BARD1 Participates with BRCA1 in Homology-Directed Repair of Chromosome Breaks (original) (raw)
Abstract
The BRCA1 tumor suppressor has been implicated in the maintenance of chromosomal stability through homology-directed repair of DNA double-strand breaks. Much of the BRCA1 in cells forms a heterodimeric complex with a structurally related protein BARD1. We report that expression of truncated mouse or human BARD1 peptides capable of interacting with Brca1 results in a homologous-repair deficiency. Repair is mildly reduced in Brca1 wild-type cells and severely reduced in cells that harbor a Brca1 splice product deleted for exon 11. Nuclear localization of the Brca1 or BARD1 peptides is not compromised, implying that the repair deficiency is caused by a more direct effect on repair. The tumor suppressor activity of BRCA1 may require the participation of BARD1 to maintain chromosome integrity through the homologous-repair pathway.
Built-in safeguards of genetic stability include the intrinsic structure of the DNA double helix and controlled access through chromatin, as well as multiple damage-specific repair pathways. One pathway of repair is homologous recombination (also termed homology-directed repair [HDR]), a process that can both generate genetic diversity during germ cell development and maintain genetic stability through the precise repair of chromosome breaks. Chromosomal breaks, including double-stand breaks (DSBs), can occur spontaneously as a result of stalled replication forks during S phase and after exposure to DNA-damaging agents (27).
Individuals who carry mutations in the hereditary breast cancer gene, BRCA1, are predisposed to early onset breast and ovary cancer (30). The BRCA1 protein is implicated in varied cellular functions that include DNA repair, transcriptional regulation, ubiquitination, and chromatin remodeling, although its precise role in tumor suppression remains speculative (2, 46). Cells that are deficient in Brca1 readily develop spontaneous chromosome abnormalities and are deficient in HDR of chromosome DSBs, suggesting that this repair defect may promote ongoing genetic instability in cells ultimately leading to tumorigenesis (19, 31).
Despite its overall poor sequence conservation, the N-terminal 100 amino acids of BRCA1 are highly conserved with 97% similarity to the murine sequence, including an identical 42-amino-acid RING motif (47). RING motifs are cysteine-rich zinc-binding domains that are known to mediate protein-protein interactions (43). Although the function of many RING motif-containing proteins is not clear, several have recently been implicated in the ubiquitin pathway, including the BRCA1 RING domain, which was shown to facilitate E2-dependent ubiquitination in vitro (25). The BRCA1-associated RING domain protein BARD1 is a protein of 777 amino acids that also has a highly conserved RING motif at the N terminus, as well as three internal ankyrin repeats and, like BRCA1, two C-terminal BRCT repeats (59). BARD1 interacts with BRCA1 over a larger N-terminal region than that which encompasses the RING motif to form a stable heterodimer (6, 29, 59).
Much of the BRCA1 in the cell is in a heterodimeric complex with BARD1 (61). Evidence suggests that the BRCA1-BARD1 heterodimer is the physiologically relevant form of BRCA1, since this interaction has been shown to be important for ubiquitin ligase activity (8, 15) and mutual protein stability (15, 22, 60). Furthermore, tumor-associated missense mutations located in the BRCA1 RING confer radiation sensitivity (45). Importantly for DNA repair, BARD1 colocalizes with BRCA1 and the HDR protein RAD51 in S-phase cells and in cells after DNA damage (21, 44). Phenotypically, Xenopus oocytes made deficient for xBARD1 by the introduction of antisense molecules show developmental problems similar to those seen with an xBRCA1 deficiency (22). Furthermore, _Bard1_-null mice generated by targeted mutagenesis display a characteristic phenotype, including early embryonic lethality that is essentially indistinguishable from that of _Brca1_-null animals (28). These results suggest an obligatory role for the BRCA1-BARD1 heterodimer for the function of either molecule.
As for BRCA1, the functional analysis of BARD1 is not yet complete. BARD1, in association with BRCA1, has been shown to reduce mRNA processing during the DNA damage response by directly binding and inhibiting CstF-50, a factor that normally stimulates 3′ cleavage of mRNA precursors (24). Also, multiple studies have confirmed that the BRCA1-BARD1 heterodimer functions as an E2-dependent E3 ubiquitin ligase (2, 8, 15, 26, 60). However, the significance of this activity to in vivo processes is not yet clear, especially since the functional consequences of ubiquitination are extremely varied; examples of this include protein degradation, repair activation, transcription regulation, and cell cycle control (36). Given the colocalization of BARD1 with BRCA1 and Rad51 in S-phase nuclei and their redistribution to common nuclear sites after DNA damage (21, 44), we hypothesize that BARD1 may also play a role in the HDR of spontaneous and induced DNA DSBs.
