A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination (original) (raw)
. Author manuscript; available in PMC: 2008 Jul 1.
Summary
In Caenorhabditis elegans, the Gli-family transcription factor TRA-1 is the terminal effector of the sex determination pathway. TRA-1 activity inhibits male development and allows female fates. Genetic studies have indicated that TRA-1 is negatively regulated by the fem-1, fem-2, and fem-3 genes. However, the mechanism of this regulation has not been understood. Here, we present data that TRA-1 is regulated by degradation mediated by a CUL-2-based ubiquitin ligase complex that contains FEM-1 as the substrate-recognition subunit, and FEM-2 and FEM-3 as cofactors. CUL-2 physically associates with both FEM-1 and TRA-1 in vivo, and cul-2 mutant males share feminization phenotypes with fem mutants. CUL-2 and the FEM proteins negatively regulate TRA-1 protein levels in C. elegans. When expressed in human cells, the FEM proteins interact with human CUL2 and induce the proteasome-dependent degradation of TRA-1. This work demonstrates that the terminal step in C. elegans sex determination is controlled by ubiquitin-mediated proteolysis.
Introduction
The nematode C. elegans has two sexes: male and hermaphrodite. The hermaphrodite has a female somatic body plan but produces both sperm and oocytes. The sexes in C. elegans are highly differentiated, displaying sexual specialization in 40% of male cells and 30% of hermaphrodite cells (Sulston and Horvitz, 1977). Most tissues and organs differ between males and hermaphrodites, and therefore the sex determination pathway has an integral role in nematode development.
The primary signal for determining sex is the ratio of X chromosomes to autosomes (X:A) (for review see Zarkower, 2006). The X:A ratio determines the expression of the initial pathway component XOL-1 in XO males but not in XX hermaphrodites. XOL-1 expression leads through a series of negative genetic interactions to the final global sex determination regulator TRA-1: active TRA-1 promotes hermaphrodite development and inhibits male development (Fig. 3A). TRA-1 is the only C. elegans member of the Gli family of transcriptional regulators, which includes Drosophila Cubitus interruptus (Ci) and the vertebrate Gli1, Gli2, and Gli3 transcription factors.
Fig. 3. CUL-2 negatively regulates TRA-1 levels in the soma and germ line.
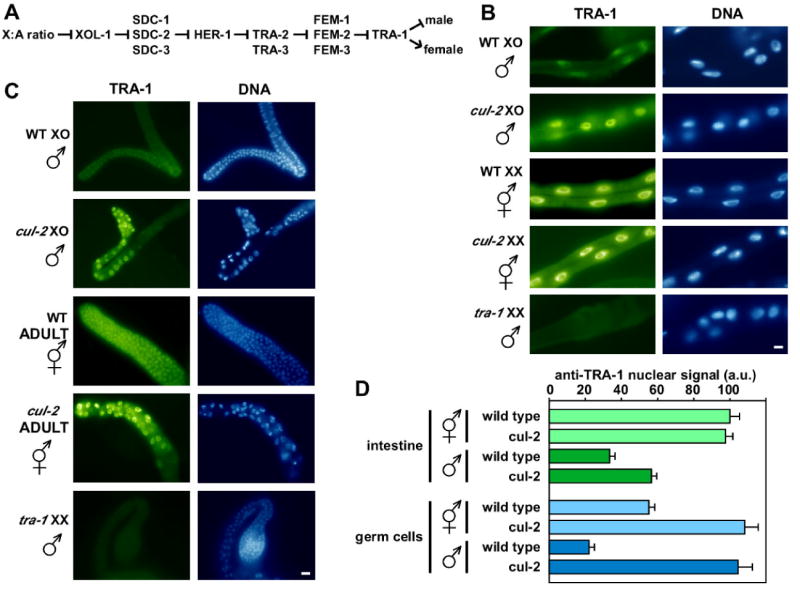
(A) Simplified genetic pathway for C. elegans somatic sex-determination (Zarkower, 2006). Arrows indicate positive interactions and bars indicate negative interactions.
(B-C) Anti-TRA-1 immunofluorescence staining of dissected adult intestine (B) and gonads (C) from the genotypes and sexes indicated. Note that germ cells in cul-2 mutants undergo a G1 phase arrest and are therefore larger than wild-type germ cells (Feng et al., 1999). Scale bars, 10 μm.
(D) Graph of anti-TRA-1 signal derived from confocal microscopy of intestine (top) and distal germ cells (bottom). anti-TRA-1 signals (arbitrary units ± s.e.m.) for all genotypes/sexes were normalized to 100 a.u. for wild-type intestine nuclei.
tra-1 expresses two differentially-spliced transcripts: tra-1A, which encodes a protein of 1109 amino acids that contains five C2H2 zinc fingers and binds DNA; and tra-1B, which encodes a protein of 287 amino acids that contains only the first two zinc fingers and does not bind DNA (Zarkower and Hodgkin, 1992; Zarkower and Hodgkin, 1993). To date, TRA-1A has been shown to regulate the transcription of three genes (egl-1, mab-3, and fog-3) to effect aspects of sex determination (Chen and Ellis, 2000; Conradt and Horvitz, 1999; Yi et al., 2000). In all three cases, TRA-1A acts as a transcriptional repressor in hermaphrodites, suggesting that the repression of male-specific genes is the primary means by which TRA-1A mediates sex determination. TRA-1B has not been linked to sex determination.
Immediately upstream of TRA-1 in the genetic sex determination pathway are the three fem genes, which promote the male fate by inhibiting TRA-1. The primary structure of the FEM proteins does not provide direct insights into how they regulate TRA-1. FEM-1 contains Ankyrin repeats that may participate in protein-protein interactions (Spence et al., 1990); FEM-2 is a putative serine/threonine type 2C phosphatase, however, its endogenous substrate has not been determined (Chin-Sang and Spence, 1996; Pilgrim et al., 1995); and FEM-3 is a novel protein with no obvious motifs (Ahringer et al., 1992).
A central unresolved question is how TRA-1 activity is regulated. Based on the observation that tra-1 is transcriptionally expressed at similar levels in both sexes, it was suggested that tra-1 must be regulated post-transcriptionally (Zarkower and Hodgkin, 1992). Segal and co-authors reported similar overall levels of TRA-1 protein in males and hermaphrodites (Segal et al., 2001). At the same time, the authors found sex-specific differences in the nuclear levels of TRA-1, with higher levels in hermaphrodites than in males, implying that TRA-1 activity is regulated by nuclear localization (Segal et al., 2001). In contrast, Schvarzstein and Spence reported that TRA-1 is primarily nuclear in both sexes but that overall TRA-1 levels are higher in hermaphrodites, implying that TRA-1 is regulated by changes in protein level (Schvarzstein and Spence, 2006).
In this study, we present evidence that TRA-1 is regulated by ubiquitin-proteasome degradation mediated by a CUL-2 complex containing the FEM proteins. CUL-2 belongs to the cullin family, whose members function as central components in multi-subunit cullin-RING ubiquitin ligase (E3) complexes (Petroski and Deshaies, 2005). CUL2 complexes consist of: CUL2, which forms a rigid scaffold; the Rbx1/Roc1 RING finger subunit that binds the C-terminus of CUL2; the adaptor protein Elongin C, which is bound to the N-terminus of CUL2 and to the ubiquitin-like protein Elongin B; and a variable substrate recognition subunit (SRS) that binds to Elongin C (Petroski and Deshaies, 2005). The SRS binds substrates and positions them for ubiquitination by ubiquitin-conjugating enzymes (E2s), which bind to the complex through interaction with Rbx1. CUL2-based E3 complexes will be referred to as CBC complexes, for the components CUL2, Elongin B, and Elongin C.
In C. elegans, CUL-2 regulates a number of critical cell cycle and developmental processes, including: the G1-to-S phase progression; meiotic progression; establishment of anterior-posterior polarity; chromosome condensation; and mitotic progression (DeRenzo et al., 2003; Feng et al., 1999; Liu et al., 2004; Sonneville and Gonczy, 2004). This study reveals a new process regulated by CUL-2, namely, the control of sex determination through the proteasome-mediated degradation of TRA-1. We demonstrate that FEM-1 functions as the SRS in a CBCFEM-1 complex that targets TRA-1 for degradation. We further show that FEM-2 and FEM-3 are cofactors that assemble with the complex and promote the efficient ubiquitination and degradation of TRA-1.
