A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane (original) (raw)
Abstract
The type II inner nuclear membrane protein emerin is a component of the LINC complex that connects the nuclear lamina to the actin cytoskeleton. In emerin-null or -deficient human dermal fibroblasts we find that the centrosome is detached from the nucleus. Moreover, following siRNA knockdown of emerin in wild-type fibroblasts, the centrosome also becomes detached from the nucleus. We show that emerin interacts with tubulin, and that nocadozole-treated wild-type cells phenocopy the detached centrosome characteristic of emerin-null/deficient cells. We also find that a significant fraction of emerin is located at the outer nuclear membrane and peripheral ER, where it interacts directly with the centrosome. Our data provide the first evidence in mammalian cells as to the nature of the linkage of the centrosome, and therefore the tubulin cytoskeleton, with the outer nuclear membrane.
Introduction
Emerin is a type II integral membrane protein residing principally at the inner nuclear membrane (INM) (Manilal et al., 1998), where it interacts with a number of other proteins such as lamin A/C (Vaughan et al., 2001) barrier-to-autointegration factor (Lee et al., 2001) and β-catenin (Markiewicz et al., 2006). Emerin has also been shown to interact with proteins that are principally found at the outer nuclear membrane (ONM), namely nesprin 1α (Mislow et al., 2002) and nesprin 2 (Zhang et al., 2005). Emerin was identified by positional candidate cloning as the gene responsible for the X-linked form of Emery Dreifuss muscular dystrophy (EDMD) (Bione et al., 1994). The autosomal-dominant form of the disease is caused by mutations in the gene LMNA, which encodes lamins A and C (Bonne et al., 1999). Two hypotheses have been formulated to explain why ubiquitously expressed proteins such as emerin, lamin A, and lamin C should cause such highly tissue-specific diseases. These are referred to as the “structural” and the “gene expression” hypotheses. The gene expression hypothesis proposes that emerin and lamins are involved in tissue-specific gene expression and disease may arise from the downstream effects of mutations on chromatin structure or gene expression (Cohen et al., 2001). The structural hypothesis proposes that emerin and lamins contribute to the structural integrity of the cell by acting as a load-bearing center underneath the nuclear envelope (NE) (Hutchison, 2002). In this hypothesis, absence of emerin or lamins in disease would render contractile cells like skeletal and cardiac muscle vulnerable to damage, leading to cell death and tissue damage (Hutchison et al., 2001).
Supporting the structural hypothesis, there is accumulating evidence that the NE is closely linked and connected to its surrounding cytoskeleton. Work on two protein families, the nesprins and Sun proteins, reveals the existence of “bridging” complexes, referred to as the LINC complexes, which span the NE, thus connecting, the INM with the actin cytoskeleton (Crisp et al., 2006). A recent study showed that disorganization of the actin, vimentin, and tubulin cytoskeletons arose as a consequence of the absence of lamins A and C in mouse embryonic fibroblasts (Broers et al., 2004), directly supporting the idea that the NE is a load-bearing center in animal cells. Here, we investigate how absence of emerin in human fibroblasts affects cytoskeleton organization. We show that emerin interacts with β-tubulin to anchor the centrosome at the ONM. This unexpected finding provides further support for the structural hypothesis and provides the first clue as to how the tubulin cytoskeleton is connected to the NE.
Results and discussion
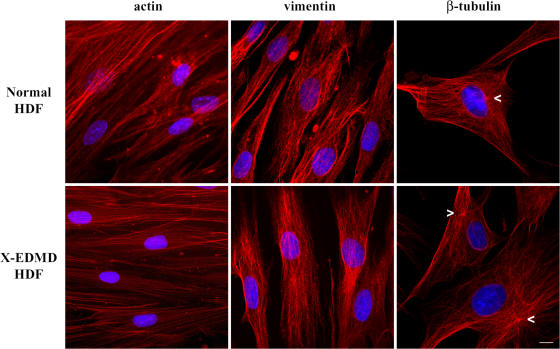
We wished to investigate whether cytoskeletal abnormalities are induced by the absence of emerin in cells. To this end, human dermal fibroblasts (HDF) from healthy individuals and from X-EDMD patients (which were null for emerin) were investigated for possible abnormalities in the actin, vimentin, and tubulin cytoskeleton (Fig. 1). In stark contrast to findings in lamin A/C-null mouse embryonic fibroblasts, there were little or no differences in the organization of any of the cytoskeletal elements in emerin-null HDFs. Surprisingly, however, we did observe that the centrosome was not positioned next to the nucleus in emerin-null HDFs (Fig. 1, arrowheads).