We have previously examined DSB repair at the molecular level in wild-type murine ES cells and in cells that are hypomorphic for Brca1 and found that HDR of chromosome DSBs is reduced in the Brca1 mutant cells (32, 33). We now report that transient expression of a truncated Bard1 peptide capable of interacting with Brca1 can decrease HDR of an induced chromosome break in wild-type ES cells. Expression of the truncated Bard1 peptide further diminishes HDR in the Brca1 mutant cells, which are deleted for exon 11, suggesting that much of the residual HDR activity in these cells is mediated by the Brca1 exon 10-exon 12 spliced product. In addition, a truncated human BARD1 peptide produces a similar dominant-negative effect on repair. These results implicate the BRCA1-BARD1 heterodimer as a functional unit in homologous recombination and DNA repair.
MATERIALS AND METHODS
Plasmid construction.
The hBARD1 and mBard1 expression vectors were constructed by modifying the pCAGGS vector (35) to contain an additional polylinker, pCAGGS-Not. All Bard1 expression plasmids encode an amino-terminal FLAG epitope. _Not_I/_Kpn_I fragments containing the full-length mBard1 cDNA or a truncated mBard1 encoding residue 1 to 301 _Not_I/_Mlu_I fragments containing the wild type or mutated full-length human BARD1 cDNA were inserted into the pCAGGS-Not vector to generate pCAG-mBard1, pCAG-mB301, and pCAG-hBARD1, respectively (in the text and the subsequent methods the reference to the pCAG plasmid backbone for all vectors is deleted). hBARD1-C83G and hBARD1-Q564H that contain the Cys83Gly and the Gln564His missense mutations, respectively, were generated from wild-type human BARD1 sequences by using the Chameleon double-stranded site-directed mutagenesis kit (Stratagene). A _Bgl_II digestion and religation of hBARD1 and hBARD1-C83G generated the hB202 and hB202-C83G expression vectors, respectively, by eliminating amino acids 203 to 777 of hBARD1. hB202-L107P was generated by using the Chameleon double-stranded site-directed mutagenesis kit. A gene-targeting construct, p59x-hB202, was created by inserting a _Hin_cII/_Stu_I fragment from hB202 into the _Sma_I site in the pim1 targeting vector p59x (34).
Cell lines, transfections, and nucleic acid analysis.
Brca1+/+ and Brca1−/− (14) embryonic stem (ES) cell lines containing the direct repeat-green fluorescent protein (DR-GFP) reporter targeted to the pim1 locus have been described (33). To generate ES cell lines stably expressing the truncated hBARD1 peptide, hB202, E14-DRGFP cells (37) were electroporated with 70 μg of linear p59x-hB202 targeting fragment and selected in 110 μg of hygromycin per ml 48 h after transfection. Individual clones were expanded, and targeted clones were identified by Southern blotting of genomic DNA digested and hybridized with a radiolabeled pim1 probe (34). For DSB repair assays, actively growing cells were electroporated at 250 V and 950 μF with 30 μg of the I-_Sce_I expression vector, pCβASce (40), and 60 μg of empty vector DNA (pCAGGS) or BARD1 expression plasmids (mBard1, mB301, hBARD1, hBARD1-C83G, hBARD1-Q564H, hB202, hB202-C83G, and hB202-L107P). Transfections were also performed with FuGene6 transfection reagent (Roche). Typically, 1.5 to 3 μg of the I-_Sce_I expression vector was cotransfected with 3 to 6 μg of control DNA or BARD1 expression plasmids at a ratio of 2.4 μl of FuGene6 to 1 μg of total DNA. Electroporation and FuGene6 transfection gave comparable results. To assess transfection efficiencies pNZE-CAG (38), a plasmid that expresses GFP was transfected as a positive control. Cells were collected by trypsinization 36 to 72 h after transfection and analyzed by flow cytometry in a Becton Dickinson FACScan by two-color fluorescence analysis on a green (FL1) versus orange (FL2) plot. The data were analyzed by using CellQuest software (Becton Dickinson).
Protein manipulations.
