Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells (original) (raw)
Abstract
So far, POM121 and gp210 are the only known anchoring sites of vertebrate nuclear pore complexes (NPCs) within the lipid bilayer of the nuclear envelope (NE) and, thus, are excellent candidates for initiating the NPC assembly process. Indeed, we demonstrate that POM121 can recruit several nucleoporins, such as Nup62 or Nup358, to ectopic assembly sites. It thus appears to act as a nucleation site for the assembly of NPC substructures. Nonetheless, we observed functional NPCs and intact NEs in severely POM121-depleted cells. Double knockdowns of gp210 and POM121 in HeLa cells, as well as depletion of POM121 from human fibroblasts, which do not express gp210, further suggest that NPCs can assemble or at least persist in a POM121- and gp210-free form. This points to extensive redundancies in protein–protein interactions within NPCs and suggests that vertebrate NPCs contain additional membrane-integral nucleoporins for anchorage within the lipid bilayer of the NE. In Stavru et al. (on p. 509 of this issue), we describe such an additional transmembrane nucleoporin as the metazoan orthologue of yeast Ndc1p.
Introduction
The nuclear envelope (NE) divides eukaryotic cells into a nuclear and a cytoplasmic compartment. It comprises two lipid bilayers, the inner and the outer nuclear membrane. Local fusions between both membranes create giant aqueous channels (nuclear pores), through which all nucleocytoplasmic exchange proceeds. These pores are embedded into elaborate protein structures of eightfold rotational symmetry called the nuclear pore complexes (NPCs; Suntharalingam and Wente, 2003; Wozniak and Clarke, 2003; Drummond and Allen, 2004; Rabut et al., 2004).
NPCs are built by multiple copies of ∼30 different nucleoporins (Nups), which form the central, proteinaceous NPC structure and thus maintain the very special membrane topology and curvature at the pore. In addition, they create a selective permeability barrier that controls the fluxes of material through the central channel.
Membrane-integral Nups anchor NPCs within the nuclear membrane. Two of them, gp210 and POM121, were previously identified in vertebrate NPCs. gp210 forms homodimers and possesses a cleavable signal sequence, an ∼200-kD luminal domain, followed by a stop-transfer sequence that serves as membrane anchor and a short cytoplasmic tail (Gerace et al., 1982; Wozniak et al., 1989; Greber et al., 1990; Favreau et al., 2001). gp210 is evolutionary well conserved and is found in metazoans, such as vertebrates (Gerace et al., 1982), insects (Berrios et al., 1995), or nematodes (Cohen et al., 2003), in several protozoa, such as Dictyostelium discoideum, and in plants (Mans et al., 2004). Nevertheless, fungi apparently lost the gp210 gene, and it has been reported that several cell types of mouse do not express the gp210 protein (Eriksson et al., 2004; Olsson et al., 2004). It therefore appears that, at least under some circumstances, cells can bypass the requirement for gp210.
POM121 (Hallberg et al., 1993) is less conserved than gp210 and is found only in vertebrates. This may indicate that NPC assembly and maintenance does not necessarily require a membrane-anchored POM121 orthologue. POM121 shows a topology opposite to that of gp210. It comprises an NH2-terminal signal anchor and an ∼120-kD COOH-terminal domain that faces the NPC channel (Soderqvist and Hallberg, 1994). The membrane anchor, however, is not required for assembling POM121 into NPCs. Instead, the central POM121 domain is necessary and sufficient for incorporation into NPCs (Soderqvist et al., 1997). The COOH-terminal part contains FG repeats and therefore might contribute to the permeability barrier of NPCs.
Results and discussion
Development of an optical assay for probing interactions between Nups
As a membrane-integral Nup, POM121 provides static anchoring sites of NPCs within the lipid bilayer of the NE. Beyond this, POM121 might also be involved in the dynamic process of assembling this structure, e.g., by recruiting other Nups to the nuclear membrane. This is, however, difficult to test in the highly complex in vivo situation because many Nups assemble nearly simultaneously into the highly complicated NPC structure.
Anchoring POM121 individually to an ectopic site should simplify this situation. Such an ectopic POM121 derivative would then constitute an isolated “seed” for Nup recruitment. The artificial assembly site should be easy to recognize but distinct from NE or endoplasmic reticulum to avoid confusion with the formation of bona fide NPCs or their cytoplasmic counterparts, annulate lamellae.
