterially elicited ROS mediates cullin deneddylation and modulates epithelial signaling
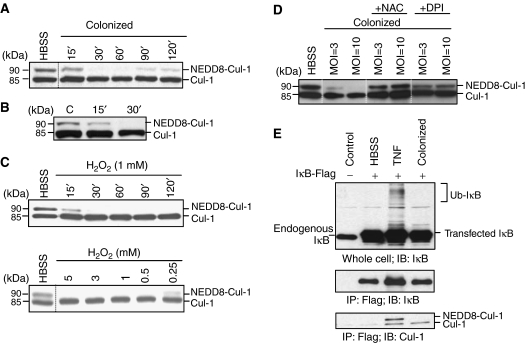
Based on the recent finding that ROS mediates inhibitory effects on the related sumoylation pathway (Bossis and Melchior, 2006), we sought to determine whether induction of ROS in response to commensal bacteria mediated similar effects on the neddylation pathway. In in vitro experiments utilizing epithelial coculture with Lactobacillus, loss of Cul-1 neddylation occurred within 30 min and was reversible (Figure 3A), a time course consistent with the observed generation of ROS. We next evaluated cullin neddylation in the mucosa of living mice inoculated with Lactobacillus. In these in vivo experiments, Lactobacillus was instilled into intact murine ileal loops and loss of Cul-1 neddylation in mucosal lysates was observed within 30 min of bacterial–epithelial contact, thus confirming that this biochemical change occurs in vivo with identical kinetics as that observed in vitro (Figure 3B).
Figure 3.
Loss of cullin-1 neddylation by bacterially mediated ROS generation. (A) Bacteria cause a time-dependent loss of Cul-1 neddylation that is reversible. Cul-1 is detected in whole-cell protein extracted from HeLa cells incubated with Lactobacillus and immunoblotted with Cul-1-specific antiserum. (B) Bacteria cause a time-dependent loss of Cul-1 neddylation in vivo. Western blot analysis with Cul-1-specific antisera of proteins extracted from mouse intestinal epithelial cells treated with bacteria via an ileal loop. (C) H2O2 causes time- and dose-dependent loss of Cul-1 neddylation. HeLa cells treated with H2O2 were analyzed as in panel A. (D) DPI and NAC prevent bacterially mediated loss of Cul-1 neddylation. HeLa cells pretreated with NAC or DPI for 30 min and colonized with Lactobacillus for 30 min were analyzed as in panel A. (E) Un-neddylated Cul-1 associates with IκB-α and does not support ubiquitination. Immunoprecipitation of IκB-Flag with anti-Flag agarose and immunoblots with IκB- and Cul-1-specific antisera of lysates from transfected HeLa cells challenged with TNF-α for 30 min or colonized by bacteria in the presence of the proteasome inhibitor MG-262. Note that the hyperneddylation of IκB-associated Cul-1 in TNF-α-treated cells is consistent with Cul-1 being in an active state to support ubiquitination of IκB and allow subsequent NF-κB activation.
To test for direct effects of ROS on cullin neddylation, we treated cultured epithelial cells with H2O2 and observed a loss of Cul-1 neddylation in a dose- and time-dependent manner (Figure 3C), indicating that ROS could recapitulate the cullin deneddylation first observed with bacterial colonization. To determine if ROS elicited endogenously within cells by bacterial contact could mediate cullin deneddylation, we pretreated epithelial cells with NAC or DPI (which prevented the bacterially induced increase in ROS; Supplementary Figure S2) and found these agents also prevented the loss of Cul-1 neddylation when cells were colonized by commensal bacteria (Figure 3D). These results indicate that bacterial-elicited ROS are the direct mediators of deneddylation of Cul-1. Bacterial colonization resulted in loss of IκB-α ubiquitination (Figure 3E); interestingly, this unmodified and thus inactive IκB was exclusively physically associated with deneddylated Cul-1, consistent with past observations showing deneddylation of Cul-1 temporally corresponding with inactivation of NF-κB.
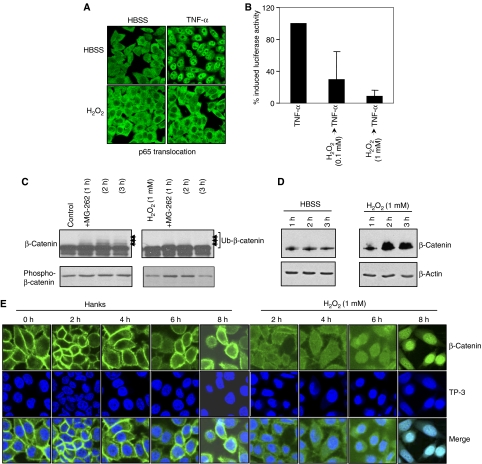
To demonstrate that ROS had functional effects on E3-SCFβTrCP-dependent pathways, HeLa cells were treated with H2O2 at concentrations that resulted in Cul-1 deneddylation, and effects on NF-κB- and Wnt-signaling pathways were evaluated. H2O2 treatment prevented TNF-α-induced nuclear translocation of the p65 subunit of NF-κB (Figure 4A), and repressed the TNF-α-stimulated activity of an NF-κB-dependent reporter gene (Figure 4B); this was identical to our previous findings with bacterial coculture (Neish et al, 2000). Consistently, the same treatment caused blockade of ubiquitination (Figure 4C) and accumulation of total β-catenin protein (Figure 4D). Further, in untreated cells β-catenin localized along the cell membrane and was excluded from the nuclei (Figure 4E). Within 2 h of treatment with 1 mM H2O2, membranous β-catenin diminished and progressively appeared within the nucleus (Figure 4E). We have also shown activation of the Wnt-responsive TCF reporter constructs by colonization of epithelial cells with non-pathogenic bacteria (Sun et al, 2004). Collectively, these results show that cells treated with live commensal bacteria or H2O2 similarly modulate E3-SCFβ−TrCP-controlled epithelial signaling pathways.