Figure 1.
Organization of the cytoskeleton in X-EDMD cells. Normal and emerin-null HDFs were fixed with methanol/acetone (1:1) and stained for β-actin, vimentin, and β-tubulin. No differences in the organization of the cytoskeleton between normal and emerin-null cells were observed, except for the position of the centrosome (arrowheads) relative to the NE. While in normal cells the centrosome is positioned next to the NE, in X-EDMD cells it localizes some distance from the NE. Chromatin was stained with DAPI. Bar, 10 μm.
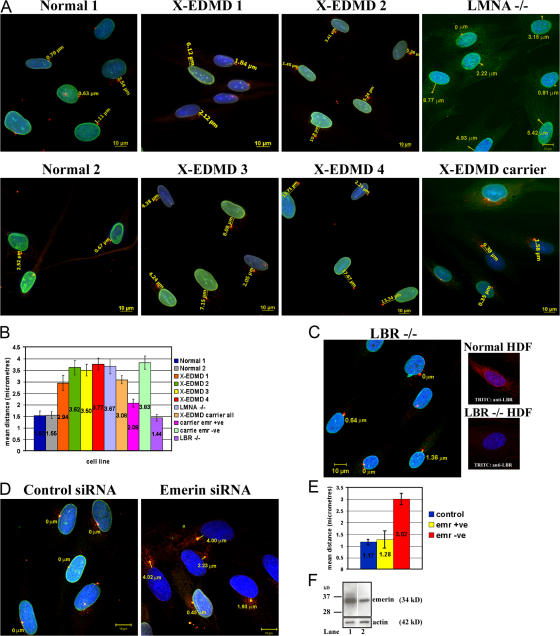
To confirm this finding, we used an antibody against pericentrin to specifically investigate the position of the centrosome. In normal HDF the centrosome was positioned next to or within 1.5 μm of the NE. In contrast, in four independent emerin-null HDF cell lines the centrosome was >3.0 μm distant from the NE (Fig. 2 A). To further investigate this phenomenon we looked at centrosome position in a cell line from an X-EDMD carrier, which has approximately equal numbers of emerin-positive and emerin-null cells. We found that in emerin-positive cells the centrosome was positioned next to the NE, whereas in emerin-negative cells the centrosome was >3.0 μm away from the NE. Finally, we investigated the centrosome position in a lamin A/C–null HDF line in which emerin was located entirely within the endoplasmic reticulum (Muchir et al., 2000). In the lamin A/C–null cell line the centrosome was also >3.0 μm away from the NE, indicating that absence of emerin from the NE was the cause of the centrosome mislocalization (Fig. 2 B). To confirm that mislocalization of the centrosome was specific to absence of emerin from the NE, we investigated centrosome positions in a fibroblast cell line from a patient with Greenberg dysplasia, which were null for the INM protein lamin B receptor (LBR) (Waterham et al., 2003). Like EDMD, Greenberg dysplasia and the related disorder Pelger-Huey anomaly are characterized by nuclear morphological defects (Hoffman et al., 2002). However, in LBR-null fibroblasts, emerin was located at the NE and similarly the centrosome was positioned adjacent to the NE (Fig. 2, B and C), suggesting that centrosome mislocalization is specific to loss of emerin from the NE. To verify these results, we performed knockdown of emerin by siRNA in normal HDFs (Fig. 2, D–F). In HDFs transfected with the scrambled siRNA, centrosomes were found adjacent to the NE. In contrast, in HDFs that were transfected with siRNA specific for emerin, the centrosome was located at a distance of >3.0 μm away from the NE, similar to the distances observed in X-EDMD cells.
Figure 2.