Results
FEM-1 is a substrate recognition subunit of a CUL-2-based ubiquitin ligase
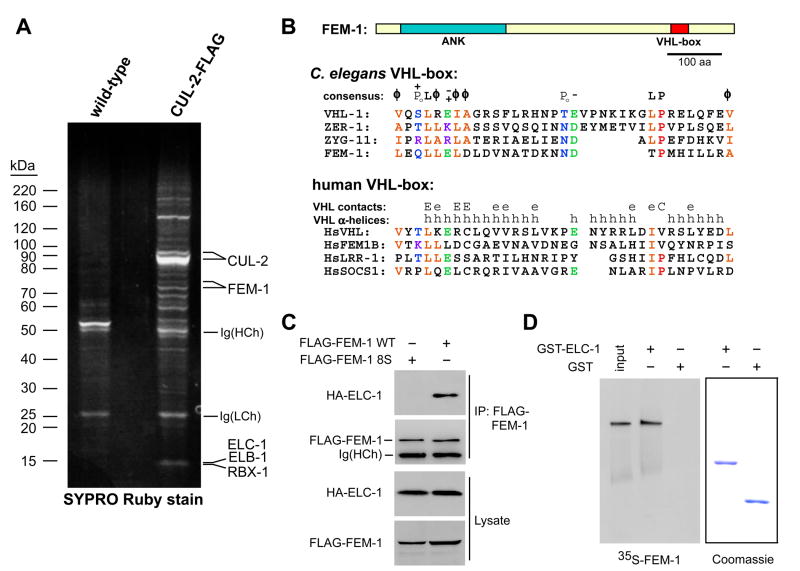
We identified CUL-2-interacting proteins in C. elegans using an affinity purification strategy in which proteins that associate with transgenic CUL-2-FLAG protein in vivo were identified by mass spectrometry. All of the expected core CBC components were identified: Elongin C (ELC-1), Elongin B (ELB-1), and RBX-1 (Fig. 1A). We also identified the cullin inhibitor CAND-1, and the CBC SRS proteins VHL-1, ZYG-11, and ZER-1 (Vasudevan et al., 2007). Additionally, we identified FEM-1 as two relatively abundant CUL-2-FLAG-associated protein bands of 72 and 75 kDa (Fig. 1A).
Fig. 1. FEM-1 associates with CUL-2 in C. elegans and interacts with the CBC core adapter ELC-1 through a VHL-box motif.
(A) FEM-1 associates with CUL-2 in C. elegans. Anti-FLAG affinity purifications from wild type (left lane) or a strain expressing the cul-2::FLAG transgene (right lane). Purified proteins were separated by SDS-PAGE, stained with SYPRO Ruby, and identified by MALDI-TOF MS (indicated on the right). Immunoglobulin heavy chain, Ig(HCh); or light chain, Ig(LCh).
(B) On the top is a schematic of FEM-1 that indicates N-terminal Ankyrin repeats (blue) and a C-terminal VHL-box (red). Below is an alignment of C. elegans and human VHL-box sequences from CBC SRSs. Residues conserved in at least half of the sequences are highlighted: orange, aliphatic residues; blue, polar; green, negatively charged; purple, positively charged; red, proline. A consensus of the C. elegans VHL-box is given above the sequences, with the following symbols for amino acid groups: φ, aliphatic; PO, polar; +, positively charged; −, negatively charged. α-helices in the human VHL-box are marked (h); residues that contact Elongin C (E for major and e for minor contacts) and CUL2 (C) (Stebbins et al., 1999).
(C) FEM-1 interacts with ELC-1 through the VHL-box motif. FLAG-tagged wild-type FEM-1 (WT) or mutant FEM-1 8S (containing serines for the first eight amino acids of the VHL-box, LEQLLELD) were co-expressed in 293T cells with HA-tagged ELC-1. Anti-FLAG immunoprecipitations were analyzed by immunoblotting for HA-ELC-1 and FLAG-FEM-1.
(D) FEM-1 binds ELC-1 in vitro. In vitro translated 35S-labeled FEM-1 was incubated with GST or GST-ELC-1 purified on glutathione beads, and bound protein was analyzed by SDS-PAGE/autoradiography (left). Input lane is 10% of the amount in the binding reaction.
Analysis of CUL-2-interacting proteins from our affinity purification revealed that several of the proteins, including FEM-1, VHL-1, ZYG-11, and ZER-1, contained a consensus C. elegans VHL-box motif that is similar to, yet distinct from, that found in mammalian VHL-box proteins (Kamura et al., 2004; Vasudevan et al., 2007) (Fig. 1B). The VHL-box motif is required for the SRS to bind to the CBC adaptor Elongin C (Kamura et al., 2004; Stebbins et al., 1999). Upon co-expression in human HEK 293T cells, HA-ELC-1 and FLAG-FEM-1 co-precipitated, indicating that the two proteins physically associate (Fig. 1C). This interaction was dependent on the VHL-box of FEM-1, as HA-ELC-1 did not bind to a FEM-1 mutant in which the first eight residues in the VHL-box motif were substituted by serines (Fig.1B, C). In vitro translated FEM-1 bound to purified GST-ELC-1, suggesting that the interaction is direct (Fig. 1D).
C. elegans FEM-1, therefore, has the hallmarks of a CBC complex SRS: 1) association with CUL-2 in vivo; 2) the presence of a VHL-box motif and an additional protein-interaction domain (Ankyrin repeats); and 3) physical association with ELC-1 in a VHL-box dependent manner.
cul-2 mutant males exhibit a feminization phenotype
C. elegans FEM-1 is required for the masculinization of the soma in males and the germ line in males and hermaphrodites (Doniach and Hodgkin, 1984; Kimble et al., 1984). fem-1 null mutants (XX or XO) that lack maternal product develop as complete females, which have a hermaphrodite (female) body structure but do not produce sperm. fem-1 null mutants that have maternal product (provided by a heterozygous parent) exhibit partial feminization phenotypes: XX animals develop as hermaphrodites but with reduced sperm number, while XO animals exhibit an intersex phenotype of a male somatic body plan with deformed male tails (including stunted and/or reduced numbers of rays), and a male-type, one-armed gonad that contains oocyte-like cells in addition to sperm (Doniach and Hodgkin, 1984).
If CUL-2 functions with FEM-1 to promote masculinization then we would expect a similar feminization phenotype in cul-2 mutants. cul-2 mutant larvae can only be obtained with maternal product, as the absence of cul-2 maternal product causes an early embryonic arrest (Feng et al., 1999). Analysis of homozygous cul-2(ek1) null mutant males revealed fully penetrant feminization phenotypes that are similar to the intersex phenotypes of fem-1 mutants with maternal product. In particular, cul-2(ek1) males exhibit defective male tails with variably degenerate morphology and a lower number of rays, and a feminized germ line containing oocyte-like cells within a one-armed male structure (Fig. 2). This indicates that CUL-2, like FEM-1, negatively regulates female fates.
Fig. 2. cul-2 mutant males exhibit a feminization phenotype.
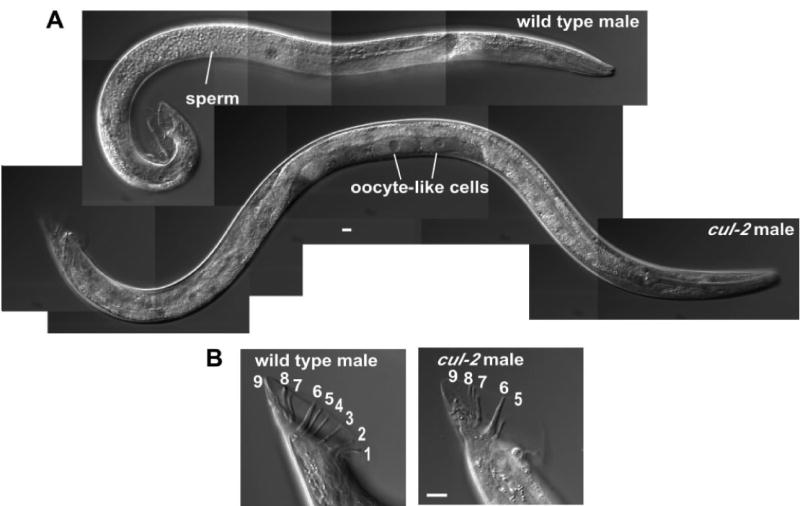
(A) DIC images of wild-type (top) and cul-2(ek1) mutant (bottom) XO males. Images are composites of overlapping pictures.
(B) DIC images of wild-type and cul-2 mutant male tails. Tail rays are numbered. Scale bars, 10 μm.
CUL-2 negatively regulates TRA-1 levels
The discovery of the CBCFEM-1 complex led us to the question of the identity of the critical substrate(s) for sex determination. TRA-1 was a likely candidate, as genetic analysis suggested that fem-1 is a negative regulator of tra-1 in males (Doniach and Hodgkin, 1984; Hodgkin, 1986) (Fig. 3A). To test the hypothesis that TRA-1 is a target of the CBCFEM-1 complex, we asked whether CUL-2 negatively regulates TRA-1 protein levels. We analyzed TRA-1 protein levels in the intestine (a somatic tissue) and the germ line, both of which are sexually dimorphic. In the intestine, TRA-1 is predominantly nuclear localized in both males and hermaphrodites. However, overall TRA-1 protein levels are significantly lower in wild-type male intestine cells (33 ± 3 arbitrary units vs. 100 ± 5 a.u. for hermaphrodites; p < 10−15, Student's t-test) (Fig. 3B, D). Intestinal TRA-1 levels are ∼70% higher in cul-2 mutant males than in wild-type males (p < 10−5).