Considering these constraints, mitochondria appeared to be the ideal platform for an ectopic POM121 bait. As a control, we first fused EGFP alone behind the NH2-terminal part of TOM20 (residues 1–70), which comprises an anchor for insertion into the outer mitochondrial membrane, followed by a very hydrophilic segment that prevents mistargeting to the endoplasmic reticulum (Kanaji et al., 2000). The resulting fusion (“Mito-GFP”) localized correctly to the outside of mitochondria, as judged by colocalization of the GFP signal with mitotracker-stained mitochondria (unpublished data; see the following paragraph).
POM121 can attract “soluble” Nups to ectopic assembly sites
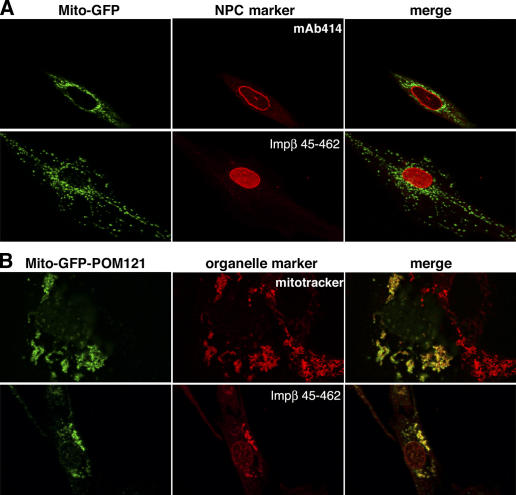
The Mito-GFP signal showed no overlap with any NPC marker (Fig. 1 A, top). However, when the central POM12173–797 domain (lacking membrane anchor and FG repeats) was added as a third module to the Mito-GFP fusion, mitochondria efficiently recruited NPC constituents (Fig. 1 B and Fig. 2). This was evident from stains with mAb414, which recognizes several FG-repeat Nups (Sukegawa and Blobel, 1993) or with the Impβ45–462 fragment that detects FG- and GLFG-repeat Nups (Kutay et al., 1997). A more detailed analysis revealed that not all Nups were attracted to the ectopic POM121 sites. In particular, Nups from the nuclear NPC side, namely, Nup50, Nup153, and TPR (translocated promoter region; Sukegawa and Blobel, 1993; Guan et al., 2000; Krull et al., 2004), remained absent from the structures (Fig. 2), possibly because they are actively imported into nuclei and thereby depleted from the cytoplasm and, thus, not available at the ectopic assembly sites.
Figure 1.
Ectopic expression of POM121 at mitochondria. (A) EGFP was fused behind residues 1–70 of TOM20, expressed in HeLa cells, and thereby anchored to the outer mitochondrial membrane (Mito-GFP). (top) Colocalization of the fusion protein with the NPC marker mAb414. (bottom) Staining of NPCs with the Alexa 568–labeled Impβ45–462 after digitonin permeabilization. Signals for mitochondria and NPCs do not overlap. (B) The POM12173–797 fragment, lacking its natural membrane anchor and the FG repeat domain, was fused behind the Mito-GFP module (Mito-GFP-POM121) and expressed in HeLa cells. (top) Colocalization of the Mito-GFP-POM121 signal with mitotracker-stained mitochondria (images depict a transfected and a nontransfected cell). (bottom) Bright staining with Impβ45–462 indicates recruitment of FG or GLFG repeat Nups to the ectopic POM121 fragment at mitochondria. Clustering of mitochondria is a side effect of this ectopic expression. The weak Mito-GFP-POM121 signal at the NE reflects the fact that targeting of the fusion protein to mitochondria is in competition with incorporation into bona fide NPCs.