Figure 4.
Oxidant modulation of E3-SCFβTrCP mediated signaling. (A) H2O2 prevents TNF-α-induced p65 nuclear translocation. Immunofluorescent staining of p65 in HeLa cells preincubated with 1 mM H2O2 for 45 min and treated with TNF-α for 30 min. (B) H2O2 inhibits activity of a NF-κB-luciferase reporter. Lysates from HeLa cells transfected with a NF-κB-luciferase reporter plasmid, preincubated with H2O2 for 45 min, and treated in the presence of H2O2 with TNF-α for 5 h were assayed for luciferase activity. Data represent the mean of three independent assays and are shown as % TNF-α-induced NF-κB-Luc. (C) H2O2 prevents ubiquitination of β-catenin. Western blot analysis with anti-β-catenin and anti-phospho-β-catenin Abs of whole-cell protein extracted from epithelial cells pretreated 1 mM H2O2 for 1 h, washed, and treated with MG-262 (500 nM) for the indicated time. (D) H2O2 causes increased accumulation of β-catenin. Western blot analysis with anti-β-catenin and anti-β-actin Abs of whole-cell protein extracted from HeLa cells treated with HBSS±1 mM H2O2 for the indicated times. (E) H2O2 induces nuclear translocation of β-catenin. Confocal microscopy of HeLa cells treated with HBSS±1 mM H2O2 for the indicated times, and stained with the anti-β-catenin antibody (green) and nuclei counterstained with To-Pro-3 iodide (TP-3, blue). Merged images display sites of co-labeling.
Bacterially induced oxidant stress causes inactivation of the neddylation machinery
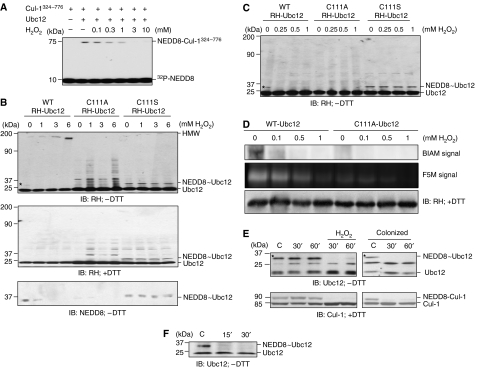
H2O2/ROS signals can be transduced via the transient oxidative inactivation of catalytic cysteine residues present in certain regulatory enzymes (Barford, 2004). These redox-sensitive cysteines are present under physiological conditions in a limited but increasingly recognized subset of enzymes including protein tyrosine phosphatases and MAPK phosphatases (Barford, 2004; Kamata et al, 2005; Rhee et al, 2005; Tonks, 2005), as well as the SUMO-conjugating enzyme Ubc9 (a member of the class of Ub-like conjugating enzymes (Ubcs) or E2s) (Bossis and Melchior, 2006). Ubc9 is paralogous with the NEDD8 E2 enzyme Ubc12 (Hochstrasser, 2000) that catalyzes the transfer of a thioesterified NEDD8 moiety from its own active site to the Lys720 of cullin substrates, resulting in neddylation via an isopeptide bond (Pan et al, 2004). We had shown that knockdown of Ubc12 had suppressive effects on NF-κB activation, similar to the effects of bacterial colonization (Collier-Hyams et al, 2005). Thus, we sought to characterize the effects of oxidant stress on Ubc12 activity. In an in vitro cell-free assay of neddylation with purified Ubc12, a complex containing Roc1 and the Cul-1 C-terminal fragment spanning amino acids 324–776, and 32P-NEDD8 revealed that H2O2 inhibited the formation of neddylated Cul-1(324−776) in a dose-dependent manner (Figure 5A).
Figure 5.
Bacterially induced oxidant stress causes inactivation of the neddylation machinery. (A) Inhibition of Cul-1 neddylation in vitro by H2O2. In vitro neddylation was carried out in the presence of H2O2 using purified Ubc12, a complex containing Roc1 and the C-terminal portion (residues 324–776) of Cul-1, and 32P-NEDD8. (B) H2O2 inhibits the formation of the NEDD8∼Ubc12 thioester bond and leads to the formation of a DTT-sensitive HMW Ubc12 species. Western blot analysis with RH- and NEDD8-specific antisera of whole-cell protein extracted from HeLa cells transfected with WT or mutant (C111A or C111S) RH-Ubc12, treated with H2O2 for 1 h at the indicated dose, and lysed in SDS lysis buffer without (−DTT) or with (+DTT) reducing agents. The star marks the 30-kDa H2O2-sensitive NEDD8∼Ubc12 thioester form. (C) H2O2 induces the loss of NEDD8∼Ubc12 thioester bond at low doses. HeLa cells transfected with RH-Ubc12 (WT, C111A or C111S) were treated with increasing H2O2 concentrations for 1 h, lysed in SDS lysis buffer without DTT and immunoblotted with anti-RH antibody. (D) In vitro treatment of purified Ubc12 with H2O2 results in the oxidation of Ubc12. Purified Ubc12 (WT) and Ubc12 (C111A) were treated with H2O2 (0.1–1 mM) and labeled with the thiol-sensitive dyes BIAM or F5M. Samples and control were separated on a 15% PAGE and labeled protein was detected as described. A parallel gel was immediately probed with anti-RH antibody to identify specific protein band. (E) Bacteria or H2O2 inhibit the formation of the NEDD8∼Ubc12 thioester bond of endogenous Ubc12 in vitro. Western blot analysis with Ubc12- and Cul-1-specific antisera of whole-cell protein extracted from HeLa cells treated with 1 mM H2O2 or Lactobacillus and lysed in SDS lysis buffer without (−DTT, top panels) or with (+DTT, bottom panels) reducing agents. The stars mark the 30-kDa H2O2- and colonization-sensitive NEDD8∼Ubc12 thioester forms. (F) Bacteria inhibit the formation of the NEDD8∼Ubc12 thioester in vivo. Western blot analysis with Ubc12-specific antisera of proteins extracted from mouse intestinal epithelial cells treated with bacteria via an ileal loop.