The hermaphrodite intestine has the female-specific function of yolk production, which occurs during the adult stage (Kimble and Ward, 1988). TRA-1 levels are lower in wild-type hermaphrodites during the L4 larval stage (prior to full yolk production) relative to the adult stage (Figs 3B and S1). L4-stage cul-2 hermaphrodites have noticeably higher level of intestinal TRA-1 relative to L4 wild-type hermaphrodites (Fig S1). In the adult stage, cul-2 mutant and wild-type hermaphrodites have similar intestinal TRA-1 levels (Fig. 3B, D). These results indicate that CUL-2 negatively regulates somatic TRA-1 levels in males and in L4-stage hermaphrodites.
In the germ lines of both sexes, TRA-1 is also predominantly nuclear localized, with higher levels in the distal gonad regions (Figs 3C and S1). However, the male germ line has significantly lower levels of TRA-1 compared to hermaphrodites (22 ± 3 a.u. vs. 55 ± 3 a.u., respectively; p < 10−9) (Fig. 3C, D). The mean intensity levels of TRA-1 staining in the germ cell nuclei of cul-2 mutant adult males and hermaphrodites are similar to each other, but are approximately twice the level observed in wild-type hermaphrodites (p < 10−6), suggesting that CUL-2 negatively regulates TRA-1 levels in the germ lines of both sexes.
Analysis of TRA-1 expression by immunoblotting confirmed the negative regulation of TRA-1 levels by CUL-2, and provided new information on the regulation of TRA-1 isoforms. TRA-1 is expressed in three isoforms: a protein with an electrophoretic mobility of ∼35 kDa corresponding to the alternatively spliced isoform TRA-1B; a full-length TRA-1A doublet of ∼132/145 kDa; and an ∼95 kDa protein (Fig. 4). Both sexes have low levels of full-length TRA-1A, but hermaphrodites accumulate a cleaved isoform of ∼95 kDa, which is essentially absent from males (Fig. 4) (Schvarzstein and Spence, 2006). The cleaved TRA-1A isoform is analogous to the cleaved forms of the Gli family members in Drosophila and vertebrates (Kasper et al., 2006). In dominant tra-1(gf) XO mutants, which exhibit full feminization (Hodgkin, 1987), the cleaved TRA-1A level is markedly increased relative to wild-type males, correlating with the acquisition of female fates.
Fig. 4. TRA-1 isoforms accumulate in cul-2, fem-1, fem-2, and fem-3 loss-of-function mutants, and a tra-1 gain-of-function mutant.
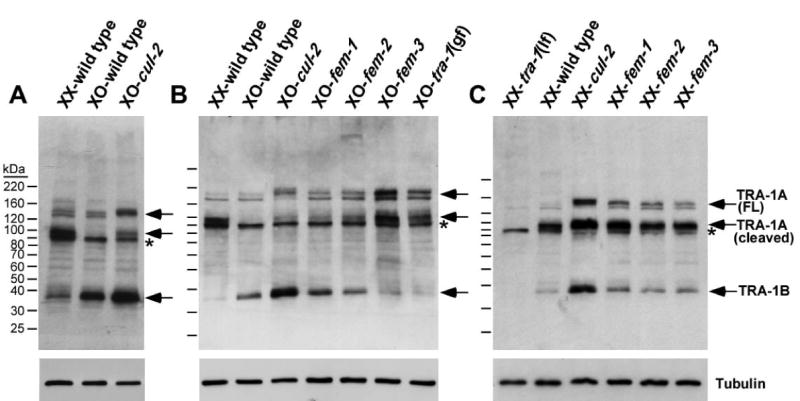
(A-C) Anti-TRA-1 western blots of whole-animal lysates of adults of the indicated genotypes and karyotypes. Anti-α-tubulin western blots are shown as loading controls. The location of full-length TRA-1A (FL), cleaved TRA-1A, and TRA-1B are marked by arrows and labeled on the right. Asterisk denotes a non-specific band. For all panels, the mutant alleles used are: loss-of-function cul-2(ek4), fem-1(hc17ts), fem-2(b245ts), fem-3(e1996) for XO, fem-3(e2006ts) for XX, and tra-1(e1099); and dominant, gain-of-function tra-1(e1575)/+. In tra-1(e1099) loss-of-function (lf) XX mutants, all three isoforms of TRA-1 are absent, while two non-specific protein bands remain, including a faint band that overlaps with the lower band of the TRA-1A full-length doublet.
In contrast to wild-type XO males, the cleaved TRA-1A isoform is present in cul-2 null XO mutant animals, although at a lower level than in hermaphrodites (cul-2 mutants have maternal product and so may have only partial loss of CUL-2) (Fig. 4A, B). The full-length TRA-1A is also increased relative to wild-type males. In XX cul-2 mutant hermaphrodites, both the full-length and cleaved TRA-1A isoforms accumulate to higher levels than in wild-type hermaphrodites (Fig. 4C). fem-1 mutant XO and XX animals have similarly increased levels of both TRA-1A isoforms (Fig. 4B, C). Interestingly, full-length TRA-1A in cul-2 mutants is shifted noticeably higher on the gel relative to the wild-type full-length TRA-1A, potentially reflecting post-translational modification (Fig. 4). This shift is more noticeable in cul-2 mutants than in fem-1 mutants and may reflect the loss of an additional FEM-1-independent CUL-2 function. Finally, we observe that cul-2 mutants have significantly higher levels of TRA-1B when compared to wild type, indicating that CUL-2 negatively regulates TRA-1B levels (Fig. 4).
Genetic interaction between cul-2 and tra-1
If the negative regulation of TRA-1 levels by CUL-2 is important for regulating TRA-1 activity, then we would expect that cul-2 inactivation would suppress tra-1 hypomorphic alleles, as this would lead to higher levels of partially-active TRA-1 protein. We tested for genetic interaction with two tra-1 hypomorphic alleles that exhibit partial masculinization of XX animals: tra-1(e1929) and tra-1(e1488). tra-1(e1929) homozygous XX mutants have a body plan that resembles that of hermaphrodites except for a male-type one-armed gonad that produces sperm and oocytes. The number of oocytes in tra-1(e1929) XX mutants is very small, and few of these attain a size comparable to wild-type oocytes (Fig. S2) (Hodgkin, 1987). cul-2 RNAi depletion generated significantly more oocytes, of which a higher percentage had wild-type size and shape (Fig. S2). tra-1(e1488) homozygous XX mutants exhibit strong masculinization of the non-gonadal soma, but their gonad has the two-armed hermaphrodite shape with oocytes that are rarely of wild-type morphology (Hodgkin, 1987). Those tra-1(e1488) XX mutant animals that are fertile are egg-laying defective, and their progeny hatch within the body cavity. After cul-2 RNAi treatment, most tra-1(e1488) XX animals had oocytes that were indistinguishable from wild type, and were able to lay eggs (Fig. S2).
We also observed that cul-2 RNAi partially suppressed other aspects of masculinization in the hypomorphic tra-1 mutant XX animals, namely the absence of hermaphrodite-specific neurons (HSNs) and the presence of male-specific cephalic companion neurons (CEMs) (Conradt and Horvitz, 1999; Sulston et al., 1983). The numbers of HSNs are severely reduced in XX tra-1(e1929) and tra-1(e1488) mutants, but are increased after cul-2 RNAi treatment (Table S1). tra-1(e1929) XX animals had only 23% of the HSNs found in wild-type XX hermaphrodites, but after cul-2 RNAi the percentage increased to 65% (p < 0.001). Analogously, tra-1(e1929) XX mutants had 13% of wild-type HSN numbers, and this increased to 33% after cul-2(RNAi) (p < 0.05). cul-2 RNAi treatment produced weak but statistically significant suppression of CEM numbers in tra-1(e1929) XX animals, with CEM numbers decreasing from 97% of wild-type male levels to 80% upon cul-2 RNAi; p < 10−4 (Table S1). For tra-1(e1488) mutants, we did not observe a significant change in CEM numbers upon cul-2 RNAi. It is possible that the general recalcitrance of C. elegans neurons to RNAi limited the effectiveness of cul-2 RNAi in CEM neurons (Simmer et al., 2002). Nevertheless, the overall analysis of HSN and CEM cells reinforces the conclusion that depletion of cul-2 can suppress tra-1 hypomorphic mutant phenotypes.