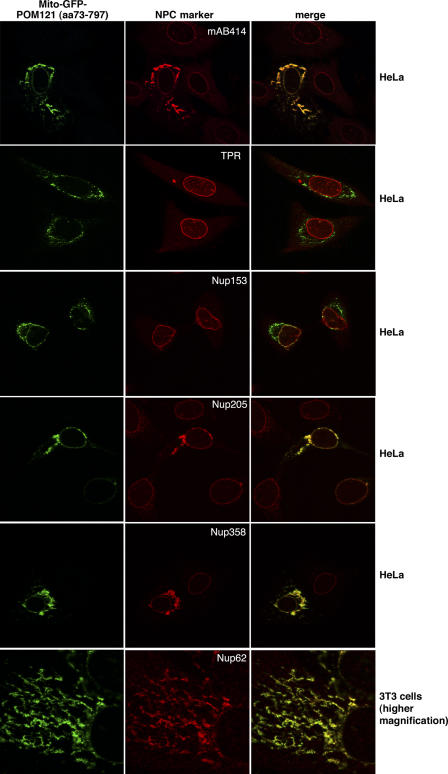
Figure 2.
The central POM121 domain is sufficient to recruit a subset of Nups. Images show immunofluorescence colocalization of Mito-GFP-POM12173–797 with indicated NPC markers. The ectopic assembly sites clearly recruited FG-repeat Nups (detected by mAb414) and stained positive for Nup358, Nup205, and Nup62 but remained negative for Nup153 and TPR. As indicated, expression was either in HeLa cells or in 3T3 fibroblasts, whose mitochondria show less clustering upon Mito-GFP-POM12173–797 expression.
The ectopic POM121 fragment most efficiently recruited the centrally located Nup62 (Guan et al., 1995; Grandi et al., 1997) and Nup205 (Krull et al., 2004). At later times of expression, we also observed recruitment of Nup98 (Radu et al., 1995) and even of Nup358/RanBP2 (Fig. 2) from the cytoplasmic NPC filaments (Wu et al., 1995; Yokoyama et al., 1995). The large distance (>20 nm) between the central Nups and the anchoring site for Nup358 within bona fide NPCs suggests that POM121 not only attracts its nearest neighbors but might even be sufficient to initiate the assembly of large substructures of the NPC.
POM121 appears nonessential for the biogenesis of mammalian NPCs
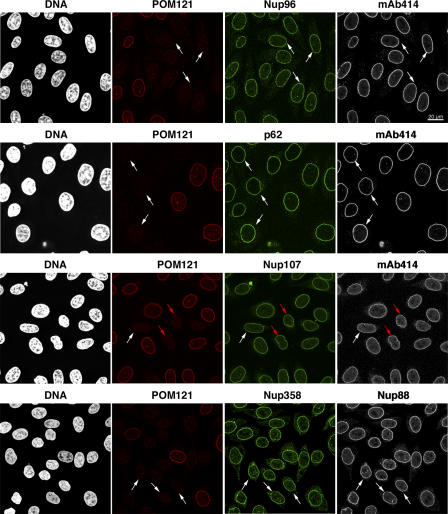
So far, POM121 and gp210 are the only known membrane-spanning Nups in vertebrates. If they constituted the primary anchors of NPCs within the pore membrane, they would be expected to be essential for the assembly of bona fide NPCs. To test this, we used RNAi against the POM121 mRNA (Elbashir et al., 2001). The depletion of POM121 was efficient, but it did not produce any obvious phenotype (Fig. 3). Pore recruitment of the Nup107-160 complex (Belgareh et al., 2001; Vasu et al., 2001) and of mAb414-reactive Nups, including Nup62 and Nup358, remained normal. Assembly of gp210 (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200601002/DC1) and Nup153 (not depicted) into NPCs was not affected either. Nuclei of POM121-depleted cells also efficiently accumulated an IBB–GFP fusion protein (Fig. S1), indicating that their NE was sufficiently intact to prevent an uncontrolled leakage of the previously imported reporter to the cytoplasm. In addition, POM121-depleted cells remained viable and divided at a rate similar to that of cells that were not transfected with the anti-POM121 siRNA duplexes (unpublished data). Finally, light-microscopical analysis of POM121-deficient cells in the various cell cycle stages did not reveal any signs of aberrant progression through mitosis or cytokinesis (Fig. 3 and not depicted).
Figure 3.