Distance of the centrosome from the nucleus in normal and EDMD cells. (A) The position of the centrosome relative to nucleus was determined in nine cell lines: two normal, four emerin null, one X-EDMD carrier, one AD-EDMD cell line (LMNA−/−), and (C) one LBR−/− cell line. Centrosomes were visualized in methanol/acetone-fixed cells with a rabbit polyclonal pericentrin antibody (TRITC) and NEs were stained with mAb Lamin A/C (FITC), except LMNA−/− X-EDMD carrier and LBR−/− cells, which were stained with mAb emerin (FITC) or anti-LBR (TRITC). Chromatin was stained with DAPI. The distance from the center of each centrosome to the NE was measured and images representative of each cell line using AxioVision (Carl Zeiss MicroImaging, Inc.) software are shown. Bars, 10 μm. (B) Nuclear–centrosome distances were measured in 200 cells for each cell line in triplicate experiments. The mean distances (μm) ± SEM are displayed in a bar chart. (D) HDFs were treated with siRNA against emerin or as a control scrambled siRNA. 48 h later, fixed cells were stained for pericentrin (TRITC) and emerin (FITC). After specific knockdown, 70% of HDFs were negative for emerin. Nuclear–centrosome distances were measured in cells using AxioVision (Carl Zeiss MicroImaging, Inc.) software as described above. For statistical analysis (E) the mean distance (μm) ± SEM was determined in samples of 200 cells in triplicate experiments for control, siRNA emerin +ve, or siRNA emerin −ve HDFs. (F) Western blotting was performed on siRNA-treated HDFs to compare the relative amounts of emerin in control (lane 1) and knock-down cultures (lane 2).
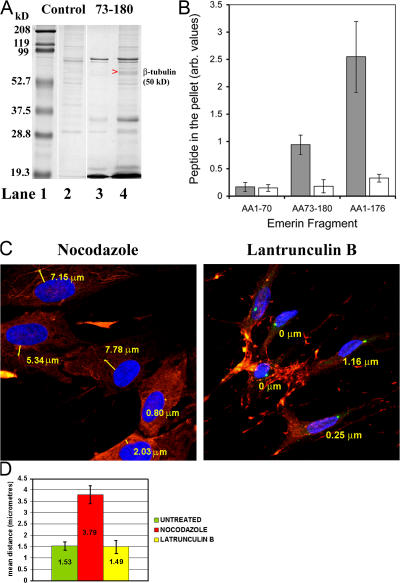
This very intriguing result raised the important question of how a protein that is localized in the INM could affect the position of the centrosome, an organelle that is localized at the ONM. To investigate whether a yet-unidentified emerin binding partner could help explain the observed phenomenon, we used recombinant emerin peptides in coprecipitation experiments to identify new emerin binding partners. The peptide was used as a bait to precipitate interacting partners from the Xenopus egg extracts, which were in turn chosen because they store very large quantities of cytoskeleton and centriolar proteins in a soluble form (Fig. 3 A). Bands that coprecipitated with emerin were cut from the gel and identified by mass spectrometry. Interestingly, β-tubulin was identified as the most consistent emerin binding protein in this assay. To confirm that emerin is a microtubule (MT) binding protein, MT cosedimentation experiments were performed in which purified MTs were polymerized by taxol and incubated with the same emerin peptide (aa 73–180) or two different emerin peptides corresponding to its chromatin binding domain (aa 1–70) or most of the nucleoplasmic domain (aa 1–176) (Fig. 3 B). Emerin 73–180 and 1–176 efficiently cosedimented with MTs, whereas emerin 1–70 did not bind to MTs. To estimate the stoichiometry of emerin/microtubule interactions we calculated the tubulin/emerin binding ratios. Emerin 1–176 bound to tubulin at an approximate ratio of 1:8, which is close to the binding ratios of known microtubule-associated proteins (MAPs) (e.g., Enconsin; Bulinski and Bossler, 1994). Emerin 73–180 bound to tubulin at an approximate ratio of 1:24, and this weaker interaction is likely due to misfolding of this peptide. Collectively, these data suggest that emerin is a novel MT-interacting protein.
Figure 3.
Emerin–β-tubulin interaction and its implications for centrosome position. (A) A His-tagged recombinant emerin peptide corresponding to aa 73–180 was immobilized on Ni2+-beads and used to pull down binding partners from cytosolic fractions of Xenopus egg extracts. Proteins that coeluted with emerin after treatment of the beads with Ni2+ were resolved by SDS-PAGE (lane 4). As a control, Xenopus cytosol was incubated with the beads in absence of emerin peptides (lane 2). The purified emerin peptide on its own is shown in lane 3. Molecular weight markers are shown in lane 1. Arrowhead indicates the band that was selected as a potential emerin binding partner and was identified by Maldi-TOF mass spectroscopy as β-tubulin (Mascot score = 166). (B) MTs were polymerized by taxol and incubated with emerin peptides 1–70, 73–180, or 1–176. The relative proportion of emerin collected in pellet fractions after centrifugation at 200,000 g, in the presence (gray bars) or absence (white bars) of MTs, was determined by densitometry performed on Coomassie-stained SDS-PAGE and is presented as the mean ± SEM of three independent experiments in a bar chart. (C) Normal HDFs were treated either with nocodazole or with latrunculin B to depolymerize MTs and actin, respectively. Cells were then fixed and stained with anti-pericentrin (FITC) and anti–β-tubulin (Cy3) or anti-actin (TRITC) antibodies. Chromatin was visualized with DAPI. Nuclear–centrosome distances were measured in 200 cells in triplicate experiments for each cell line. Results (D) are expressed as mean distances (μm) ± SEM in bar charts.