The hermaphrodite-specific development of HSNs is controlled by TRA-1A through the direct transcriptional repression of the pro-apoptotic gene egl-1 (Conradt and Horvitz, 1999). The suppression of HSN apoptosis in tra-1 hypomorph XX mutants by cul-2 RNAi, suggests that CUL-2 indirectly regulates egl-1 transcription. We tested two other direct targets of TRA-1 transcriptional repression in hermaphrodites, mab-3 and fog-3, to determine if they are regulated by CUL-2 activity. As expected, mab-3 and fog-3 transcripts were observed in male but not hermaphrodite wild-type adults (Chen and Ellis, 2000; Yi et al., 2000). In cul-2 males, we detected significantly less RT-PCR products of both genes compared to wild-type males, consistent with increased TRA-1 activity in cul-2 males (Fig. S3). In total, these results suggest that CUL-2 is an upstream negative regulator of TRA-1, and support the model that TRA-1 activity is regulated by protein degradation.
CBCFEM-1 mediates the proteasome-dependent degradation of TRA-1
To obtain molecular insights into the degradation of TRA-1, we performed experiments in human 293T cells. Transfection of 293T cells with a full-length TRA-1A cDNA generated two TRA-1A protein bands of ∼135 and ∼90 kDa, analogous to what was observed in C. elegans (Fig. 5A). When TRA-1A was co-expressed with FEM-1, both TRA-1A isoforms were significantly reduced or eliminated from the cells (Fig. 5A). To determine if the disappearance of TRA-1A resulted from proteasome-mediated degradation, we co-expressed TRA-1A and FEM-1 in the presence of the proteasome inhibitor LLnL. Full-length TRA-1A was detected when the proteasome was inactivated, indicating that FEM-1 co-expression (in the absence of LLnL) induces the proteasome-mediated degradation of TRA-1A (Fig. 5A). The failure to observe the ∼90 kDa (presumably cleaved) isoform upon treatment with LLnL may reflect that the cleavage to generate this isoform is proteasome-dependent, similar to the proteasome-dependent cleavage of Gli homologs in Drosophila and humans (Kasper et al., 2006).
Fig. 5. The CBCFEM-1 complex with the cofactors FEM-2 and FEM-3 promotes TRA-1A degradation.
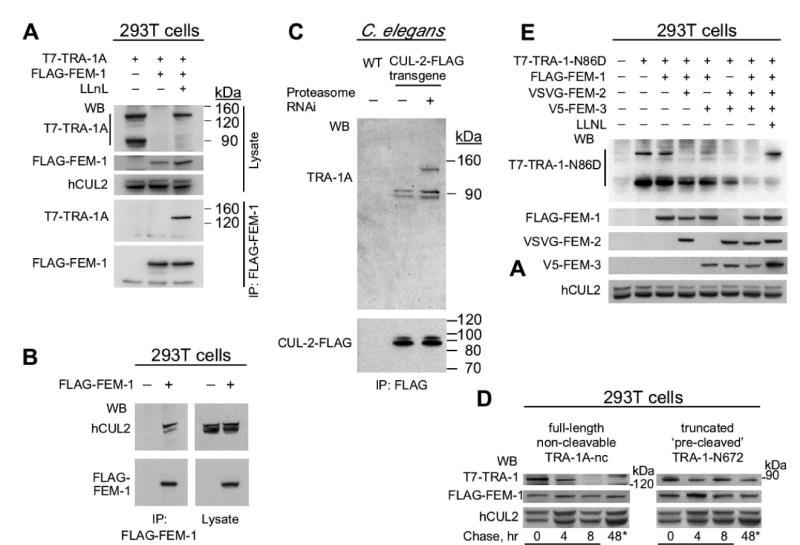
(A) FEM-1-mediated degradation of TRA-1A. T7-TRA-1A was expressed with or without FLAG-FEM-1 in human 293T cells in the presence or absence of the proteasome inhibitor LLnL (listed at the top). Cell lysate and an immunoprecipitation with anti-FLAG antibody were analyzed by western blotting with anti-T7, anti-FLAG, and anti-human CUL2 antibodies.
(B) C. elegans FEM-1 co-immunoprecipitates human CUL2 in 293T cells.
(C) TRA-1A physically associates with CUL-2 in C. elegans when the proteasome is inactivated. CUL-2-FLAG was affinity purified from whole-worm extract of wild type (WT) or a him-8 mutant strain expressing the cul-2::FLAG transgene grown in the presence or absence of pbs-4 RNAi bacteria to inactivate the proteasome, and further analyzed by western blot with anti-TRA-1 (top) or anti-FLAG (bottom) antibodies.
(D) Pulse chase experiment demonstrates that FEM-1 preferentially induces the degradation of the full-length TRA-1A isoform. A non-cleavable full-length T7-TRA-1A-nc mutant and a ‘pre-cleaved’ T7-TRA-1-N672 mutant (containing the N-terminal 672 residues) were co-expressed with FLAG-FEM-1 in the presence of LLnL. At time 0, LLnL was removed and further protein synthesis was blocked by cycloheximide. The final lanes (asterisk) are for a 48 hr time point without LLnL or cycloheximide.
(E) The gain-of-function TRA-1-N86D protein is fully degraded only upon expression of all three FEM proteins. Combinations of the FEM proteins were co-expressed to determine their effect on TRA-1-N86D degradation. The last lane represents expression of all FEM proteins plus TRA-1-N86D in the presence of LLnL. The transfected tagged FEM proteins and human CUL2 were detected as expression and loading controls.
The full-length TRA-1A co-precipitated with FEM-1 in the presence of LLnL, indicating that the two proteins physically associate (Fig. 5A). Human CUL2 also co-precipitated with FEM-1 (Fig. 5B). The CBC adaptor protein ELC-1 was similarly able to co-precipitate human CUL2, but could only associate with TRA-1A when FEM-1 was co-expressed (Figs 6E and S6). This suggests that the FEM-1-mediated degradation of TRA-1A occurs through association with human CBC components and that FEM-1 is required for the CBC complex to bind TRA-1A.
Fig. 6. The three FEM proteins bind to TRA-1A and to each other in the context of a CBC complex.
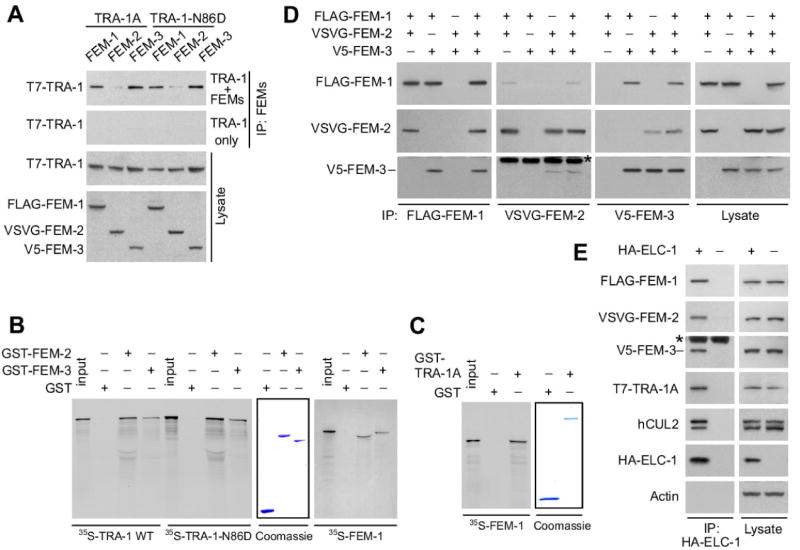
(A) Each of the three FEM proteins interacts with both TRA-1A and TRA-1-N86D. FLAG-FEM-1, VSVG-FEM-2, and V5-FEM-3 were co-expressed in 293T cells with either wild-type T7-TRA-1A or the gain-of-function T7-TRA-1-N86D in the presence of LLnL. The individual FEM proteins were immunoprecipitated and then analyzed by immunoblotting for T7-TRA-1. The top panel is a western of immunoprecipitates from cells co-expressing FEM and TRA-1A proteins; while the second panel shows immunoprecipitates from cells expressing TRA-1A proteins alone. Western blots of cell lysates are shown in the bottom two panels.
(B) in vitro translated TRA-1A and FEM-1 bind recombinant GST-fusion FEM-2 and FEM-3. 35S-TRA-1A, 35S-TRA-1A-N86D, and 35S-FEM-1 were incubated with GST, GST-FEM-2, and GST-FEM-3 purified on glutathione Sepharose beads, and the beads were analyzed by SDS-PAGE/autoradiography. The middle panel is a Coomassie R250-stained gel showing the relative amounts of GST and GST-fusion proteins used in the reactions.
(C) in vitro translated 35S-FEM-1 binds recombinant GST-TRA-1A in vitro.