POM121 is not limiting for NPC assembly. HeLa cells were transfected with POM121-specific siRNAs and fixed 60 h later. Triple immunofluorescences were performed with combinations of rabbit anti-POM121; mouse mAb414 or anti-Nup88; and guinea pig anti-Nup107, -Nup96, -Nup62, or -Nup358. Bright NE staining for POM121 is visible only in nontransfected cells that are shown for reference. POM121-depleted NPCs still contained wild-type levels of Nup107, Nup88, Nup358, Nup96, and Nup62 and stained normally with mAb414. White arrows indicate cells with <5% residual POM121. Red arrows label a virtually POM121-free pair of cells that just completed cytokinesis.
This unexpected lack of phenotype could be explained formally by a catalytic action of the depleted factor, whereby minute residual amounts suffice for normal function. POM121, however, possesses no known enzymatic activity and associates so stably with NPCs that the half-time for its dissociation from NPCs is longer than a typical cell cycle (Daigle et al., 2001). It is therefore hard to imagine how a POM121 molecule from one NPC could possibly catalyze the assembly of another NPC that is devoid of POM121.
The knockdown could reduce the POM121 signal at NPCs at least 20-fold without affecting the NPC localization of other Nups (Fig. 3). After such depletion, some NPCs still gave a faint POM121 signal, whereas others appeared POM121 negative (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200601002/DC1). Therefore, a substantial proportion of NPCs and the vast majority of asymmetric NPC units assembled independently of POM121 under these conditions, suggesting that POM121 is not essential for the process.
In contrast, it has been proposed that POM121 is essential for assembling an NPC-perforated NE from components of the Xenopus laevis egg extract (Antonin et al., 2005). This reported requirement for POM121 might be specific for very fast embryonic cell cycles during amphibian embryogenesis. For somatic mammalian cells, however, our RNAi data suggest that POM121 is either not limiting or even fully dispensable for the formation of the NE and NPCs. Phylogenetic data also argue against a unique and indispensable role for POM121 in NPC/NE assembly. Neither yeasts, insects, nematodes, nor, indeed, any nonvertebrate eukaryote contains a recognizable, membrane-anchored POM121 orthologue, yet they have functional NPCs and an intact NE.
gp210 is not essential for NPC formation and function in human cells
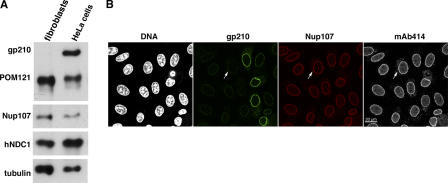
gp210 is far better conserved than POM121. It probably already existed in the earliest eukaryotes, and it might, therefore, be less dispensable for NPC assembly. However, mouse gp210 is absent from many mesenchymal cell types of kidney, teeth, and lung as well as from several epithelial and fibroblast cell lines (Eriksson et al., 2004; Olsson et al., 2004). To determine whether this represents a mouse-specific phenomenon or applies to other mammals as well, we raised antibodies against human gp210, analyzed several human cell lines, and observed that human primary fibroblasts also lack gp210 (Fig. 4 A).
Figure 4.
Assembly of human NPCs in the absence of gp210. (A) Primary neonatal fibroblasts (5 × 104 cells) and HeLa cells (2.5 × 104 cells) were analyzed by immunoblotting with the indicated antibodies. Note that fibroblasts appear gp210 negative. (B) HeLa cells were transfected with a gp210-specific siRNA and fixed 60 h later. DNA was stained with Hoechst 33342. Triple immunofluorescence was with rabbit anti-gp210, guinea pig anti-Nup107, and mouse mAb414. Arrows exemplify a transfected cell with <5% residual gp210. The gp210 depletion did not impair assembly of Nup107 or mAb414-reactive Nups into NPCs.
Having found that NPCs of human fibroblasts naturally operate without gp210, we studied the consequences of gp210 depletion from HeLa cells, a cell type that normally expresses this protein. RNAi against gp210 mRNA could drastically reduce gp210 levels at NPCs. Yet, other Nups assembled normally into NPCs (Fig. 4 B), as judged by immunofluorescence detecting the Nup107-160 subcomplex and the FG Nups.