Whole-cell extracts were prepared from cells 24 to 36 h after transfection. The total protein content was determined by Bio-Rad protein assay. For Western blot analysis of mBard1 and hBARD1 proteins, 30 μg of lysate was separated by Tris-glycine-8% polyacrylamide gel electrophoresis (PAGE). Western analysis of Brca1 was performed by 6% PAGE. Cytoplasmic and nuclear protein was prepared 24 h after transfection by using NE-PER nuclear and cytoplasmic extraction reagents (Pierce). A total of 30 μg of fractionated protein lysate was separated by 8% PAGE for full-length hBARD1 protein, 6% PAGE for endogenous Brca1 protein, and 10% PAGE for truncated hBARD1 peptides. Immunoprecipitations were performed with 400 μg of whole-cell extracts and 35 μl of anti-FLAG-M2 agarose affinity gel (A-2220; Sigma) overnight at 4°C. Immune complexes were washed with lysis buffer and separated by Tris-HCl-4 to 20% PAGE. The membranes were probed with anti-FLAG-M2 antibody (A-8592; Sigma), anti-BARD1 antibody (H-300; Santa Cruz), and mouse Brca1 GH118 antibody (a gift from S.Ganesan, Dana-Farber Cancer Institute, Boston, Mass.). Anti-tubulin antibody (T-9026; Sigma) was used to assess protein loading.
MMC survival assay.
Actively growing cells were exposed to various mitomycin C (MMC) concentrations for 4 h, followed by three rinses with phosphate-buffered saline. The cells were replated at low density and allowed to grow undisturbed for 8 to 10 days. Clones were stained and counted. Survival experiments were performed in triplicate.
Two-hybrid analysis.
Mammalian expression plasmids that encode the DNA-binding domain of GAL4 fused to residues 1 to 202 of human BARD1 were constructed by inserting wild-type and mutant BARD1 cDNA fragments into pGAL4-A, a derivative of the pCMV-GAL4 vector (59). Each of the three resulting plasmids was then used for mammalian two-hybrid analysis, together with a plasmid (BR304/pVP-Flag) encoding the VP16 transactivation domain fused to BRCA1 residues 1 to 304 (59). Mammalian two-hybrid assays were conducted in 293 cells as described previously (54).
Statistical analysis.
The difference in the cell survival rate after exposure to MMC between the parental cell line and the combined data from two hB202 stable cell lines was assessed by using the Wilcoxon rank sum test. Because we expected the parental line to be less sensitive, a one-sided test was used. For transient cotransfections of the I-_Sce_I expression vector and the mBard1 or hBARD1 expression plasmids, differences in percent GFP-positive cells between expression of the full-length mBard1 or hBARD1 and the truncated and/or mutated mBard1 or hBARD1 peptides, respectively, were also quantitated by using the same test. All of these tests were two sided. The cutoff for significance for all tests was 0.05.
RESULTS
HDR of a chromosomal DSB is diminished by expression of a truncated Bard1.
Full-length cDNAs encoding murine Bard1 (mBard1) and a truncated mBard1 containing the first 301 amino acids (mB301) were constructed with an N-terminal FLAG epitope and expressed in ES cells (Fig. 1A). The products of these cDNAs are expected to interact with Brca1 at the N-terminal RING and flanking sequences. However, since the ankyrin and BRCT repeats of Bard1 have been deleted from the mB301 construct, we expect that expression of the truncated mB301 peptide will substitute for endogenous Bard1 in the formation of the Brca1-Bard1 heterodimer and may thereby function as a dominant-negative peptide. Specifically, if the Brca1-Bard1 interaction is important for HDR, we should observe a decrease in the ability of cells to repair chromosome breaks.
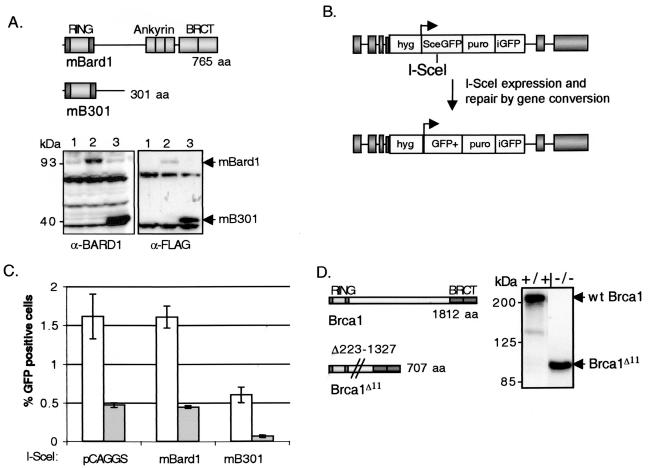
FIG. 1.