(D) Reciprocal co-immunoprecipitations of the FEM proteins co-expressed in 293T cells. Epitope-tagged FEM proteins (noted at the top) were co-expressed in 293T cells in double combinations or all together. Each FEM protein was immunoprecipitated (noted at the bottom) and analyzed by western blotting for co-precipitated proteins (noted on the left side). The last panels on the right are westerns of the lysates.
(E) Co-immunoprecipitation of human and C. elegans CBCFEM-1 components. Anti-HA immunoprecipitation from 293T cells (treated with LLnL) co-expressing FLAG-FEM-1, VSVG-FEM-2, V5-FEM-3, and T7-TRA-1A with or without HA-ELC-1, followed by western blot to detect the tagged proteins and human CUL2. Actin was probed as a negative control. For (D) and (E), asterisk denotes immunoglobulin heavy chain.
To further investigate if TRA-1 is directly targeted for degradation by a CBCFEM-1 complex in C. elegans, we asked whether inactivation of the proteasome would allow us to detect the physical association of CUL-2 and TRA-1 in C. elegans. We expressed cul-2::FLAG in a him-8 mutant strain, which produces ∼40% male progeny, and the proteasome was inactivated by RNAi depletion of the core proteasome subunit pbs-4. CUL-2-FLAG was affinity purified and analyzed by immunoblotting with anti-TRA-1 antibody or by mass spectrometry. Both methods revealed that full-length TRA-1A stably associated with CUL-2-FLAG in C. elegans only when the proteasome was inactivated (Fig. 5C; data not shown). Interestingly, the cleaved form of TRA-1A associated with CUL-2-FLAG both in the presence or absence of pbs-4 RNAi (Fig. 5C), implying that it has a more stable association than the full-length isoform in the presence of active proteasome. We did not observe TRA-1B in the CUL-2-FLAG pull-down samples, suggesting that it may not be a direct substrate of a CUL-2 complex.
To clarify which of the TRA-1A isoforms is targeted for degradation by CBC FEM-1, we performed pulse-chase experiment in 293T cells. We followed the degradation of both a non-cleavable full length TRA-1A (which is missing residues required for SCFβ-TrCP-mediated cleavage to generate the 90 kDa isoform; Wang and Li, 2006), and a truncated ‘pre-cleaved’ TRA-1A (TRA-1-N672). Non-cleavable full-length TRA-1A was degraded within 8 hrs (similar to wild type full length TRA-1A), while the ‘pre-cleaved’ TRA-1-N672 was stable up to 48 hrs (Fig. 5D; data not shown). A second type of experiment, in which FEM-1 was transfected into cells that already expressed both isoforms of wild-type TRA-1A, also suggests that the full-length TRA-1A isoform is the preferred target, with the cleaved isoform resistant to FEM-1-mediated degradation (Fig. S4).
FEM-2 and FEM-3 are cofactors for the CBCFEM-1 E3 complex
Genetic studies of the sex determination pathway have placed two other genes, fem-2 and fem-3, in the same position as fem-1, as negative regulators of TRA-1 (Hodgkin, 1986) (Fig. 3A). We conducted experiments to address whether FEM-2 and FEM-3 function in the same pathway as CUL-2 and FEM-1 to regulate TRA-1 protein levels.
In both XO and XX fem-2 and fem-3 mutants, full-length and cleaved forms of TRA-1A were increased relative to wild-type animals (Fig. 4). The extent of TRA-1A accumulation differed somewhat between the fem and cul-2 mutant strains examined. The highest TRA-1A levels in XO animals were observed in fem-3(e1996) null mutants, which correlated with their stronger feminization phenotype relative to the other fem mutants analyzed: fem-3(e1996) XO null mutants are fertile females, while fem-1(hc17ts) and fem-2(b245ts) XO hypomorph mutants exhibit intersex phenotypes (Hodgkin, 1986; Kimble et al., 1984; Nelson et al., 1978). We suggest that differences in TRA-1A levels among the fem and cul-2 mutants are likely due to residual gene activity (due to hypomorphic alleles or the perdurance of maternal product); however, we cannot rule out the existence of TRA-1A regulatory pathways that involve only a subset of these genes.
To assess the contributions of FEM-2 and FEM-3 to CBC FEM-1 mediated degradation of TRA-1A, we used the human expression system. A gain-of-function TRA-1A mutant protein (with an N86D point mutation, corresponding to the tra-1(e1575) allele; de Bono et al., 1995), demonstrates significantly higher stability than the wild-type TRA-1A when co-expressed with FEM-1 (Figs 5E and S5). Co-expression of double combinations of FEM proteins increased TRA-1A-N86D degradation, however, complete degradation was only achieved upon expression of all three FEM proteins (Figs 5E and S5). Inhibition of the proteasome by LLnL blocked the degradation of the full-length TRA-1-N86D induced by the three FEM proteins, indicating that this degradation is also proteasome-dependent (Fig. 5E).
We found that FEM-1, FEM-2, and FEM-3 proteins each physically associate with full-length TRA-1A when co-expressed in human 293T cells (in the presence of the proteasome inhibitor LLnL), although the FEM-2–TRA-1A interaction is weak (Fig. 6A). Interestingly, the FEM proteins interacted with similar efficiency with the gain-of-function TRA-1A mutant protein (Fig. 6A). In vitro binding assays show that the interaction of FEM-1, FEM-2, and FEM-3 with TRA-1A or the gain-of-function TRA-1-N86D is direct (Fig. 6B, C; data not shown). Overall, these experiments indicate that each of the FEM proteins can independently bind TRA-1A, and that a gain-of-function mutation that stabilizes TRA-1A does not block interaction with the CBCFEM-1 complex.
Reciprocal co-immunoprecipitation experiments with FEM proteins expressed in 293T cells showed that each of the FEM proteins interacts with each of the other FEM proteins (Fig. 6D). Moreover, when all three FEM proteins were co-expressed, each of the FEM proteins co-precipitated the other two FEM proteins. We observed that in vitro translated 35S-FEM-1 bound to purified recombinant GST-FEM-2 and GST-FEM-3, demonstrating direct interaction between these proteins (Fig. 6B). Direct interaction between FEM-2 and FEM-3 had been shown earlier (Chin-Sang and Spence, 1996). Therefore, FEM-1, FEM-2, and FEM-3 each directly bind to each other. Moreover, FEM-1, FEM-2, FEM-3, human CUL2, and the substrate TRA-1A, all co-precipitated with the CBC adaptor ELC-1 in the 293T system (in the presence of LLnL) (Fig. 6E).
We next sought to determine the role of the FEM proteins in facilitating the ubiquitination of TRA-1A. We found that in 293T cells, FEM-1 induced the in vivo ubiquitination of full-length TRA-1A, and that the ubiquitination was enhanced upon co-expression of all three FEM proteins compared to FEM-1 alone (Fig. 7A). In these experiments, we used the TRA-1 gain-of-function mutant containing an additional mutation that made it non-cleavable (a deletion of residues 650-680) in order to eliminate the endogenous ubiquitination of TRA-1 that is associated with its processing to the cleaved 90 kDa form. The deletion of residues 650-680 of TRA-1A was used in order to eliminate the presumed interaction of TRA-1A with the ubiquitin ligase SCFβ-TrCP, which is known to induce the ubiquitin-dependent cleavage of mammalian Gli3 protein (Wang and Li, 2006). We also observed that the CBCFEM-1 complex was capable of the in vitro ubiquitination of wild-type TRA-1A, and again found that the ubiquitination was significantly enhanced in the presence of all three FEM proteins compared to FEM-1 alone (Figs 7B and S6). Taken together, our results suggest that FEM-2 and FEM-3 bind the CBCFEM-1 complex to promote the ubiquitin-mediated proteolysis of TRA-1A.
Fig. 7. FEM-2 and FEM-3 enhance CBCFEM-1-mediated ubiquitination of TRA-1A.