The interaction of emerin with β-tubulin led us to investigate whether MTs are involved in the attachment of the centrosome to the NE. To investigate this possibility, normal and X-EDMD fibroblasts were treated with nocodazole and its effects on MT organization and centrosome position were investigated (Fig. 3, C and D). As expected, nocodazole treatment led to the depolymerization of the MT network. Interestingly, when normal HDFs were treated with nocodazole the centrosome was observed to be located >3.0 μm away from the NE, just as was observed in emerin-null fibroblasts. As a control, normal HDFs were treated with latrunculin B to depolymerize the actin cytoskeleton. In latrunculin B–treated HDFs the centrosome was located adjacent to the NE, implying that only disruption of the tubulin cytoskeleton leads to an emerin-null phenocopy. We confirmed this finding using biochemical fractionation to determine whether centrosomes cosedimented with the nucleus in a range of HDF lines (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200702026/DC1). In normal HDFs, nuclei and centrosomes cosedimented at a 1:1 ratio. In emerin-null HDFs or normal HDFs treated with nocodazole, nuclei and centrosomes cosedimented at a 1:0.4 ratio, again showing that centrosomes were detached from the NE.
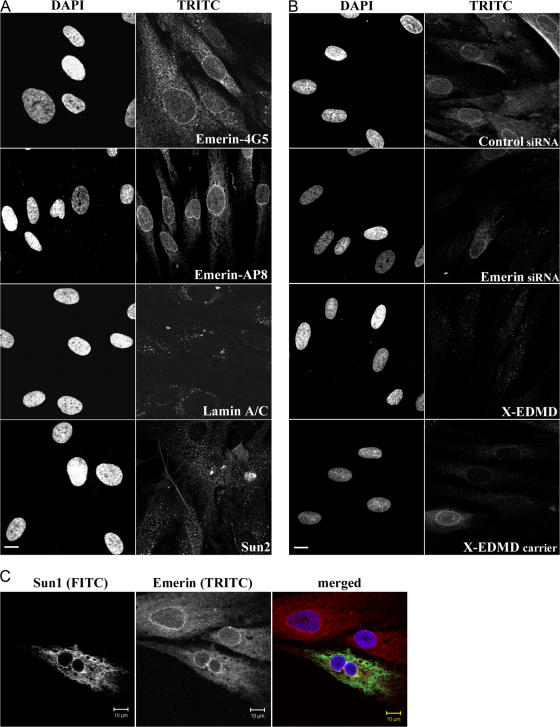
All the above experiments provide strong evidence that emerin links centrosomes to the NE via a MT association. Given that emerin is a protein of the INM, this raised the question as to whether emerin acts via another protein that crosses the NE. We therefore investigated whether either SUN domain proteins or one of the nesprins is also mislocalized in emerin- null HDFs as a first step to determining whether these proteins might be involved in centrosome localization. We could not detect any change in the distribution of SUN1 or nesprin 1 (not depicted) or SUN2 or nesprin 2 (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200702026/DC1) in these cells, suggesting that these proteins were not involved in centrosome localization. As a result of this finding we decided to reinvestigate the localization of emerin in normal fibroblasts. Using digitonin permeabilization, we showed that a considerable fraction of emerin was concentrated at the ONM, with a further dispersed fraction in the peripheral ER, with two independent anti-emerin antibodies, whereas lamins A/C and SUN2 were undetectable under similar conditions and therefore located exclusively at the INM (Fig. 4 A). The anti-Sun2 antibody used in this assay recognizes a luminal domain, indicating that not only is the INM intact, but also the ONM, strengthening the finding that a fraction of emerin resides at the ONM. To confirm that the protein detected at the ONM was indeed emerin, we performed siRNA knockdown of emerin on control fibroblasts and again stained the cells with anti-emerin antibodies after digitonin permeabilization. In these experiments knockdown was ∼70% efficient, and whereas emerin was detected at the ONM in fibroblasts treated with scrambled siRNA, staining was eliminated in cells transfected with siRNA specific to emerin (Fig. 4 B). As a further control we also stained X-EDMD fibroblasts, (which are null for emerin) or an X-EDMD carrier in which emerin is absent from ∼50% of cells. We found that staining of the ONM was undetectable in emerin-null fibroblasts, further supporting the presence of emerin at the ONM. This finding implies that emerin residing at the ONM can interact directly with centrioles via MTs. It has recently been reported that localization of emerin at the INM is dependent upon the presence of both nesprin1α and 2β (Wheeler et al., 2007). To investigate whether nesprins might also be involved in localization of emerin to the ONM, we transfected normal HDF with a dominant-negative Sun1 mutant that causes loss of the ONM form of nesprin 2 (Crisp et al., 2006). We found that emerin was still detected at the ONM in the presence of this mutant, suggesting that nesprin2 does not cause the localization of emerin to the ONM (Fig. 4 C).