Mammalian genomes encode a second gp210 paralogue (gp210L) that shares 41% sequence identity with the canonical gp210 (unpublished data). We can rule out the possibility that gp210L compensates for any loss of gp210 function because gp210L is expressed in neither HeLa cells nor fibroblasts (unpublished data). Collectively, this body of data suggests that gp210 paralogues are not essential for the NPC assembly process in mammalian cells. This conclusion is in line with the observations that gp210 appears dispensable for nuclear assembly in the X. laevis egg extract system (Antonin et al., 2005) and that NPCs in gp210-deficient Caenorhabditis elegans cells appear structurally intact at the EM level and stain positive for FG-repeat Nups (Cohen et al., 2003).
NPC assembly appears normal after codepletion of POM121 and gp210
That POM121 and gp210 can be singly depleted from HeLa cells without disturbing NPC assembly might indicate that the two proteins can substitute for each other in the assembly process. In the simplest of such scenarios, POM121 should become essential in the absence of gp210, and vice versa. In light of the diametrically different membrane topology of POM121 and gp210, such a scenario might appear counterintuitive. Nevertheless, the hypothesis had to be tested.
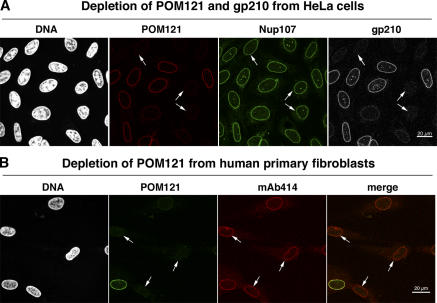
In simultaneous knockdowns, POM121 and gp210 could be depleted from HeLa cells to <5–10% of their original levels (Fig. 5 A). Nonetheless, the assembly of other Nups into NPCs appeared undisturbed and the cells remained viable. Likewise, no defects in NPC assembly became apparent when POM121 was depleted from primary human fibroblasts (Fig. 5 B), which are already devoid of gp210 (Fig. 4 A).
Figure 5.
NPCs appear normal even in cells lacking gp210 and depleted of POM121. (A) HeLa cells were treated simultaneously with POM121- and gp210-specific siRNAs and fixed 60 h later. Triple immunofluorescence was with Alexa 647–labeled anti-POM121, Alexa 488 anti-Nup107, and Alexa 568 anti-gp210. Arrows point to cells almost completely depleted of gp210 and POM121. Comparison with nontransfected cells indicates that this codepletion affected neither NE assembly nor assembly of Nup107 into NPCs. (B) POM121 was depleted by RNAi from human primary fibroblasts that already lack gp210 (Fig. 4 A). Arrows indicate cells whose POM121 staining was reduced to background levels. NE or NPC assembly remained unaffected as judged by their normal mAb414 signal.
An anchorage of NPCs within the NE without a membrane-integral protein is unlikely. The assembly of functional NPCs in cells devoid of gp210 and depleted of POM121 therefore predicts the existence of at least one additional human membrane-integral Nup, which promotes NPC biogenesis also in the absence of POM121 and gp210. Indeed, in the accompanying article, we describe the metazoan orthologue to yeast Ndc1p as a novel, six times membrane–spanning constituent of animal NPCs and demonstrate that it crucially contributes to the NPC assembly process (Stavru et al., 2006).
We also report in the accompanying article that none of the membrane-integral Nups, not even NDC1, are universally required for NPC formation (Stavru et al., 2006). Crucial cellular functions are often backed by multiple players, and this recurring theme apparently also applies to the membrane anchorage of NPCs. This yields the experimental problem that such redundancies easily obscure the function of a given protein. POM121 is a good example of this: even though it is apparently nonessential for NPC formation and maintenance, we could demonstrate that the central POM121 domain can recruit other Nups to an ectopic assembly site and thus appears capable of initiating at least some steps of the NPC assembly process.
Materials and methods
Antibody production
Antibodies were newly raised in rabbits against human POM121448–647 and gp2101828–1887. Antibodies against human Nup50, Nup62, Nup96, Nup98, Nup107, Nup153, Nup205, Nup358, and TPR have been described (Hase and Cordes, 2003). All polyclonal antibodies were affinity purified on the respective antigen columns. mAb414 was obtained from Eurogentec and the mAb against Nup88 from BD Biosciences.