In cell-based experiments, immunoblots from cells transfected with wild-type (WT) Ubc12 prepared under non-reducing conditions (without DTT) revealed an NEDD8∼Ubc12 thioester form (Figure 5B, star, top left panel), whereas under standard reducing conditions (with DTT) a single lower molecular weight band was observed at the expected 21 kDa (Figure 5B, middle left panel). The identity of the higher molecular weight band as the NEDD8∼Ubc12 form was confirmed by immunoprecipitation of transfected myc-NEDD8 followed by immunoblots for transfected myc-NEDD8 and RH-Ubc12 (Supplementary Figure S3). Significantly, the NEDD8∼Ubc12 thioester form was totally abolished in a dose-dependent manner in cells treated with H2O2 and corresponded to the appearance of high-molecular-weight (HMW) oxidized species (Figure 5B, top left panel), because these forms were abolished when analyzed under reducing conditions (Figure 5B, middle left panel). The identity of the NEDD∼Ubc12 thioester form was also confirmed with an NEDD8 immunoblot under non-reducing conditions (Figure 5B, bottom left panel). A mutant Ubc12 bearing a cysteine to alanine substitution (C111A) in the active site exhibited no 30-kDa conjugate at all, with RH-Ubc12 or endogenous NEDD8 immunoblots, indicting that Cys111 is the site of the thioester modification, and did not show the accumulation of HMW oxidized species in response to H2O2, suggesting redox sensitivity of the WT Ubc12 catalytic Cys111. Finally, a Ubc12 mutant with a cysteine to serine substitution (C111S) was used, which is known to form stable oxyester bond with NEDD8 rather than the DTT-sensitive thioester bond (Wada et al, 2000). This neddylated Ubc12(C111S) was stable in cells treated with H2O2 (Figure 5B, middle right panel) and did not show the formation of the Ubc12-HMW forms (Figure 5B, top right panel). Importantly, loss of the Ubc12∼NEDD8 thioester form was also observed with 0.25 mM H2O2 (Figure 5C, star), a concentration that also caused loss in neddylation of Cul-1 (Figure 3C). Also as observed before, the mutant Ubc12 carrying C111A substitution exhibited no 30-kDa conjugate, whereas mutant Ubc12 carrying C111S substitution exhibited a DTT-insensitive NEDD8 conjugate (Figure 5C).
To confirm the oxidant sensitivity of Ubc12, we utilized biotinylated iodoacetamide (BIAM) and fluorescein-5-maleimide (F5M). Iodoacetamide and maleimide irreversibly modify reactive protein thiols using different chemistry; derivatives of these molecules are commonly employed as a detection reagent for proteins that contain redox-sensitive cysteines (Richard, 1998; Leichert and Jakob, 2006). After affinity purification of transfected WT Ubc12 and Ubc12(C111A), proteins were treated with BIAM or F5M, with and without prior oxidation with H2O2. Without addition of H2O2, only the WT form of Ubc12 was significantly modified by BIAM or F5M, indicating that Cys111 is a reactive cysteine (Figure 5D). WT Ubc12 treated with H2O2 before BIAM/F5M addition showed a loss of BIAM/F5M reactivity, showing that the reactive Cys111 is redox sensitive (Figure 5D).
Next, we sought to evaluate the effects on endogenous Ubc12. In cell lysates isolated under non-reducing conditions, we found that the 30-kDa NEDD8∼Ubc12 thioester adduct was abolished in cells treated with H2O2 (Figure 5E, upper left panel) and, importantly, by coculture with commensal bacteria (Figure 5E, upper right panel) at the same times and MOI that resulted in cullin deneddylation. Finally, we evaluated Ubc12 in the mucosa of living mice into which Lactobacillus was instilled via intact ileal loops (Figure 5F). Scrapings of intestine were subjected to immunoblot showing loss of the 30-kDa NEDD8∼Ubc12 thioester adduct that also temporally corresponded to the loss of neddylated Cul-1 shown earlier (Figure 3B). Collectively, these data indicate the NEDD8∼Ubc12 thioester is abolished in cells exposed to oxidant stress or colonized with commensal bacteria secondary to the oxidation of the active site cysteine.