Overexpression of truncated mBard1 decreases HDR in Brca1+/+ and Brca1−/− cells. (A) Schematic drawing of wild-type murine Bard1 and an N-terminal fragment of mBard1 (mB301). The RING domains of both Bard1 and Brca1 consist of a central RING motif, which binds two zinc atoms flanked by antiparallel α-helices, which are indicated by the darker shading. Bard1, like Brca1, contains an N-terminal RING domain and two C-terminal BRCT repeats. Bard1 also has three central tandem ankyrin repeats. mB301 contains amino acids 1 to 301 of the Bard1 protein, which retains the N-terminal RING interacting domain but deletes the C-terminal domains. Both mBard1 constructs have an N-terminal FLAG epitope (not depicted). Representative Western blots of Brca1+/+ cell extracts after transient transfection of the FLAG-tagged mBard1 and mB301 expression plasmids are shown. In the left panel, an anti-BARD1 antibody was used to demonstrate endogenous mBard1expression (lane 1) compared to overexpression of a full-length 93-kDa mBard1 protein (lane 2) and a 40-kDa truncated mB301 peptide (lane 3). The amount of endogenous mBard1 protein does not appear to be affected by expression of the truncated mB301 peptide (lane 3). An anti-FLAG-M2 antibody was also used to detect exogenous mBard1 peptide expression (right panel). Lane 1 depicts cell lysates transfected with the empty vector, pCAGGS. The expression levels of the peptides were similar for both Brca1+/+ and Brca1−/− cells (see Fig. 3). (B) The gene conversion repair substrate, p59x-DR-GFP6 (34), is gene targeted to chromosome 17 at the pim1 locus in both the Brca1+/+ and Brca1−/− cells. After in vivo expression of the I-_Sce_I endonuclease, a chromosome DSB is introduced at the I-_Sce_I site in SceGFP. If the break is repaired by HDR with the donor iGFP gene, a functional GFP+ gene is created. This repair event can be observed by using cellular green fluorescence and then quantitated by flow cytometry. (C) mBard1 and mB301 constructs were transiently cotransfected with an I-_Sce_I expression vector in Brca1+/+ (open bars) or Brca1−/− (gray bars) cells. Green fluorescent cells that have undergone HDR by gene conversion can be detected by two-color fluorescence analysis 48 h after transfection. When mB301 is expressed in Brca1+/+ cells HDR, as deduced from the percentage of GFP-positive cells, was reduced 3.3-fold (P = 0.03). The Brca1−/− cells are deficient in HDR (33) due to the targeted disruption of full-length Brca1. The ability to perform HDR was further decreased 7.3-fold when mB301 was expressed compared to the Brca1−/− cells expressing full-length mBard1 (P = 0.03) and 24-fold compared to the Brca1+/+ cells transfected with control DNA. Error bars represent the standard error from four replicate experiments. (D) Schematic drawing of wild-type murine Brca1 and an exon 11-deleted Brca1 splice product with the remaining N-terminal RING domain and C-terminal BRCT repeats. In Brca1−/− cells, exon 10 sequences are spliced directly to exon 12, deleting exon 11, which encodes amino acids 223 though 1327. A Western blot of whole-cell extracts from Brca1+/+ (+/+) and Brca1−/− (−/−) ES cells depicts expression of the 210-kDa wild-type (wt) Brca1 and the 92-kDa Brca1Δ11 spliced product with a Brca1 antibody.
To test the hypothesis that Bard1, through its heterodimeric partnering with Brca1, has a role in HDR, we used a previously reported in vivo repair assay that measures gene conversion repair of an induced chromosome break. The DR-GFP reporter substrate is composed of two differentially mutated GFP genes oriented as a direct repeat (38). A site for the rare cutting endonuclease I-_Sce_I mutates the upstream GFP (SceGFP) gene, and a GFP gene fragment (iGFP) truncated at both the 5′ and 3′ ends is located downstream (Fig. 1B). Expression of I-_Sce_I in cells that have the DR-GFP substrate integrated into their genome results in a DSB in the chromosome at the position of the I-_Sce_I site. Repair of the induced DSB in SceGFP by noncrossover gene conversion with iGFP reconstructs a functional GFP+ gene that can be scored by flow cytometry. Although other DSB repair events at the I-_Sce_I site are possible, they are not detected since the 11-bp substitutions in the SceGFP gene cannot be restored to the wild-type GFP sequence except through a templated gene conversion event (34, 38).