Cell culture
Primary human neonatal fibroblasts (Hs27 cells) were obtained from the European Collection of Cell Cultures and cultivated for no more than 20 passages in Dulbecco's modified Eagle's medium high glucose, supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Human HeLa cells (European Collection of Cell Cultures) were maintained in Dulbecco's modified Eagle's medium low glucose, supplemented with 10% FCS, 1× nonessential amino acids (Sigma-Aldrich), 100 U/ml penicillin, and 100 μg/ml streptomycin.
RNAi
Transfection of cultured human cells with siRNAs was performed essentially as described earlier (Hase and Cordes, 2003). Annealed siRNAs were purchased from Dharmacon Research. Antisense strands were complementary to the following nucleotide positions of the respective open reading frames: POM1211389–1409, POM1213207–3227, and gp2105562–5580.
Knockdown efficiency was quantitated at the single-cell level by immunofluorescence, whereby the staining intensities at the NE were scored by false-color representation (in NIH Image J) and by numerical integration of pixel values. Nontransfected cells served as a reference.
DNA transfections
DNA transfections were performed with Fugene6 (Roche) according to the manufacturer's instructions. Expression of the Mito-EGFP and the Mito-EGFP-POM12173–797 fusion was driven by a doxycycline-regulatable promoter system (pRevTRE2; CLONTECH Laboratories, Inc.) at 10 ng/ml doxycycline for 24–60 h.
Immunofluorescence and microscopy
For immunofluorescence, cultured cells were washed briefly with PBS; fixed for 4 min in 3% paraformaldehyde; freshly dissolved in PBS; washed in PBS followed by PBS + 50 mM NH4Cl (5 min); permeabilized with 0.25% Triton X-100 in PBS; and blocked for at least 30 min in 1% BSA, 10% goat serum, and 0.1% Triton X-100. Primary antibodies were applied for 60 min in blocking buffer. Nonbound antibodies were washed off with PBS. Alexa-labeled secondary antibodies were obtained from Invitrogen and used at 1:250 dilution. The secondary antibodies and the DNA stain Hoechst 33342 were applied for 30–60 min in blocking solution, followed by extensive washing and mounting in Vectashield (Vector Laboratories).
For anti-Nup205 stains, conditions were modified as previously described (Krull et al., 2004). Confocal microscopy was performed with a laser-scanning microscope (SP2; Leica) using the 405-, 488-, 561-, or 633-nm laser lines for excitation. All pictures were taken with PlanApo oil objectives (100× NA 1.4 and 63× NA 1.32; Leica); for scans with the 405-nm laser, λ blue objectives were used. Figures were assembled in Photoshop or Illustrator (Adobe).
Online supplemental material
Fig. S1 shows an intact NE in POM121-depleted cells. Fig. S2 shows distribution of residual POM121 after depletion by RNAi. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200601002/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
We thank Petra Rübmann and Ursula Jäkle for excellent technical help, the members of our laboratory for stimulating discussions, and Bastian Hülsmann for critical reading of the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 638) and the Alfried Krupp Foundation and by fellowships from the Boehringer Ingelheim Fonds (to F. Stavru) and the Carlsberg Foundation (to G. Nautrup-Pedersen).
Abbreviations used in this paper: NE, nuclear envelope; NPC, nuclear pore complex; Nup, nucleoporin.