Discussion
In this report we have described important eukaryotic regulatory processes profoundly influenced by interaction with commensal/probiotic bacteria. Bacteria induced rapid, transient ROS generation in epithelia in vitro and in vivo, which resulted in the loss of neddylated Cul-1 and consequent modulation of SCF-dependent pathways. The rapid loss of cullin neddylation was mediated by oxidative inactivation of Ubc12, the cellular NEDD8-conjugating enzyme (E2). These results indicate that bacterial members of the normal intestinal flora, including a widely used probiotic agent, can influence important mammalian signaling pathways by modulating the activity of an enzymatic regulatory control point of the Ub–proteasome system.
Neddylation of E3-SCF Ub ligases is emerging as a key regulatory event in cellular processes that are controlled by protein degradation (Pan et al, 2004; Parry and Estelle, 2004; Petroski and Deshaies, 2005). It is a reaction that is apparently specific to substrates of the cullin family and thus acts as a post-translational modification specific to E3 Ub ligases of the SCF class. The process begins when NEDD8 forms a thioester bond with a heterodimeric ATP-dependent activation enzyme (E1) designated APP-BP1/Uba3. NEDD8 is then linked to Cys111 of Ubc12 via a transthiolation reaction. Recent structural studies have shown that Uba3 and Ubc12 form a dynamic complex where the catalytic cysteines of both enzymes are in close approximation during transthiolation (Dye and Schulman, 2007). NEDD8-thioesterified Ubc12 then dissociates from the complex and can mediate cullin neddylation, defined as an isopeptide bond linking the C-terminal Gly76 of NEDD8 to the ɛ-amino group of a conserved cullin lysine residue.
Cullin neddylation is generally conceptualized to be cyclical, with iterative rounds of cullin neddylation and deneddylation necessary for each consecutive Ub added to the target substrate (e.g., IκB-α) by the SCF complex. The reverse reaction, deneddylation, is catalyzed by the COP9 signalosome (CSN), an evolutionarily conserved multisubunit protein complex that possesses an ATP-independent metalloprotease activity (CSN5) responsible for cullin deneddylation (Lyapina et al, 2001; Cope and Deshaies, 2003). Consistently, we have shown that knockdown of csn5 was a positive activator of both basal and induced NF-κB activity (Khoury et al, 2007). Thus, oxidative inactivation of the forward neddylation reaction would permit unopposed deneddylation by the CSN, and repression of E3 activity.
ROS production in the gut
Deliberate generation of ROS in non-phagocytic mammalian cells can be achieved by enzymes homologous to phagocytic NADPH oxidase (NOX), by 5′-lipoxygenase, or by mitochondrial sources (Lambeth, 2004; Chiarugi and Fiaschi, 2007). In our experiments, the inhibitor DPI suppressed ROS generation and cullin deneddylation, suggesting that the NOX enzymes are involved. In humans, seven homologues of phagocytic NOX ((NOX 1–5) and dual oxidase (DUOX1 and DUOX2)) have been identified (Lambeth, 2004). These enzymes are expressed in a variety of tissues and are activated by varying (though still ill defined) stimuli (Geiszt and Leto, 2004; Lambeth, 2004; Geiszt, 2006), suggesting that the intentional generation of ROS, rather than being a unique feature of phagocytes, is a general feature of many cells. Interestingly, Nox1 and DUOXs, are highly expressed in colonic epithelial cells (Cheng and Lambeth, 2005; El Hassani et al, 2005; Rokutan et al, 2006). These enzymes have been suggested to play a role in epithelial host defense in Drosophila, although their physiological functions in vertebrates are not fully established (Ha et al, 2005a). Direct implication of the NOX enzymes in bacterial induction of ROS, and the specific eliciting signals involved are actively under investigation.
Oxidative regulation of Ub-like conjugating enzymes
ROS have traditionally been viewed as unavoidable byproducts of aerobic metabolic processes, or as consequences of pathological states such as hypoxia. However, evidence is emerging that certain ROS species such as H2O2 possess essential signaling roles. Indeed, several redox-sensitive proteins have been shown to be regulated by reversible H2O2-mediated oxidation of their active-site cysteines, thus allowing these proteins to act as sensors of intracellular H2O2 concentration and control critical steps in several signal transduction pathways. The labile nature of H2O2, the number and diversity of enzymes that generate it, and the reversibility of the active-site inactivation of target proteins are all consistent with a role in signal transduction. In most proteins, cysteine residues are protonated at physiological pH (Cys-SH), whereas so-called low p_K_a cysteines exist as thiolate anions (Cys-S−) and are more readily oxidized by H2O2. These redox-sensitive thiolates are present under physiological conditions in a limited but increasingly recognized subset of enzymes (Barford, 2004; Kamata et al, 2005; Rhee et al, 2005; Tonks, 2005).
Bossis and Melchior (2006) proposed that ROS play a regulatory role in the sumoylation reaction, by showing the rapid H2O2-induced formation of a disulfide bridge between the SUMO E1 (Uba2) and E2 (Ubc9), resulting in inactive dimer formation. Significantly, NEDD8 E1 (Uba3) and E2 (Ubc12) are paralogous with the sumoylation enzymes, with greater than 30% primary sequence homology (Kerscher et al, 2006). We did not observe oxidant-induced disulfide linkage between Ubc12 and Uba3. This may reflect intrinsic differences in the two complexes or differences in experimental design. Instead, H2O2 caused loss of the thioester form of Ubc12. This is consistent with data of Bossis and Melchior showing loss of the SUMO thioester form of Uba2 at H2O2 doses of 0.01 mM concentration, an order of magnitude lower than H2O2 concentration necessary to induce dimer formation in their experimental systems.