Figure 4.
Digitonin permeabilization of normal and X-EDMD HDFs. (A) Normal HDFs were permeabilized with digitonin and stained for emerin with two antibodies: a mouse monoclonal (4G5) and a rabbit polyclonal (AP8) antibody. Cells were also stained for lamin A/C and for Sun 2. Digitonin treatment selectively permeabilizes the plasma membrane, leaving the NE intact, therefore rim staining with emerin represents staining of the ONM. Bar, 10 μm. (B) Normal HDFs that were used to knock down emerin by siRNA were stained for emerin (mAb 4G5) after digitonin permeabilization. As a control, cells treated with scrambled siRNA are shown. Also X-EDMD cells, which are emerin null and cells from an X-EDMD carrier, which are a mixture of emerin +ve and −ve cells, were also permeabilized with digitonin and stained with emerin mAb 4G5. Bar, 10 μm. (C) Normal HDFs were transfected with a Sun1 construct that encodes the soluble Sun1 lumenal domain and displaces Nesprin 2 from the ONM, were permeabilized with digitonin, and were stained for Sun1 (FITC) and emerin (TRITC). Emerin localization at the ONM does not seem to depend on Nesprin 2.
Overall, our results show that emerin interacts directly with MTs and that emerin and MTs both are necessary for the association of the centrosome with the NE. Our findings are supported by previous work, which showed a colocalization of emerin with β-tubulin in mitotic cells (Dabauvalle et al., 1999) and an enrichment of emerin at the kinetochores, near the spindle poles, during NE reassembly (Haraguchi et al., 2000). Recent evidence has demonstrated how complexes involving lamins A/C, the SUN domain proteins, and the nesprins link the actin and intermediate filament cytoskeletons to the NE in mammalian cells (Wilhelmsen et al., 2005; Crisp et al., 2006). Our current data now reveal how the tubulin cytoskeleton via the centriole interacts with the NE in human fibroblasts.
Materials and methods
Cell culture
HDF cells and LBR-null cells (provided by K. Hoffmann, Charité Humboldt University, Berlin, Germany) were grown in DME supplemented with 10 U/ml penicillin, 50 μg/ml streptomycin, and 10% vol/vol NCS, and maintained at 37°C in a humidified atmosphere containing 5% CO2 until 80% confluence. Serial passage was performed in the presence of trypsin and 0.5% EDTA.
Immunofluorescence and confocal microscopy
HDFs were fixed with methanol/acetone (1:1) on ice for 5 min or with 4% paraformaldehyde for 12 min at RT followed by permeabilization either with 1% Triton X-100 (for 5 min at RT) or with Digitonin (40 μg/ml for 2 min, on ice). Primary antibodies used and their dilutions are described in Table I. Anti-Sun2 was provided by B. Burke (University of Florida, Gainesville, FL) and anti-Nesprin 2 K1 antibody was provided by I. Karakesisoglou (University of Cologne, Cologne, Germany). FITC- or TRITC-conjugated secondary antibodies were obtained from Stratech and chromatin was visualized with DAPI/Mowiol.
Table I.