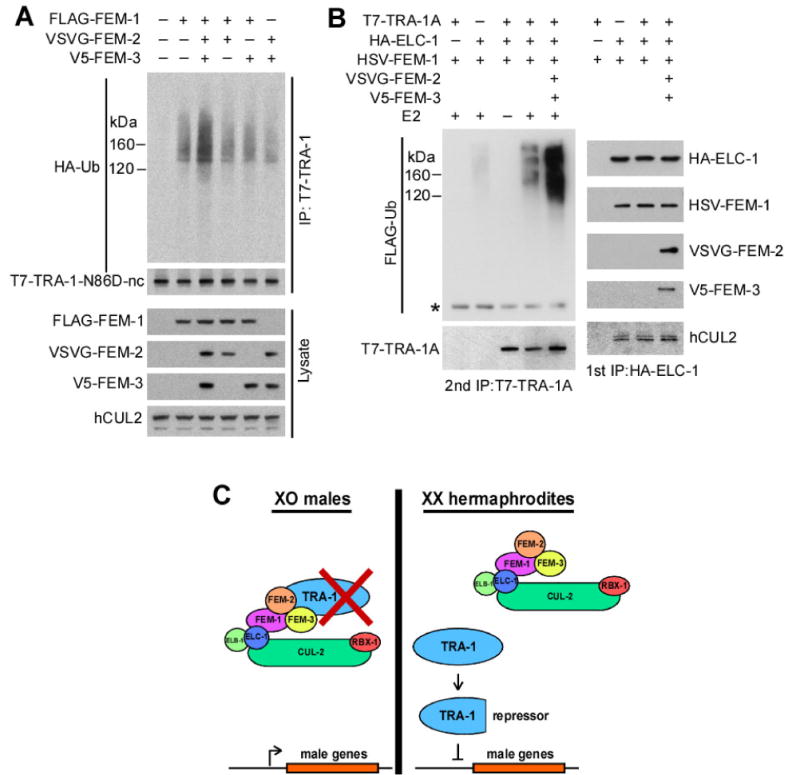
(A) The ubiquitination of TRA-1A in 293T cells is stimulated by the FEM proteins. A non-cleavable gain-of-function TRA-1A (T7-TRA-1-N86D-nc) was co-expressed with HA-ubiquitin, and with or without FEM proteins (noted at the top). T7-TRA-1-N86D-nc was immunoprecipitated in the presence of 0.1% of SDS, followed by anti-HA and anti-T7 immunoblotting. Whole cell lysates were blotted to assess protein expression.
(B)in vitro ubiquitination of TRA-1A by the CBCFEM-1 complex. Epitope-tagged ELC-1, FEM-1, and TRA-1A were co-expressed in 293T cells with or without FEM-2 and FEM-3 (noted at the top) in the presence of LLnL. The CBCFEM-1 complex was immunoprecipitated with anti-HA antibody (1st IP). The immunocomplex (with associated T7-TRA-1A) was used in an in vitro ubiquitination reaction. T7-TRA-1A was subsequently immunoprecipitated in the presence of 0.1% SDS (2nd IP), followed by immunoblotting for FLAG-ubiquitin and T7-TRA-1A. Samples from the 1st immunoprecipitation were analyzed by western blot with antibodies to the respective epitope tags and human CUL2 to reveal the level of proteins in the immunocomplex used in the ubiquitination reaction.
(C) Model of CBCFEM-1 regulation of TRA-1A. (left) In XO males, the CBCFEM-1 complex is active and targets TRA-1A for degradation. FEM-1 binds full-length TRA-1A with the assistance of FEM-2 and FEM-3, which bind to each other and to TRA-1A. TRA-1A is polyubiquitinated by the complex and is subsequently degraded by the proteasome. In the absence of TRA-1A, genes that promote male fates are expressed. (right) In XX hermaphrodites, the ability of CBCFEM-1 to degrade TRA-1A is restricted [possible mechanisms include inhibitory binding of FEM proteins by the upstream regulator TRA-2 (Mehra et al., 1999), or post-translational modification of TRA-1A]. In the absence of degradation, the full-length TRA-1A protein is proteolytically processed to form the cleaved isoform, which represses genes that promote male fates.
Discussion
FEM-1, FEM-2, and FEM-3 function in a CBC complex to promote TRA-1 degradation
We identified FEM-1 in an affinity purification screen for proteins that physically associate with CUL-2 in C. elegans. CUL-2 functions in CBC ubiquitin-ligase complexes, which employ variable SRS components to recognize distinct subsets of substrates (Petroski and Deshaies, 2005). FEM-1 contains a C-terminal VHL-box motif, which is a feature of SRSs that mediates binding to the adaptor protein Elongin C (Kamura et al., 2004; Stebbins et al., 1999). Consistent with a role as an SRS, FEM-1 binds to Elongin C in a VHL-box-dependent manner, and contains an additional protein-interaction motif (N-terminal Ankyrin repeats).
Inactivation of an SRS is expected to produce a subset of the phenotypes associated with the loss of core CBC components. cul-2 mutant XO animals exhibit a feminized intersex phenotype similar to that of fem-1 XO animals with maternal product, indicating that the two genes share a common sex determination phenotype. Consistent with FEM-1 functioning as an SRS for the CBC complex, cul-2 mutants have a number of additional phenotypes that are not shared by fem-1 mutants (DeRenzo et al., 2003; Feng et al., 1999; Liu et al., 2004; Sonneville and Gonczy, 2004). This partial phenotypic overlap, combined with the interaction of FEM-1 and CUL-2 in C. elegans, and the binding FEM-1 to Elongin C, supports the conclusion that FEM-1 is an SRS for a CBCFEM-1 complex.
Our work indicates that TRA-1 is the critical substrate of the CBCFEM-1 complex. Inactivation of cul-2 leads to an increase in TRA-1 levels in the soma and germ line of XO males. In XX hermaphrodites, inactivation of cul-2 leads to increases in TRA-1 levels in the germ line, and during the L4 stage, in the soma. Loss of cul-2 partially suppresses the masculinization associated with tra-1 hypomorphic alleles. cul-2 RNAi also decreases the expression of genes in males that are direct targets of TRA-1 repression in hermaphrodites. Both of these results imply an increase in TRA-1 activity upon inactivation of CUL-2. Our overall analyses, therefore, suggests that CUL-2 inhibits TRA-1 activity by negatively regulating TRA-1 levels.
We show that TRA-1A expressed in human cells can be degraded by a CBCFEM-1 complex. TRA-1A degradation is stimulated by co-expression of FEM-1, which physically binds TRA-1A and induces its ubiquitination. The core CBC component ELC-1 will co-precipitate TRA-1A, but only when FEM-1 is present. A CBC complex containing ELC-1, FEM-1, and human CUL2 associates with TRA-1A, and can mediate the in vitro ubiquitination of TRA-1A. The observation that CUL-2 and TRA-1A physically associate with each other in C. elegans upon inactivation of the proteasome, implies that the CBCFEM-1 complex directly mediates TRA-1A degradation in C. elegans.
Genetic studies have identified fem-1, fem-2, and fem-3 as immediate upstream negative regulators of tra-1 (Doniach and Hodgkin, 1984; Hodgkin, 1986; Kimble et al., 1984). Loss of fem-1, fem-2, or fem-3 leads to increased TRA-1A levels, indicating that all three FEM proteins are required to negatively regulate TRA-1 levels in C. elegans. When expressed in human cells, each of the FEM proteins can physically bind to TRA-1A and to each other. Immunoprecipitation of the core CBC component ELC-1 pulls down FEM-1, FEM-2, and FEM-3, along with human CUL2 and the substrate TRA-1A. Significantly, the association of FEM-2 and FEM-3 with the CBCFEM-1 complex expressed in human cells enhances the ubiquitination of TRA-1A in vitro and in vivo. We therefore propose that FEM-2 and FEM-3 are cofactors for the CBCFEM-1 complex that increase the efficiency of ubiquitination of the substrate TRA-1A.
The CBCFEM-1 complex appears to be conserved in other metazoa. While our project was ongoing, it was shown that FEM1B is a component of a human CBC complex, although the function of this complex is not known (Kamura et al., 2004). FEM-3 appears to be a nematode-specific protein that is not widely conserved (Haag et al., 2002); however, it has been shown to interact with FEM-2 in other nematodes (Stothard and Pilgrim, 2006). Finally, the human ortholog of FEM-2, while it has not been linked to ubiquitin-mediated proteolysis, interacts with the human FEM-1 homolog FEM1B (Tan et al., 2001), suggesting that the association of the CBCFEM-1 complex with cofactors is conserved.
CBCFEM-1 controls TRA-1 repressor activity through degradation of full-length TRA-1A
Drosophila and vertebrate TRA-1 homologs are proteolytically-processed to form cleaved proteins that function as transcriptional repressors, while the full-length forms are activators (Kasper et al., 2006). In C. elegans, cleaved TRA-1A is the most abundant form in hermaphrodites, while it is missing in males (Schvarzstein and Spence, 2006; and this study). Significantly, all three of the identified downstream target genes of TRA-1A (egl-1, mab-3, and fog-3) are transcriptionally repressed by TRA-1A in hermaphrodites (Chen and Ellis, 2000; Conradt and Horvitz, 1999; Yi et al., 2000). In XO animals lacking cul-2, fem-1, fem-2, or fem-3, the cleaved form of TRA-1A accumulates, and these animals exhibit female traits rather than the male traits normally associated with the XO karyotype. Consistently, the mRNA levels for mab-3 and fog-3 decrease dramatically in cul-2 mutant XO males, presumably resulting from increased repression mediated by the cleaved TRA-1A isoform. Therefore, the ultimate focus of the sex determination pathway in males appears to be the CBCFEM-1-mediated degradation of TRA-1A to prevent the accumulation of the cleaved, transcriptional-repressor isoform (Fig. 7C).