Interestingly, the reactive cysteines of the general class of Ub-like conjugating enzymes are not predicted to exist as low p_K_a in isolation (Tolbert et al, 2005). It is possible that the ROS sensitivity of the Ubc enzymes is restricted to the in vivo complex, which is characterized by close physical approximation of catalytic cysteines on the respective E1 and E2 molecules (Haas, 2007). Thus, the rapid and cyclical transthiolation between the closely adjacent catalytic cysteine of Uba3 and Ubc12 could result in the exposure of the transient thiolate anions to oxidative signals once the NEDD8 moiety was passed. In any event, the similarity of loss of E2 thioester formation in the absence of low-p_K_a cysteines in both the NEDD8 and SUMO conjugation enzymes is highly consistent with the notion that these enzymes (Ubcs) are sensitive to regulation by ROS.
Effects of ROS generation on intestinal biology
We propose that ROS generation can influence the equilibrium between neddylated and un-neddylated Cul-1 in cells of the gut in contact with the normal flora (Figure 6), and thus modulate the activity of the SCF ligases and the pathways these enzymes control. In vivo, epithelial ROS generation could be elicited by substantial changes in the gut luminal ecology resulting from abrupt dietary changes, antibiotic administration, probiotic use, or the developmental acquisition of the normal flora during the neonatal period. Additionally, there is a wide range in the abundance of prokaryotes in the gut, ranging in number from 104 CFU/ml in the small bowel, to 1012 CFU/ml in the cecum and ascending colon and markedly less in the descending colon (O'Hara and Shanahan, 2006). Different anatomical regions of the gut may thus experience different levels of ROS. Of note, there is an inverse correlation between bacterial numbers in the colon with the pathological distribution of inflammatory ulcerative colitis, with the disease characteristically most predominant in the rectum and ascending colon, regions with the lowest bacterial numbers.
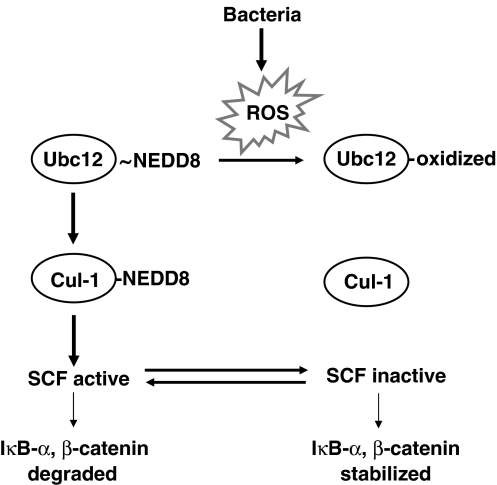
Figure 6.
Model of bacterially induced ROS generation on signaling pathways. Summary diagram of bacterial inhibition of SCF signaling. Bacteria elicit ROS generation in epithelial cells. ROS inactivate the Ubc12 enzyme, preventing the neddylation of cullin-1. Un-neddylated cullin in the E3-SCFβ−TrCP complex renders it unable to carry out ubiquitination and is thus inactive.
We also show that certain bacteria (i.e., Lactobacillus) are more potent inducers of ROS. Possibly, the relative abundance of members of the gut flora with different capacity to stimulate ROS could have functional consequences at the organism level, and may contribute to the concept of a ‘eubiotic' or ‘dysbiotic' flora associated with health or inflammatory disease, respectively. Our data suggest that dynamic changes in ROS production may be involved in suppression of NF-κB, an effect that may account for the usual inflammatory tolerance of the intestinal mucosal, but in cases of excessive ROS generation may result in deleterious suppression of NF-κB-mediated survival factors and contribute to tissue injury. Finally, E3-SCFβTrCP is required for the regulation of the Snail, Twist, and Hedgehog pathways (Zhou and Hung, 2005) as well as others, suggesting additional targets whereby microbially mediated modulation of SCF E3s could potentially influence many aspects of mucosal homeostasis. Functional analysis of the consequences of physiochemical effects on multiple pathways in vivo will be challenging future work.
Materials and methods
Reagents
H2O2, luminol, peroxidase, NAC, and DPI were purchased from Sigma-Aldrich. MG-262 was purchased from Biomol. Recombinant human TNF-α was purchased from R&D Systems.
Bacterial strains
L. rhamnosus GG purchased from the American Type Culture Collection (ATCC cat. no. 53103) was cultured in 100 ml LBS and grown 16 h at 37°C under non-agitated microaerophilic conditions. B. thetaiotaomicron was obtained from the American Type Culture Collection (ATCC cat. no. 29148) and was grown in trypticase–yeast extract glucose medium for 24 h under anaerobic conditions. E. coli human commensal strains were grown in Luria–Bertani broth in a rotary shaker at 37°C for 6–8 h.
Bacterial suspensions for colonization experiments were prepared by centrifugation of 100 ml culture and resuspension in a small volume of Hank's balanced salt solution (HBSS). Multiplicity of infection (MOI) was calculated by estimating epithelial cell density by counting and measuring bacterial culture density using spectrophotometry (OD550). Unless otherwise noted, Lactobacillus was used at an MOI of 1 and other bacteria were used at a MOI of 10.
Epithelial cell culture
Human (Caco-2, HeLa, T-84) or rat (IEC-6) epithelial cells were grown in standard tissue culture vessels and maintained as previously described (Collier-Hyams et al, 2005).
Glutathione and thioredoxin-1 redox measurements
Following treatment samples were assayed for the concentrations of glutathione and glutathione disulfide as the concentration of _S_-carboxymethyl, _N_-dansyl derivatives by HPLC, using γ-glutamylglutamate as an internal standard (Jones, 2002).