To assay the effect of the truncated Bard1 peptide on HDR, an I-_Sce_I expression vector was transiently cotransfected with the mB301 expression vector, or as controls, with the full-length mBard1 expression vector or the empty vector (pCAGGS), into wild-type Brca1+/+ ES cells containing a DR-GFP repair substrate (33). Expression was verified by Western blotting with both anti-BARD1 to detect endogenous and transfected mBard1 proteins and anti-FLAG to assess only the transfected mBard1 peptides (Fig. 1A, left and right panels). After transfection with molar equivalents of each expression plasmid, the full-length 93-kDa mBard1 and the 40-kDa mB301 peptide were overexpressed compared to endogenous Bard1 (Fig. 1A, left panel). The truncated mB301 peptide was typically expressed at higher levels than the full-length 93-kDa mBard1 (Fig. 1A, left and right panels).
Electroporated cells were typically examined 48 h after transfection by flow cytometry. GFP-positive cells were not detected or rarely detected (<0.01%) in any of the transfected cell lines in the absence of I-_Sce_I expression (data not shown), indicating that spontaneous intrachromosomal gene conversion is rare. After coexpression of I-_Sce_I and the empty vector, GFP-positive cells were readily detected in the Brca1+/+ cells, indicating robust HDR of the induced DSB. Coexpression of I-_Sce_I and full-length mBard1 had no effect on the frequency of HDR compared to I-_Sce_I coexpression with the empty vector (Fig. 1C). However, coexpression of I-_Sce_I and the truncated mB301 in Brca1+/+ cells resulted in a 3.3-fold decrease in the number of GFP-positive cells observed compared to expression of full-length mBard1, indicating a decrease in HDR at the induced chromosome break (P = 0.03) (Fig. 1C).
There were no cell growth changes observed by either cell counts or cell morphology by light microscopy at the time of flow cytometric analysis (data not shown). Repression of Bard1 expression by antisense approaches has been noted to alter cell morphology and inhibit S-phase progression of the cell cycle, and BARD1 overexpression has been shown to increase apoptosis in several cell lines (17, 18). As a further check for cellular effects, therefore, we also transiently cotransfected either the full-length Bard1 or truncated Bard1 expression vector with a GFP expression vector. This vector utilizes the same control elements to express GFP as the I-_Sce_I and Bard1 expression vectors. We did not observe any decrease in the number of GFP positive cells during the time course of the HDR analysis, indicating no significant effect on cellular viability or proliferation (data not shown).
HDR of a chromosomal DSB in Brca1−/− cells is diminished further by expression of truncated Bard1.
Using the same assay system described above, we previously demonstrated that HDR of a chromosome DSB is reduced in the Brca1 mutant ES cell line 236.44 (33). This Brca1 mutant cell line, like other BRCA1 mutant cells, is hypersensitive to DNA-damaging agents, including cisplatin and MMC, and incurs spontaneous chromosome damage (3, 33). This cell line, formally designated Brca1Δ223-763/Δ223-763 (14) but simplified here as Brca1−/−, expresses an alternatively spliced Brca1 transcript that skips exon 11 and which is predicted to encode an internally deleted peptide (Fig. 1D) (48). Therefore, both the Brca1 N-terminal RING domain, which heterodimerizes with Bard1, and the C-terminal BRCT repeats would remain intact in the Brca1Δ11 peptide.
HDR of an induced break, as well as damage-induced Rad51 focus formation, is reduced about five- to sixfold in the Brca1−/− cell line compared to wild-type cells but is not completely abolished (3, 32, 33). Thus, either the Brca1Δ11 product retains residual repair activity or the residual HDR arises from Brca1-independent homologous repair pathways. To determine whether the residual HDR activity detected in the Brca1−/− cells could be further diminished by alterations to the Brca1-Bard1 complex, cotransfection of the I-_Sce_I and the mBard1 expression vectors was performed. Relative to the Brca1+/+ cells, the Brca1−/− cells exhibited a decrease in GFP-positive cells after I-_Sce_I coexpression with empty vector and mBard1 (Fig. 1C), in concordance with our prior reports with this repair substrate (33). The number of GFP-positive cells was not further reduced by expression of full-length mBard1. However, coexpression of the mB301 peptide and I-_Sce_I reduces HDR to an even greater extent in these cells (7.3-fold) compared to coexpression of full-length mBard1. Overall, a 24-fold reduction in HDR was observed in the Brca1−/− cells expressing a truncated Bard1 peptide compared to Brca1+/+ cells transfected with the empty vector (Fig. 1C). Since the RING domain of the Brca1Δ11 peptide is intact in the Brca1−/− cells, these results are consistent with mB301 inhibition of HDR through its interaction with Brca1.