Antibodies used in this study
| Antibody | Antigen | Host | IF dilution | IB dilution | Source/Reference |
|---|---|---|---|---|---|
| Emerin clone 4G5 | Emerin | Mouse | 1:30 | 1:250 | Vector Inc. |
| Emerin AP8 | Emerin | Rabbit | 1:250 | – | ImmuQuest |
| Anti-tubulin | β-tubulin | Mouse | 1:100 | – | Sigma-Aldrich |
| Pericentrin | Pericentrin | Rabbit | 1:400 | – | Abcam |
| JOL2 | Lamin A/C | Mouse | 1:30 | – | Dyer et al., 1997 |
| AC-40 | β-actin | Mouse | 1:300 | 1:2,000 | Sigma-Aldrich |
| Vimentin clone v9 | Vimentin | Mouse | 1:100 | – | Sigma-Aldrich |
| Anti-Sun 2 | Sun 2 | Mouse | 1:100 | – | Crisp et al., 2006 |
| Anti-Nesprin 2 K1 | Nesprin 2 | Rabbit | 1:50 | – | Libotte et al., 2005 |
| Anti-LBR | Lamin B receptor | Rabbit | 1:250 | – | Abcam |
| Anti-HA | Hemagglutinin | Mouse | 1:50 | – | Abcam |
For imaging, a confocal microscope (LSM510 META; Carl Zeiss MicroImaging, Inc.) with LSM510 image browser software (Carl Zeiss MicroImaging, Inc.) was used at ambient temperature, equipped with 40×/1.3 and 63×/1.4 oil-immersion lenses and a non-imaging photodetection device (photomultiplier tube; Carl Zeiss MicroImaging, Inc.). A dynamic range adjustment was used to optimize the signal for the fluorophores, and images were collected in multitrack mode. Any brightness and contrast adjustments were performed in Adobe Photoshop.
Gel electrophoresis and immunoblotting
One-dimensional SDS-PAGE was performed according to Laemmli (1970). For immunoblotting, proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell) using the Mini Trans-Blot system (Bio-Rad Laboratories). HRP-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories. Bands were visualized by enhanced chemiluminescence (ECL reagents; GE Healthcare).
siRNA
Emerin-specific siRNA duplexes were obtained from Ambion. The sequence of sense nucleotides was as follows: 5′-GGUGGAUGAUGACGAUCUUtt-3′. RNAi transfection procedure was performed as in Harborth et al. (2001). Specific silencing of emerin was confirmed by three independent experiments.
Protein purification and coprecipitation experiments
Histidine-tagged emerin peptides were expressed in bacteria, extracted as soluble peptides, and purified on Ni-NTA–Agarose beads (QIAGEN) by immobilized metal affinity chromatography according to the QIAGEN protocol.
For the pull-down experiments, purified emerin peptide 73–180 was immobilized on Ni-beads and incubated with the cytosolic fraction of Xenopus egg extracts for 4 h at 4°C, on a roller. Non-specific binding was removed with 250-mM NaCl washes. Emerin together with co-eluting proteins were resolved by SDS-PAGE. Xenopus egg extracts were prepared and fractionated to generate membrane-free cytosol as described by Drummond et al. (1999).
MT-binding assays
Tubulin was purified from bovine brain according to the method of Shelanski et al. (1973). Microtubules were polymerized in the presence of taxol as described previously (Smertenko et al., 2004). 10 μg of purified emerin peptides 1–70 and 73–180 were mixed with 20 μg of taxol-stabilized MTs and centrifuged at 200,000 g. No MTs were added to control samples. Samples were analyzed by SDS-PAGE. The amount of the emerin fragments was quantified in the gel using NIH Image software and normalized by the mean. To measure the relative binding ratio of tubulin to emerin in the microtubules, microtubule cosedimentation assays were performed as described above, then the samples were separated by SDS-PAGE and Coomassie brilliant blue R-250 was used to stain the gels. The gels were quantified using NIH Image software. The average density values were multiplied by the measured area values to get the absolute intensity, and then the background values were subtracted from the protein band values. The tubulin/emering binding ratio was calculated using the following equation assuming that tubulin is present as heterodimer and emerin fragment as monomer: R = (Itubulin × _M_remerin)/(2 × Iemerin × _M_rtubulin), where I is absolute intensity for the corresponding protein band and M r is molecular mass (50 kD for tubulin, 22 kD for aa 1–176 fragment, and 15 kD for aa 73–180 fragment).