Separation of oxidized and reduced forms of Trx1 in Caco2 or in mice tissue was based on procedures as described in Watson et al (2003) and Chen et al (2006). Redox state was calculated using the densitometric measurements of the oxidized and the reduced form in the Nernst equation.
Immunofluorescent ROS detection
Epithelial cells colonized with Lactobacillus for the indicated time were washed with HBSS and incubated in the dark with HBSS containing 5 μM of CM-H2DCF-DA (Molecular Probes) or 5 μM DHE (Molecular Probes) for an additional 5 min. To evaluate if antioxidants could block the observed ROS generation, cells were pretreated with NAC (20 mM) or DPI (40 μM) for 30 min before colonization with bacteria. All images were acquired using a confocal laser scaning microscope (Zeiss LSM 510) through a × 20 objective. For DCF imaging, images were captured using the following parameters: 488 nM excitation and 515–540 nM emission. For DHE imaging, images were captured using the following parameters: 490 nM excitation and 610 nm emission.
For in vivo ROS measurements, female mice (BALB/cj; 6–8 weeks) were starved overnight and 0.2 ml of PBS containing 108 CFU of L. rhamnosus was rectally administered with flexible No. 4 French catheters inserted 3–4 cm into the distal colon. After 30 min of bacterial administration, 0.2 ml of 5 μM DHE was administered and the mice sacrificed. Harvested colon was frozen in OCT medium (Miles Inc.) and cryostat sectioned at 5-μm thickness. Also, the same sections were further stained with DAPI (Molecular Probes). Control and treated mice were processed in parallel and images were acquired with identical parameters using a Zeiss Axiovert 135 microscope with a × 40 objective.
Luminol assay
Trypsinized HeLa cells were incubated with L. rhamnosus and ROS production was measured by luminol-dependent chemiluminescence assay as described in Cheng and Lambeth (2004). Wells without bacteria or HeLa cells served as negative controls.
Transient transfections and luciferase activity
HeLa cells were transiently transfected using Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Roche) according to the manufacturer's instructions. For luciferase reporter assays, cells seeded on 24-well plates were transfected with 20 ng pNF-κB-Luc plasmid (Stratagene), allowed 18 h for expression, preincubated for 45 min with H2O2, treated with TNF-α (20 ng/ml) for 5 h, and lysed according to the manufacturer's protocol. Luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega).
Immunofluorescence studies of p65 or _β_-catenin
Immunofluorescent labeling of adherent HeLa cells grown on 12 mm glass coverslips was performed as follows: cells were fixed for 20 min in 3.7% paraformaldehyde in PBS, washed in PBS, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and washed again. Fixed samples were incubated in blocking buffer (5% normal goat serum in PBS) overnight at 4°C. A 1-h incubation with each antibody diluted in blocking buffer was performed as follows: treatment with 1° rabbit anti-p65 (Rockland Immunochemicals) and 2° fluorescein (FITC)-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) antibodies; or with 1° anti-β-catenin monoclonal (BD Biosciences) and 2° FITC-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) antibodies. Nuclei were stained with To-Pro-3 iodide (Molecular Probes). Cells were washed three times between each antibody treatment. The coverslips were mounted on glass slides and stained cells were observed by laser confocal microscopy. Images were acquired through a × 63 objective.
Western blot analysis
Following experimental treatment, epithelial cells were washed in cold HBSS and whole-cell extracts were prepared by rapid lysis in SDS–PAGE buffer with or without DTT, as noted. Cell lysates were electrophoresed on SDS–polyacrylamide gels and transferred to nitrocellulose following standard protocols. Immunoreactive proteins were detected with antibodies to IκB-α (Santa Cruz Biotechnology Inc.), Cul-1 (Zymed), NEDD8 (Zymed), β-catenin (BD Biosciences), Ubc12 (Rockland), c-myc (Clontech), or RGS-His (RH; Qiagen) using the Amersham enhanced chemiluminescent protocol (GE Healthcare) and HRP-conjugated donkey anti-rabbit or sheep anti-mouse secondary antibody (GE Healthcare).
Immunoprecipitation
After transfection and expression of epitope-tagged proteins (Flag, c-myc) or treatment, HeLa cells were washed in HBSS and scraped from wells or plates in a small volume (<1 ml) of non-denaturing lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Nonidet P-40 or SDS, 1 mM EGTA, pH 8.0, 250 pM MG-262, Sigma protease inhibitor cocktail diluted 1:200, 10 mM NaF, 0.4 mM Na3VO4). The lysates were incubated on a shaker for 10 min at 4°C and centrifuged at 15 000 g for 20 min. For Flag-IκB, the clarified lysates were precleared with IgG sepharose beads (Sigma) for 2 h, and then immunoprecipitated for 4 h at 4°C with α-Flag-conjugated agarose (Sigma). The bound agarose was washed six times in non-denaturing buffer and the immunopurified proteins were eluted with a minimum volume of denaturing SDS–PAGE buffer. Myc-NEDD8 was immunoprecipitated from lysates using an anti-c-myc immunoprecipitation kit (Sigma) according to the manufacturer's protocol.