The question arises whether CBCFEM-1 targets the degradation of full-length TRA-1A, the cleaved isoform, or both? In whole-animal lysates, loss of CBCFEM-1 components results in the accumulation of both TRA-1A isoforms. In CUL-2-FLAG affinity purifications from C. elegans, full-length TRA-1A co-purified with CUL-2-FLAG only when the proteasome was inhibited. In contrast, the cleaved form of TRA-1A associated with CUL-2-FLAG regardless of whether the proteasome was inhibited. This suggests two possibilities: 1) the proteasome-mediated degradation of the cleaved isoform is not as efficient as that of full-length TRA-1A, and it remains in contact with the CUL-2 complex for an extended period prior to degradation; or 2) the cleaved TRA-1A isoform associates with CUL-2 in a context other than that of a substrate. In the 293T system, full-length TRA-1A is efficiently degraded upon co-expression with FEM-1, while the cleaved TRA-1A isoform is not. This suggests that the CBCFEM-1 complex principally degrades the full-length TRA-1A prior to its proteolytic processing to the cleaved isoform. This implies that the absence of the cleaved TRA-1 isoform in males is a secondary consequence of the degradation of the full-length TRA-1A isoform prior to its proteolytic processing. The full-length TRA-1A isoform is predominantly expressed in the germ line (Schvarzstein and Spence, 2006), suggesting that in somatic tissues of the male, CBCFEM-1-mediated degradation effectively eliminates all TRA-1A isoforms. One speculative model is that in hermaphrodites, the accumulation of cleaved TRA-1A further reduces the degradation of full-length TRA-1A by binding the CBCFEM-1 complex and acting as a competitive inhibitor.
The regulation of TRA-1A activity by ubiquitin-mediated proteasome degradation
Our study suggests that TRA-1A activity is controlled predominantly by ubiquitin-proteasome degradation targeted by the CBCFEM-1 complex. tra-1 gain-of-function mutants have unregulated TRA-1 activity and induce the full feminization of both XO and XX animals (Hodgkin, 1987). The observation that a tra-1 gain-of-function mutant has increased TRA-1A protein levels independently reinforces the conclusion that protein degradation is the primary method of TRA-1 regulation. Twenty seven tra-1(gf) mutations have been molecularly identified, and all of these map to a small 16-amino acid region in the N-terminus of TRA-1 (de Bono et al., 1995). The TRA-1 GF mutant protein is stabilized in C. elegans males. In human 293T cells, the TRA-1A GF protein shows greater resistance to FEM-1-mediated degradation. The simplest model for how tra-1(gf) mutations prevent degradation would be that recognition of TRA-1 by the CBCFEM-1 complex requires an intact GF region. However, this model is not supported by our analysis of protein interactions, which has not revealed differences in the binding of FEM proteins to the wild-type or gain-of-function TRA-1A proteins.
FEM-2 and FEM-3 increase the efficiency of FEM-1-mediated degradation of the TRA-1A GF protein in 293T cells. Expression of FEM-2 and FEM-3 in the absence of FEM-1 can induce limited TRA-1A degradation, which we presume occurs through the substitution of a human FEM1 homolog in the complex. All three FEM proteins can bind TRA-1A. The addition of FEM-2 and FEM-3 to the CBCFEM-1 complex expressed in human cells increases the efficiency of TRA-1A ubiquitination both in vivo and in vitro.
The exact mechanism of how FEM-2 and FEM-3 enhance CBCFEM-1 ubiquitination of TRA-1A is not understood. FEM-3 contains no known motifs and does not appear to be conserved beyond nematodes (Ahringer et al., 1992). FEM-2 encodes a putative type 2C Ser/Thr phosphatase (Chin-Sang and Spence, 1996; Pilgrim et al., 1995). The TRA-1-GF region contains a predicted GSK-3 phosphorylation site (de Bono et al., 1995). A speculative model is that phosphorylation of TRA-1A inhibits CBCFEM-1-mediated degradation, but this is reversed by FEM-2, which interacts with a docking site on or near the TRA-1-GF region in order to dephosphorylate TRA-1A. Further experimentation will be required to test this model, and to determine precisely how the CBCFEM-1-mediated degradation of full-length TRA-1A is regulated by upstream sex determination pathway components.
Materials and Methods
C. elegans analysis and RNAi
To obtain pure populations of XO fem mutant animals for western blot analysis, we employed the following strains: ET286, fem-1(hc17ts); dpy-21(e428) him-5(e1490); ET287, fem-2(b245ts); dpy-21(e428) him-5(e1490); and ET288, fem-3(e1996 null)/unc-24(e138); dpy-21(e428) him-5(e1490). XO animals were isolated based on the fact that dpy-21 homozygotes manifest a Dpy phenotype in XX but not XO animals (Hodgkin, 1980). XX fem mutants were collected from strains containing ts alleles for fem-1(hc17ts), fem-2(b245ts), or fem-3(e2006ts), which were grown at the restrictive temperature of 25°C.
The numbers of CEM cells in adult animals were determined in strains carrying a pkd-2::GFP transgene, which expresses in the four CEM neurons in the head region. The numbers of HSN cells in adult animals were determined in strains carrying a cgh-3::CFP transgene, which expresses in the HSN neurons located on each lateral side of the animal. CEMs and HSNs were identified by fluorescence of the pkd-2::GFP or cgh-3::CFP transgenes, respectively.
RNAi experiments were performed by the feeding RNAi method as described (Timmons et al., 2001), using full-length cul-2 cDNA (Feng et al., 1999) or pbs-4 cDNA (encoding a β-type subunit of the 20S proteasome core particle; Fraser et al., 2000). To avoid severe embryonic lethality associated with fully penetrant cul-2 or pbs-4 RNAi, we exposed embryos or L1 larvae (which had cul-2 and pbs-4 maternal product) to RNAi and analyzed that cohort of animals.
Immunofluorescence and microscopy
Adults or L4 larvae were dissected to extrude intestines and gonads in egg salt buffer (Edgar, 1995), and then processed for immunofluorescence using the freeze-crack method (Miller and Shakes, 1995). Rabbit polyclonal anti-TRA-1 antibody was affinity purified as described (Mathies et al., 2004). Anti-rabbit Alexa Fluor 488 (Molecular Probes) was used as a secondary antibody. DNA was stained with 1 μg/ml Hoechst 33258 dye. Four independent experiments to assess TRA-1 levels in cul-2 mutants gave similar results.
A Zeiss Axioskop microscope was used for differential interference contrast (DIC) and immunofluorescence microscopy. A Hamamatsu ORCA-ER digital camera and Openlab 4.0.2 software (Improvision) were used to capture digital images. Matched images of anti-TRA-1 immunofluorescence were taken at the same exposure, and processed identically with Adobe Photoshop 7.0 (Fig. 3). Quantitation of anti-TRA-1 staining was performed by confocal microscopy using a Leica TCS SP2 Spectral Confocal Microscope with Coherent Ti:sapphire multiphoton laser. Anti-TRA-1 nuclear signal was determined as the mean signal of nuclei minus the average signal from the nuclear regions of tra-1(e1099) null mutants, which lack obvious anti-TRA-1 nuclear signal. Six to eight animals were analyzed for each genotype/sex (n = 34 cells). Quantitation was performed with ImageJ software (version 1.37) (Abramoff et al., 2004).
Affinity purification from C. elegans extract
_S_train ET099, which expresses CUL-2-FLAG, was fed the E. coli strain OP50, or, for RNAi experiments, E. coli strain HT115 expressing pbs-4 dsRNA or the empty vector pPD129.36. Late L4 larvae or young adults were collected, purified by flotation on 30% sucrose (Sulston and Hodgkin, 1988), and frozen at −80°C, then ground in liquid nitrogen, followed by suspension in Lysis buffer: 50 mM HEPES, pH7.8, 300 mM NaCl, 10% glycerol, 0.2% Triton X-100, 2 mM DTT, 1 mM EDTA, and protease inhibitor cocktail (Roche). The extract was clarified by consecutive low speed (1,500xg, 5 min) and high speed (100,000xg, 40 min) centrifugations and incubated for 2 hrs at 4°C with anti-FLAG M2 antibody coupled to agarose beads (Sigma) in the presence of 200 mM NaCl/Lysis buffer. After washing with the latter buffer, bound proteins were eluted with 0.4 mg/ml FLAG peptide (Sigma) in Elution buffer: 20 mM HEPES, pH7.8, 150 mM KCl, 15% glycerol, 0.05% NP-40, 2 mM DDT, 1 mM EDTA, protease inhibitor cocktail (Roche). The eluates were resolved on NuPAGE 4-12% Bis-Tris gels, and analyzed by western blot or stained with SYPRO Ruby (Molecular Probes) or Coomassie R-250 (Sigma) and used for MALDI-TOF MS identification of individual proteins (University of Georgia Proteomics Facility). In separate experiments, the eluates were analyzed by trypsin digestion in solution followed by LC-MS/MS using a Thermo Finnigan LTQ mass spectrometer with Nanoflow HPLC and nanospray capability. LC-MS/MS data was analyzed with Bioworks/Turbosequest software.