Similar transfection efficiencies were observed as measured by GFP expression after cotransfection with a GFP expression vector and either the mBard1 or mB301 expression vectors (data not shown). No alteration in cell recovery was noted after the control cotransfections, confirming that mBard1 or mB301 transient overexpression did not result in significant apoptosis or gross alterations in cell cycle progression (data not shown).
Truncated human BARD1 can interfere with Brca1-dependent HDR.
Overall, the human and murine BARD1 proteins are poorly conserved although, like BRCA1, the amount of identity is much higher in specific regions, i.e., the RING, ankyrin, and BRCT domains (1, 18). Both mouse Bard1 and frog BARD1 interact with human BRCA1, supporting the high degree of evolutionary conservation observed within the domains that direct heterodimerization (1, 22). To assess the ability of human BARD1 to interact and affect the function of mouse Brca1, we expressed FLAG-tagged full-length BARD1 (hBARD1) and an N-terminal BARD1 peptide that is truncated at amino acid 202 (hB202) (Fig. 2A). Expression of both peptides was verified by Western blotting with an anti-FLAG antibody (Fig. 2B) or a human BARD1 antibody (data not shown). As with mB301, the N-terminal hB202 peptide was expressed at higher levels compared to the full-length hBARD1. To confirm that the truncated and full-length hBARD1 peptides interact with endogenous Brca1, the FLAG-tagged hBARD1 peptides were immunoprecipitated with anti-FLAG antibody from extracts of Brca1+/+ and Brca1−/− cells after transient transfection and analyzed by Western blotting (Fig. 2C). Immunoblotting revealed that both wild-type Brca1 and the Brca1Δ11 peptide interact with the hBard1 and hB202 peptides.
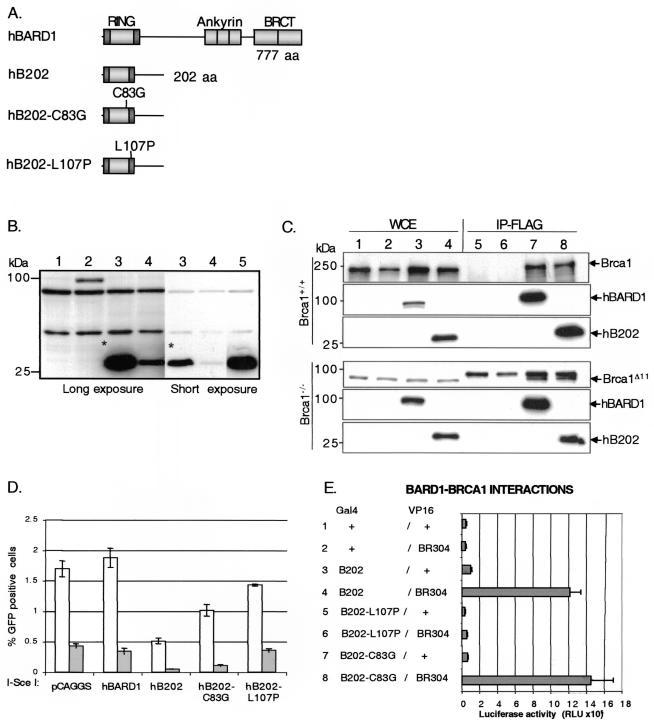
FIG. 2.