In vivo murine experiments
Female mice (BALB/cj; 6–8 weeks) were starved overnight and given a single dose of PBS (0.2 ml) containing 108 CFU of L. rhamnosus (or B. thetaiotaomicron) by gavage. After 30 min of bacterial administrations, mice were sacrificed and small intestine was harvested. The tissue was opened along the mesenteric border, and epithelial tissue scraped and assayed for changes in Trx1 or GSH as described above. Bacterial transit was monitored by concurrent administration of methylene blue. Sections of bowel with and without dye were selected for experimental and control analyses.
For ileal loop assay, 6- to 8-week-old BALB/cj mice were anesthetized, a ligated intestinal loop (2–8 cm from the ileocecal junction) prepared in situ, and Lactobacillus (108 CFU/ml) was introduced. The mice were maintained under anesthesia at 37°C for up to 30 min and then euthanized, and tissues were removed for analysis. The small intestine was opened along the mesenteric border, and epithelial tissue scraped and lysed by homogenization in RIPA buffer (100 mg tissue per milliliter of buffer) and centrifuged at 16 000 r.p.m. for 20 min at 4°C. Protein concentrations of supernatants were determined by protein assay (Bio-Rad) and equal amounts of protein were diluted with denaturing SDS–PAGE loading buffer and separated by SDS–PAGE.
In vitro neddylation assay
In vitro Cul-1 neddylation was performed in a reaction mixture (30 μl) that contained 50 mM Tris–HCl, pH 7.4, 0.6 mM DTT, 5 mM MgCl2, 2 mM ATP, 2 mM NaF, 10 nM okadaic acid, Roc1-Cul-1324–776 (5 pmol), 32P-NEDD8 (200 pmol), and APP-BP1/Uba3 (0.15 pmol; purchased from Boston Biochem), in the presence or absence of H2O2. The neddylation reaction was initiated by the addition of Ubc12 (25 pmol) and incubated at room temperature for 10 min. The reaction was terminated with SDS loading buffer (10 μl) containing DTT and the products were separated on a 4–20% SDS–PAGE gradient denaturing gel and visualized by autoradiography. Roc1-Cul-1324–776, 32P-Nedd-8, and Ubc12 were prepared as described elsewhere (Yamoah et al, 2005).
Analysis of protein oxidation
Ubc12 WT and Ubc12 C111A oxidation was monitored by labeling with the thiol-modifying reagents BIAM (Molecular Probes) and F5M (Molecular Probes). Protein was purified from HeLa cells transiently transfected with RGS epitope-tagged WT Ubc12 and Ubc12C111A using the Ni-NTA resin (Qiagen) as described in Wada et al (2000). Purified protein was quantified using the Bradford reagent (BioRad). An equal amount of the WT and the mutant protein was subjected to oxidation with H2O2 and the proteins were then labeled with either BIAM or F5M. For BIAM labeling, proteins were incubated in a BIAM buffer (50 mM bis-Tris–HCl, pH 6.5, 0.5% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, Protease inhibitor tablets (Roche), 20 μM BIAM) for 10 min at 37°C in the dark. The labeling reaction was stopped by adding IAM at a final concentration of 5 mM. Proteins were separated with SDS–PAGE, transferred to PVDF membrane, and stained with streptavidin Alexa Fluor 680 conjugate (Invitrogen), and blots were scanned in an Odyssey scanner (LiCor). A parallel gel run was probed with RH antibody to visualize equal protein loading.
For F5M labeling, protein samples were incubated in F5M buffer (20 mM Tris–HCl, pH 7.4, 0.1 mM MgCl2, 1 mM MgCl2, 0.3 mM F5M) for 5 min on ice. F5M labeling was stopped by the addition of 100 mM DTT. Proteins were separated by SDS–PAGE. Fluorescence intensities of the proteins were quantified by scanning gels at an excitation wavelength of 526 nm using a fluorescence-image analyzer Storm 7200 (GE HealthCare). The same gel was then probed with RH antibody to visualize protein load.
Supplementary Material
Supplementary Figure S1–S3
Acknowledgments
We thank J David Lambeth and Keith D Wilkinson for helpful discussions. This work was supported in part by National Institutes of Health grants DK-071604 and AI-064462 to ASN, DK-68105 to LSC-H, and DK-064399.