Transfection of normal HDF with the Sun1 construct
Normal HDFs were transfected with 2 μg of plasmid SS-HA-Sun1L-KDEL (Crisp et al., 2006) with the Amaxa Biosystems Nucleofection system using the Basic Nucleofector kit for primary mammalian fibroblasts (VPI-1002) as described by the manufacturer. Cells were fixed 24 h after transfection with 4% paraformaldehyde and underwent sequential permeabilization. Cells were initially permeabilized with digitonin and stained with the rabbit-emerin AP8 antibody. Cells were then fixed again with 4% paraformaldehyde incubated with 1% Triton X-100 and stained with a mouse-HA tag antibody to detect the Sun1 construct.
Nuclear isolation experiments
Cells were grown to 90% confluence and collected by centrifugation. Cell pellets were resuspended in Nuclear Isolation Buffer (NIB) (250 mM NaCl, 3 mM MgCl, 10 mM Tris-HCL, pH 7.6, 0.5% Nonidet, and protease inhibitor cocktail; 1 ml NIB/106 cells) and incubated on ice for 15 min. Nuclei were released with a Wheaton homogenizer and isolated on coverslips by centrifugation (4,000 rpm for 10 min at 4°C) through a 30% sucrose cushion.
Online supplemental material
Fig. S1 shows nuclear isolation experiments in normal and X-EDMD cells with increased salt concentration. Fig. S2 shows Sun 2 and Nesprin 2 staining in normal and X-EDMD HDFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200702026/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
We are grateful to Brian Burke for the supply of anti-SUN antibodies and of the SS-HA-Sun1L-KDEL construct, to Dr. Akis Karakesisoglou for the supply of nesprin antibodies, and to Dr. Katrina Hoffman for the supply of LBR-null fibroblasts.
This work was supported by the Muscular Dystrophy Campaign of Great Britain and European Union FP6.
Abbreviations used in this paper: EDMD, Emery Dreifuss muscular dystrophy; HDF, human dermal fibroblast; INM, inner nuclear membrane; LBR, lamin B receptor; MT, microtubule; NE, nuclear envelope; ONM, outer nuclear membrane.
References
- Bione, S., E. Maestrini, S. Rivella, M. Mancini, S. Regis, G. Romeo, and D. Toniolo. 1994. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 8:323–327. [DOI] [PubMed] [Google Scholar]
- Bonne, G., M.R. Di Barletta, S. Varnous, H.M. Becane, E.H. Hammouda, L. Merlini, F. Muntoni, C.R. Greenberg, F. Gary, J.A. Urtizberea, et al. 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21:285–288. [DOI] [PubMed] [Google Scholar]
- Broers, J.L., E.A. Peeters, H.J. Kuijpers, J. Endert, C.V. Bouten, C.W. Oomens, F.P. Baaijens, and F.C. Ramaekers. 2004. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 13:2567–2580. [DOI] [PubMed] [Google Scholar]
- Bulinski. J.C., and A. Bossler. 1994. Purification and characterization of ensconsin, a novel microtubule stabilizing protein. J. Cell Sci. 107:2839–2849. [DOI] [PubMed] [Google Scholar]
- Cohen, M., K.K. Lee, K.L. Wilson, and Y. Gruenbaum. 2001. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem. Sci. 26:41–47. [DOI] [PubMed] [Google Scholar]
- Crisp, M., Q. Liu, K. Roux, J.B. Rattner, C. Shanahan, B. Burke, P.D. Stahl, and D. Hodzic. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabauvalle, M.C., E. Muller, A. Ewald, W. Kress, G. Krohne, and C.R. Muller. 1999. Distribution of emerin during the cell cycle. Eur. J. Cell Biol. 78:749–756. [DOI] [PubMed] [Google Scholar]
- Drummond, S., P. Ferrigno, C. Lyon, J. Murphy, M. Goldberg, T. Allen, C. Smythe, and C.J. Hutchison. 1999. Temporal differences in the appearance of NEP-B78 and an LBR-like protein during Xenopus nuclear envelope reassembly reflect the ordered recruitment of functionally discrete vesicle types. J. Cell Biol. 144:225–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, J.A., I.R. Kill, G. Pugh, R.A. Quinlan, E.B. Lane, and C.J. Hutchison. 1997. Cell cycle changes in A-type lamin association detected in human dermal fibroblasts using monoclonal antibodies. Chromosome Res. 5:383–394. [DOI] [PubMed] [Google Scholar]
- Haraguchi, T., T. Koujin, T. Hayakawa, T. Kaneda, C. Tsutsumi, N. Imamoto, C. Akazawa, J. Sukegawa, Y. Yoneda, and Y. Hiraoka. 2000. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113:779–794. [DOI] [PubMed] [Google Scholar]