Expression of truncated human BARD1 peptides that interact with Brca1 are dominant-negative for HDR. (A) Schematic drawing of full-length human BARD1 (hBARD1), an N-terminal fragment of hBARD1 (hB202), an N-terminal fragment of hBARD1 with a point mutation in a zinc-binding residue in the RING motif (hB202-C83G), and an N-terminal fragment containing a point mutation in the second BRCA1-interacting α-helix of BARD1 (hB202-L107P). All peptides have an N-terminal FLAG epitope (not shown). (B) Western analysis with a FLAG antibody with Brca1+/+ cell lysates after transient transfection of the hBARD1 peptides. Lanes: 1, control DNA; 2, full-length hBARD1; 3, hB202; 4, hB202-C83G; 5, hB202-L107P. The hB202-C83G peptide (lane 4) was consistently expressed at lower levels, and the hB202-L107P peptide (lane 5) was consistently expressed at higher levels compared to the hB202 peptide (the asterisk denotes a comparison of short and long exposures of hB202). The peptides were expressed at similar levels in Brca1+/+ and Brca1−/− cells (see Fig. 3). (C)Whole-cell extracts (WCE) of Brca1+/+ and Brca1−/− cells were obtained after transient transfection of control DNA and hBARD1 peptides. Lanes: 1 and 5, untransfected; 2 and 6, control DNA; 3 and 7, full-length hBARD1; 4 and 8, hB202. To detect an interaction with Brca1, extracts were immunoprecipitated (lanes 5 to 8) with anti-FLAG-M2 antibody (IP-FLAG). For Western analysis, anti-FLAG antibody was used to detect the FLAG-tagged hBARD1 and hB202 peptides, and anti-Brca1 antibody (GH118) was used to detect endogenous Brca1. Immune complexes generated from untransfected (lane 5) and control DNA transfected (lane 6) cell extracts revealed no detectable Brca1 protein, whereas immune complexes generated from transfection with hBARD1 (lane 7) and hB202 (lane 8) cell extracts revealed an association with Brca1 and Brca1Δ11. A cross-reacting band that was slightly larger than the Brca1Δ11 product was observed in all extracts. (D) The hBARD1 and hB202 constructs were transiently coexpressed with an I-_Sce_I expression vector in Brca1+/+ (open bars) or Brca1−/− (gray bars) cells. HDR events were scored as GFP-positive cells 48 h after transfection as described in Fig. 1. When the N-terminal fragment of hBARD1, hB202, was expressed in Brca1+/+ cells, a 3.7-fold decrease in HDR was observed compared to expression of hBARD1 (P = 0.0012). The defect was more pronounced in Brca1−/− cells, which exhibited an 8.6-fold decrease compared to the Brca1−/− cells transfected with hBARD1 (P = 0.00018) and a 36-fold decrease compared to the Brca1+/+ control transfection. The point mutation in the RING of hB202 (hB202-C83G) showed a weaker, but significant defect in HDR in Brca1+/+ (1.8-fold, P = 0.0043) and Brca1−/− (3.3-fold, P = 0.0058) cells. The hB202-L107P point mutation that disrupts the interaction with the second Brca1 α-helix abolished the defect in HDR observed in the Brca1+/+ and Brca1−/− cells. Error bars represent the standard error. (E) Mammalian two-hybrid analysis with the GAL4 DNA-binding domain fused to B202 (BARD1 amino acids 1 to 202) and the VP-16 transactivation domain fused to BR304 (BRCA1 amino acids 1 to 304). Luciferase activity after cotransfection of the hybrid peptides and a GAL4-luciferase reporter plasmid in 293 cells was determined in triplicate experiments. Both B202 (lane 4) and B202-C83G (lane 8) mutation retain a strong interaction with BRCA1, whereas the B202-L107P mutation (lane 6) abolishes the interaction with BRCA1. Error bars represent the standard deviation of six values.
To determine whether full-length or truncated hBARD1 affects HDR in mouse cells, Brca1+/+ and Brca1−/− ES cells were cotransfected with the expression vectors for the hBARD1 peptides and I-_Sce_I. As with full-length mBard1, expression of hBARD1 had no effect on chromosome break repair by HDR or cell proliferation in either cell line (Fig. 2D and data not shown). However, the truncated hBARD1 peptide, hB202, resulted in decreased HDR in the Brca1+/+ and the Brca1−/− cells, a finding similar to that for truncated mBard1. The Brca1+/+ cells revealed a 3.7-fold decrease in the number of GFP-positive cells compared to cells transfected with full-length hBARD1. The Brca1−/− cells showed an even more pronounced defect than was seen with truncated mBard1; an 8.6-fold decrease in HDR was observed when the truncated hB202 peptide was expressed compared to expression of hBARD1. When compared to Brca1+/+ cells transfected with hBARD1, the Brca1−/− cells expressing the hB202 peptide exhibited a 36-fold reduction in the HDR of an I-_Sce_I-induced chromosome break (Fig. 2D). These results demonstrate that expression of a truncated hBARD1 peptide can significantly decrease repair of a chromosome DSB by the interruption of the murine Brca1-Bard1 heterodimer. In addition, these results reinforce the hypothesis that BARD1 sequences downstream from the RING domain are required for proper repair function by the complex.
Disruption of the BRCA1-BARD1 interaction abolishes the HDR defect.