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host–bacterial mutualism in the human intestine. Science 307: 1915–1920 [DOI] [PubMed] [Google Scholar]
- Barford D (2004) The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol 14: 679–686 [DOI] [PubMed] [Google Scholar]
- Bossis G, Melchior F (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21: 349–357 [DOI] [PubMed] [Google Scholar]
- Chen Y, Cai J, Jones DP (2006) Mitochondrial thioredoxin in regulation of oxidant-induced cell death. FEBS Lett 580: 6596–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Lambeth JD (2004) NOXO1, regulation of lipid binding, localization, and activation of Nox1 by the Phox homology (PX) domain. J Biol Chem 279: 4737–4742 [DOI] [PubMed] [Google Scholar]
- Cheng G, Lambeth JD (2005) Alternative mRNA splice forms of NOXO1: differential tissue expression and regulation of Nox1 and Nox3. Gene 356: 118–126 [DOI] [PubMed] [Google Scholar]
- Chiarugi P, Fiaschi T (2007) Redox signalling in anchorage-dependent cell growth. Cell Signal 19: 672–682 [DOI] [PubMed] [Google Scholar]
- Collier-Hyams LS, Sloane V, Batten BC, Neish AS (2005) Cutting edge: bacterial modulation of epithelial signaling via changes in neddylation of cullin-1. J Immunol 175: 4194–4198 [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ (2003) COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114: 663–671 [DOI] [PubMed] [Google Scholar]
- Dye BT, Schulman BA (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- El Hassani RA, Benfares N, Caillou B, Talbot M, Sabourin JC, Belotte V, Morand S, Gnidehou S, Agnandji D, Ohayon R, Kaniewski J, Noel-Hudson MS, Bidart JM, Schlumberger M, Virion A, Dupuy C (2005) Dual oxidase2 is expressed all along the digestive tract. Am J Physiol Gastrointest Liver Physiol 288: G933–G942 [DOI] [PubMed] [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI (1998) Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev 62: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiszt M (2006) NADPH oxidases: new kids on the block. Cardiovasc Res 71: 289–299 [DOI] [PubMed] [Google Scholar]
- Geiszt M, Leto TL (2004) The Nox family of NAD(P)H oxidases: host defense and beyond. J Biol Chem 279: 51715–51718 [DOI] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367 [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Bae YS, Lee WJ (2005a) A direct role for dual oxidase in Drosophila gut immunity. Science 310: 847–850 [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, Brey PT, Lee WJ (2005b) An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell 8: 125–132 [DOI] [PubMed] [Google Scholar]
- Haas AL (2007) Structural insights into early events in the conjugation of ubiquitin and ubiquitin-like proteins. Mol Cell 27: 174–175 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (2000) Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol 2: E153–E157 [DOI] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI (2001) Molecular analysis of commensal host–microbial relationships in the intestine. Science 291: 881–884 [DOI] [PubMed] [Google Scholar]
- Jones DP (2002) Redox potential of GSH/GSSG couple: assay and biological significance. Methods Enzymol 348: 93–112 [DOI] [PubMed] [Google Scholar]
- Jones DP (2006) Redefining oxidative stress. Antioxid Redox Signal 8: 1865–1879 [DOI] [PubMed] [Google Scholar]
- Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M (2005) Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120: 649–661 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Khoury J, Ibla JC, Neish AS, Colgan SP (2007) Anti-inflammatory adaptation to hypoxia through adenosine-mediated cullin-1 deneddylation. J Clin Invest 117: 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchoni SO, Gachomo EW (2006) The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J Biosci 31: 389–404 [DOI] [PubMed] [Google Scholar]
- Lambeth JD (2004) NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4: 181–189 [DOI] [PubMed] [Google Scholar]
- Leichert LI, Jakob U (2006) Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signal 8: 763–772 [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Macdonald TT, Monteleone G (2005) Immunity, inflammation, and allergy in the gut. Science 307: 1920–1925 [DOI] [PubMed] [Google Scholar]
- Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL (2000) Prokaryotic regulation of epithelial responses by inhibition of IkappaB–alpha ubiquitination. Science 289: 1560–1563 [DOI] [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F (2006) The gut flora as a forgotten organ. EMBO Rep 7: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K (2004) Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23: 1985–1997 [DOI] [PubMed] [Google Scholar]
- Parry G, Estelle M (2004) Regulation of cullin-based ubiquitin ligases by the Nedd8/RUB ubiquitin-like proteins. Semin Cell Dev Biol 15: 221–229 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA (2005) Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol 17: 183–189 [DOI] [PubMed] [Google Scholar]
- Richard P (1998) Handbook of Fluorescent Probes and Research Chemicals, 6th edn. pp 47–62. Molecular Probes Inc.: Eugene, OR, USA [Google Scholar]
- Rokutan K, Kawahara T, Kuwano Y, Tominaga K, Sekiyama A, Teshima-Kondo S (2006) NADPH oxidases in the gastrointestinal tract: a potential role of Nox1 in innate immune response and carcinogenesis. Antioxid Redox Signal 8: 1573–1582 [DOI] [PubMed] [Google Scholar]
- Sun J, Hobert ME, Rao AS, Neish AS, Madara JL (2004) Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol 287: G220–G227 [DOI] [PubMed] [Google Scholar]
- Terada LS (2006) Specificity in reactive oxidant signaling: think globally, act locally. J Cell Biol 174: 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert BS, Tajc SG, Webb H, Snyder J, Nielsen JE, Miller BL, Basavappa R (2005) The active site cysteine of ubiquitin-conjugating enzymes has a significantly elevated p_K_a: functional implications. Biochemistry 44: 16385–16391 [DOI] [PubMed] [Google Scholar]
- Tonks NK (2005) Redox redux: revisiting PTPs and the control of cell signaling. Cell 121: 667–670 [DOI] [PubMed] [Google Scholar]
- Wada H, Yeh ET, Kamitani T (2000) A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo. J Biol Chem 275: 17008–17015 [DOI] [PubMed] [Google Scholar]
- Wang X, Feng S, Nakayama N, Crosby WL, Irish V, Deng XW, Wei N (2003) The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson WH, Pohl J, Montfort WR, Stuchlik O, Reed MS, Powis G, Jones DP (2003) Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J Biol Chem 278: 33408–33415 [DOI] [PubMed] [Google Scholar]
- Weng M, Walker WA (2006) Bacterial colonization, probiotics, and clinical disease. J Pediatr 149: S107–S114 [Google Scholar]
- Yamoah K, Wu K, Pan ZQ (2005) In vitro cleavage of Nedd8 from cullin 1 by COP9 signalosome and deneddylase 1. Methods Enzymol 398: 509–522 [DOI] [PubMed] [Google Scholar]
- Zhou BP, Hung MC (2005) Wnt, hedgehog and snail: sister pathways that control by GSK-3beta and beta-Trcp in the regulation of metastasis. Cell Cycle 4: 772–776 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S3