- Harborth, J., S.M. Elbashir, K. Bechert, T. Tuschl, and K. Weber. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114:4557–4565. [DOI] [PubMed] [Google Scholar]
- Hoffmann, K., C.K. Dreger, A.L. Olins, D.E. Olins, L.D. Shultz, B. Lucke, H. Karl, R. Kaps, D. Muller, A. Vaya, et al. 2002. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly). Nat Genet. 31:410–414. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.J. 2002. Lamins: building blocks or regulators of gene expression? Nat. Rev. Mol. Cell Biol. 3:848–858. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.J., M. Alvarez-Reyes, and O.A. Vaughan. 2001. Lamins in disease: why do ubiquitously expressed nuclear envelope proteins give rise to tissue-specific disease phenotypes? J. Cell Sci. 114:9–19. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lee, K.K., T. Haraguchi, R.S. Lee, T. Koujin, Y. Hiraoka, and K.L. Wilson. 2001. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114:4567–4573. [DOI] [PubMed] [Google Scholar]
- Libotte, T., H. Zaim, S. Abraham, V.C. Padmakumar, M. Schneider, W. Lu, M. Munck, C.J. Hutchison, M. Wehnert, B. Fahrenkrog, et al. 2005. Lamin A/C–dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol. Biol. Cell. 16:3411–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manilal, S., T. Nguyen, and G.E. Morris. 1998. Colocalization of emerin and lamins in interphase nuclei and changes during mitosis. Biochem. Biophys. Res. Commun. 249:643–647. [DOI] [PubMed] [Google Scholar]
- Markiewicz, E., K. Tilgner, N. Barker, M. van de Wetering, H. Clevers, M. Dorobek, I. Hausmanowa-Petrusewicz, F.C. Ramaekers, J.L. Broers, W.M. Blankesteijn, et al. 2006. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 25:3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mislow, J.M., J.M. Holaska, M.S. Kim, K.K. Lee, M. Segura-Totten, K.L. Wilson, and E.M. McNally. 2002. Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 525:135–140. [DOI] [PubMed] [Google Scholar]
- Muchir, A., G. Bonne, A.J. van der Kooi, M. van Meegen, F. Baas, P.A. Bolhuis, M. de Visser, and K. Schwarz. 2000. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet. 9:1453–1459. [DOI] [PubMed] [Google Scholar]
- Shelanski, M.L., F. Gaskin, and C.R. Cantor. 1973. Microtubule assembly in absence of added nucleotides. Proc. Natl. Acad. Sci. USA. 70:765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko, A.P., H.Y. Chang, V. Wagner, D. Kaloriti, S. Fenyk, S. Sonobe, C. Lloyd, M.T. Hauser, and P.J. Hussey. 2004. The Arabidopsis microtubule-associated protein AtMAP65-1: Molecular analysis of its microtubule bundling activity. Plant Cell. 16:2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, A., M. Alvarez-Reyes, J.M. Bridger, J.L. Broers, F.C. Ramaekers, M. Wehnert, G.E. Morris, W.G.F. Whitfield, and C.J. Hutchison. 2001. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J. Cell Sci. 114:2577–2590. [DOI] [PubMed] [Google Scholar]
- Waterham, H.R., J. Koster, P. Mooyer, G.Gv. Noort, R.I. Kelley, W.R. Wilcox, R.J. Wanders, R.C. Hennekam, and J.C. Oosterwijk. 2003. Autosomal recessive HEM/Greenberg skeletal dysplasia is caused by 3 beta-hydroxysterol delta 14-reductase deficiency due to mutations in the lamin B receptor gene. Am. J. Hum. Genet. 72:1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, M.A., J.D. Davies, Q. Zhang, L.J. Emerson, J. Hunt, C.M. Shanahan, and J.A. Ellis. 2007. Distinct functional domains in nesprin-1alpha and nesprin-2beta bind directly to emerin and both interactions are disrupted in X-linked Emery-Dreifuss muscular dystrophy. Exp. Cell Res. 313:2845–2857. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen, K., S.H. Litjens, I. Kuikman, N. Tshimbalanga, H. Janssen, I. van den Bout, K. Raymond, and A. Sonnenberg. 2005. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171:799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., C.D. Ragnauth, J.N. Skepper, N.F. Worth, D.T. Warren, R.G. Roberts, P.L. Weissberg, J.A. Ellis, and C.M. Shanahan. 2005. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 118:673–687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]