Pulse-chase experiments
The non-cleavable T7-TRA-1A mutant protein, T7-TRA-1A-nc, is a full-length TRA-1A with a deletion of residues 650- 680, which is analogous to the region in Drosophila Ci that includes phosphorylation sites required for Ci binding to Slimb and its subsequent cleavage (Smelkinson and Kalderon, 2006). The analogous region in vertebrate Gli proteins is responsible for binding to β-TrCP1 (the vertebrate Slimb ortholog) and SCFβ-TrCP1-dependent ubiquitination and subsequent cleavage (Jiang, 2006; Wang and Li, 2006). The T7-TRA-1A-nc protein did not undergo cleavage when expressed in 293T cells (data not shown). A truncated ‘pre-cleaved’ TRA-1A, T7-TRA-1-N672, contained the N-terminal 672 residues and produced an ∼90 kDa protein. This truncation is analogous to the tra-1(e2272) actively feminizing allele (Lum et al., 2000; Schvarzstein and Spence, 2006). For pulse-chase experiments, each protein was co-expressed with FLAG-FEM-1 in the presence of LLnL to prevent its degradation by the proteasome. LLnL was removed at time 0, and further protein synthesis was blocked by the addition of 25 μg/ml cycloheximide. The stability of the TRA-1 mutant proteins was assessed by immunoblotting at 0, 4, and 8 hr time points. FLAG-FEM-1 and human CUL2 were analyzed as loading controls. As an additional test of the stability of the TRA-1 isoforms, each TRA-1 mutant protein was co-expressed with FLAG-FEM-1 for 48 hrs without LLnL or cycloheximide and analyzed by immunoblot of whole cell lysate.
RT-PCR
RT-PCR (reverse transcriptase-polymerase chain reaction) was performed according to the protocol provided by Dr. H. Kawahara (Shimada et al., 2006). Total RNA was isolated from 60 adult hermaphrodites and 130 wild-type or cul-2(ek4) adult males (more males were picked to achieve the same mass because males have a smaller body size). Equal portions of total RNA samples were used for RT-PCR. The PCR product of fog-3 corresponds to nucleotides 467 through 680 of coding sequence (spanning introns 5, 6 and 7 in the genomic sequence). The PCR product of mab-3 corresponds to nucleotides 527-764 of coding region (spanning intron 4). The primers for gpd-1, used as a loading and amplification control, were as described (Cram et al., 2003).
Supplementary Material
Acknowledgments
We thank Yuji Kohara, Yue Xiong, and Hiroyuki Kawahara for reagents or protocols; Hui Feng for creating the transgenic cul-2::FLAG strain; Barbara Conradt and the Caenorhabditis Genetics Center for providing strains; Christopher Dowd for technical support; and members of the Kipreos laboratory for critical reading of the manuscript. This work was supported by grants from the American Cancer Society (RSG-01-251-01-DDC) and NIH, National Institute of General Medical Sciences (R01 GM074212) to ETK.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with Image. J Biophotonics International. 2004;11:36–42. [Google Scholar]
- Ahringer J, Rosenquist TA, Lawson DN, Kimble J. The Caenorhabditis elegans sex determining gene fem-3 is regulated post-transcriptionally. Embo J. 1992;11:2303–2310. doi: 10.1002/j.1460-2075.1992.tb05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Ellis RE. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development. 2000;127:3119–3129. doi: 10.1242/dev.127.14.3119. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, Spence AM. Caenorhabditis elegans sex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes Dev. 1996;10:2314–2325. doi: 10.1101/gad.10.18.2314. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Cram EJ, Clark SG, Schwarzbauer JE. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J Cell Sci. 2003;116:3871–3878. doi: 10.1242/jcs.00705. [DOI] [PubMed] [Google Scholar]
- de Bono M, Zarkower D, Hodgkin J. Dominant feminizing mutations implicate protein-protein interactions as the main mode of regulation of the nematode sex-determining gene tra-1. Genes Dev. 1995;9:155–167. doi: 10.1101/gad.9.2.155. [DOI] [PubMed] [Google Scholar]
- DeRenzo C, Reese KJ, Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 2003;424:685–689. doi: 10.1038/nature01887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Edgar LG. Blastomere culture and analysis. Methods Cell Biol. 1995;48:303–321. doi: 10.1016/s0091-679x(08)61393-x. [DOI] [PubMed] [Google Scholar]
- Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt EK, Kipreos ET. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat Cell Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Haag ES, Wang S, Kimble J. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr Biol. 2002;12:2035–2041. doi: 10.1016/s0960-9822(02)01333-7. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. More sex-determination mutants of Caenorhabditis elegans. Genetics. 1980;96:649–664. doi: 10.1093/genetics/96.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1987;1:731–745. doi: 10.1101/gad.1.7.731. [DOI] [PubMed] [Google Scholar]
- Jiang J. Regulation of Hh/Gli Signaling by Dual Ubiquitin Pathways. Cell Cycle. 2006;5:2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- Kimble J, Edgar L, Hirsh D. Specification of male development in Caenorhabditis elegans: the fem genes. Dev Biol. 1984;105:234–239. doi: 10.1016/0012-1606(84)90279-3. [DOI] [PubMed] [Google Scholar]
- Kimble J, Ward S. Germ-line development and fertilization. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 191–213. [Google Scholar]
- Liu J, Vasudevan S, Kipreos ET. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development. 2004;131:3513–3525. doi: 10.1242/dev.01245. [DOI] [PubMed] [Google Scholar]
- Lum DH, Kuwabara PE, Zarkower D, Spence AM. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 2000;14:3153–3165. [PMC free article] [PubMed] [Google Scholar]
- Mathies LD, Schvarzstein M, Morphy KM, Blelloch R, Spence AM, Kimble J. TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development. 2004;131:4333–4343. doi: 10.1242/dev.01288. [DOI] [PubMed] [Google Scholar]
- Mehra A, Gaudet J, Heck L, Kuwabara PE, Spence AM. Negative regulation of male development in Caenorhabditis elegans by a protein-protein interaction between TRA-2A and FEM-3. Genes Dev. 1999;13:1453–1463. doi: 10.1101/gad.13.11.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence Microscopy. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego: Academic Press; 1995. pp. 365–394. [Google Scholar]
- Nelson GA, Lew KK, Ward S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. doi: 10.1016/0012-1606(78)90247-6. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pilgrim D, McGregor A, Jackle P, Johnson T, Hansen D. The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol Biol Cell. 1995;6:1159–1171. doi: 10.1091/mbc.6.9.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M, Spence AM. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Dev Cell. 2006;11:733–740. doi: 10.1016/j.devcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Segal SP, Graves LE, Verheyden J, Goodwin EB. RNA-Regulated TRA-1 nuclear export controls sexual fate. Dev Cell. 2001;1:539–551. doi: 10.1016/s1534-5807(01)00068-5. [DOI] [PubMed] [Google Scholar]
- Shimada M, Kanematsu K, Tanaka K, Yokosawa H, Kawahara H. Proteasomal ubiquitin receptor RPN-10 controls sex determination in Caenorhabditis elegans. Mol Biol Cell. 2006;17:5356–5371. doi: 10.1091/mbc.E06-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr Biol. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Sonneville R, Gonczy P. zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development. 2004;131:3527–3543. doi: 10.1242/dev.01244. [DOI] [PubMed] [Google Scholar]
- Spence AM, Coulson A, Hodgkin J. The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell. 1990;60:981–990. doi: 10.1016/0092-8674(90)90346-g. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- Stothard P, Pilgrim D. Conspecific and interspecific interactions between the FEM-2 and the FEM-3 sex-determining proteins despite rapid sequence divergence. J Mol Evol. 2006;62:281–291. doi: 10.1007/s00239-005-0084-5. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tan KM, Chan SL, Tan KO, Yu VC. The Caenorhabditis elegans sex-determining protein FEM-2 and its human homologue, hFEM-2, are Ca2+/calmodulin-dependent protein kinase phosphatases that promote apoptosis. J Biol Chem. 2001;276:44193–44202. doi: 10.1074/jbc.M105880200. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Starostina NG, Kipreos ET. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 2007;8:279–286. doi: 10.1038/sj.embor.7400895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li Y. Evidence for the direct involvement of βTrCP in Gli3 protein processing. Proc Natl Acad Sci U S A. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Zarkower D. Somatic Sex Determination. WormBook, The C elegans Research Community. 2006 doi: 10.1895/wormbook.1.84.1. WormBook, doi/10.1895/wormbook.1.84.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J. Zinc fingers in sex determination: only one of the two C. elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 1993;21:3691–3698. doi: 10.1093/nar/21.16.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.