References
- Antonin, W., C. Franz, U. Haselmann, C. Antony, and I.W. Mattaj. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell. 17:83–92. [DOI] [PubMed] [Google Scholar]
- Belgareh, N., G. Rabut, S.W. Bai, M. van Overbeek, J. Beaudouin, N. Daigle, O.V. Zatsepina, F. Pasteau, V. Labas, M. Fromont-Racine, et al. 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrios, M., V.H. Meller, M. McConnell, and P.A. Fisher. 1995. Drosophila gp210, an invertebrate nuclear pore complex glycoprotein. Eur. J. Cell Biol. 67:1–7. [PubMed] [Google Scholar]
- Cohen, M., N. Feinstein, K.L. Wilson, and Y. Gruenbaum. 2003. Nuclear pore protein gp210 is essential for viability in HeLa cells and _Caenorhabditis elegans._Mol. Biol. Cell. 14:4230–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, S., and T. Allen. 2004. Structure, function and assembly of the nuclear pore complex. Symp. Soc. Exp. Biol. 2004:89–114. [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- Eriksson, C., C. Rustum, and E. Hallberg. 2004. Dynamic properties of nuclear pore complex proteins in gp210 deficient cells. FEBS Lett. 572:261–265. [DOI] [PubMed] [Google Scholar]
- Favreau, C., R. Bastos, J. Cartaud, J.C. Courvalin, and P. Mustonen. 2001. Biochemical characterization of nuclear pore complex protein gp210 oligomers. Eur. J. Biochem. 268:3883–3889. [DOI] [PubMed] [Google Scholar]
- Gerace, L., Y. Ottaviano, and C. Kondor-Koch. 1982. Identification of a major polypeptide of the nuclear pore complex. J. Cell Biol. 95:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., T. Dang, N. Pane, A. Shevchenko, M. Mann, D. Forbes, and E. Hurt. 1997. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell. 8:2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber, U.F., A. Senior, and L. Gerace. 1990. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 9:1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, T., S. Muller, G. Klier, N. Pante, J.M. Blevitt, M. Haner, B. Paschal, U. Aebi, and L. Gerace. 1995. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell. 6:1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, T., R.H. Kehlenbach, E.C. Schirmer, A. Kehlenbach, F. Fan, B.E. Clurman, N. Arnheim, and L. Gerace. 2000. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20:5619–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg, E., R.W. Wozniak, and G. Blobel. 1993. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J. Cell Biol. 122:513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase, M.E., and V.C. Cordes. 2003. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell. 14:1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaji, S., J. Iwahashi, Y. Kida, M. Sakaguchi, and K. Mihara. 2000. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151:277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull, S., J. Thyberg, B. Bjorkroth, H.R. Rackwitz, and V.C. Cordes. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell. 15:4261–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay, U., E. Izaurralde, F.R. Bischoff, I.W. Mattaj, and D. Görlich. 1997. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans, B.J., V. Anantharaman, L. Aravind, and E.V. Koonin. 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 3:1612–1637. [DOI] [PubMed] [Google Scholar]
- Olsson, M., S. Scheele, and P. Ekblom. 2004. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp. Cell Res. 292:359–370. [DOI] [PubMed] [Google Scholar]
- Rabut, G., P. Lenart, and J. Ellenberg. 2004. Dynamics of nuclear pore complex organization through the cell cycle. Curr. Opin. Cell Biol. 16:314–321. [DOI] [PubMed] [Google Scholar]
- Radu, A., M.S. Moore, and G. Blobel. 1995. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 81:215–222. [DOI] [PubMed] [Google Scholar]
- Soderqvist, H., and E. Hallberg. 1994. The large C-terminal region of the integral pore membrane protein, POM121, is facing the nuclear pore complex. Eur. J. Cell Biol. 64:186–191. [PubMed] [Google Scholar]
- Soderqvist, H., G. Imreh, M. Kihlmark, C. Linnman, N. Ringertz, and E. Hallberg. 1997. Intracellular distribution of an integral nuclear pore membrane protein fused to green fluorescent protein–localization of a targeting domain. Eur. J. Biochem. 250:808–813. [DOI] [PubMed] [Google Scholar]
- Stavru, F., B.B. Hülsmann, A. Spang, E. Hartmann, V.C. Cordes, and D. Görlich. 2006. NDC1, a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173:509–519. [DOI] [PMC free article] [PubMed]
- Sukegawa, J., and G. Blobel. 1993. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 72:29–38. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and S.R. Wente. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 4:775–789. [DOI] [PubMed] [Google Scholar]
- Vasu, S., S. Shah, A. Orjalo, M. Park, W.H. Fischer, and D.J. Forbes. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, R., and P.R. Clarke. 2003. Nuclear pores: sowing the seeds of assembly on the chromatin landscape. Curr. Biol. 13:R970–R972. [DOI] [PubMed] [Google Scholar]
- Wozniak, R.W., E. Bartnik, and G. Blobel. 1989. Primary structure analysis of an integral membrane glycoprotein of the nuclear pore. J. Cell Biol. 108:2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., M.J. Matunis, D. Kraemer, G. Blobel, and E. Coutavas. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209–14213. [DOI] [PubMed] [Google Scholar]
- Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature. 376:184–188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]