Although the zinc binding core regions of the RING motifs have a role in BRCA1-BARD1 interaction, structural studies of the BRCA1-BARD1 heterodimeric RING domain have determined that flanking α-helices in both BRCA1 and BARD1 form a four-helix bundle which is the main interaction interface (6). To further support the hypothesis that the observed defect in HDR after expression of a truncated hBARD1 peptide is due to its interaction with endogenous Brca1, a point mutation that disrupts one of the BRCA1-interacting α-helices of BARD1 was constructed by substituting a leucine at position 107 with a helix-destabilizing proline residue (hB202-L107P). By disrupting the helix, this mutation is predicted to block formation of the BRCA1-BARD1 four-helix bundle and thereby prevent the association of hB202-L107P with Brca1. To test this prediction, we performed a mammalian-two hybrid experiment with hybrid polypeptides containing the first 304 amino acids of BRCA1 fused to the VP16 transactivation domain and the BARD1 truncations hB202 and hB202-L107P fused to a GAL4 DNA-binding domain. Cotransfection with the BRCA1 and BARD1 expression vectors in 293 cells, together with a GAL4-responsive luciferase reporter, confirmed that the wild-type GAL4-hB202 fragment interacts strongly with BRCA1 and that the L107P mutation abolishes this interaction (Fig. 2E).
To examine the effect on HDR, hB202-L107P was coexpressed with I-_Sce_I in Brca1+/+ and Brca1−/− cells. No significant decrease in HDR, as measured by cellular fluorescence, was observed (Fig. 2D). Western blot analysis revealed consistently higher expression of the hB202-L107P peptide compared to the hB202 peptide (Fig. 2B, lane 5), eliminating the possibility that poor expression or decreased stability of the hB202-L107P peptide abrogated the HDR defect.
Brca1 nuclear localization is not affected by transient overexpression of BARD1.
BRCA1 is a nuclear protein that contains two nuclear localization signals (NLSs) located within exon 11 (53). BRCA1 also contains a nuclear export signal (NES) that can function as a shuttle for the protein from the nucleus to the cytoplasm (41). The BRCA1 NES is located in the RING domain, with its function presumed to be shielded by its interaction with BARD1 (10). An NLS for BARD1 has yet to be reported. Splice variants of mouse or human BRCA1 that lack exon 11 can localize to the nucleus, implicating inefficient nuclear export of these peptides, additional NLSs, or use of alternative nuclear transport pathways (10, 16, 56, 58).
To confirm that the defects in DNA repair we observed were not due to improper localization of either endogenous Brca1 or the transfected BARD1 peptides, cellular fractionation and immunoblotting was performed on the transfected ES cells. As shown in Fig. 3, endogenous Brca1 and Brca1Δ11 were localized to the nucleus. Likewise, the transiently expressed FLAG-tagged BARD1 peptides hBARD1, hB202, and hB202-L107P were predominantly nuclear, although a small proportion of each could be observed in the cytoplasmic fraction. Importantly, expression of hBARD1 and hBARD1 truncations that interact with Brca1 (hBARD1 and hB202) and that do not interact with Brca1 (hB202-L107P) does not affect the nuclear localization of endogenous Brca1 or Brca1Δ11 (Fig. 3). These data support the interpretation that the HDR defect is a direct dominant-negative effect due to heterodimer formation between nuclear Brca1 and truncated BARD1 and not due to mislocalization.
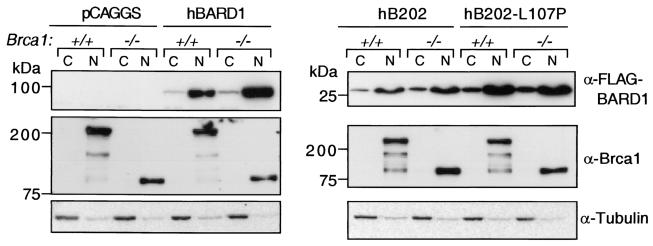
FIG. 3.
Nuclear localization of Brca1 and exogenous BARD1 peptides. Cytoplasmic (C) and nuclear (N) protein fractions were prepared from Brca1+/+ (+/+) and Brca1−/− (−/−) cells after transient transfection with control (pCAGGS), hBARD1, hB202, or hB202-L107P expression vectors. hBARD1, hB202, and hB202-L107P, as detected by anti-FLAG-M2 antibody, are predominantly nuclear in the Brca1+/+ and Brca1−/− cells (top panel). Expression or localization of endogenous Brca1 proteins, either the 210-kDa wild-type (Brca1+/+) or the 92-kDa Brca1Δ11 (Brca1−/−), was not affected by overexpression of full-length or truncated hBARD1. Brca1 was located exclusively in the nucleus. hB202-L107P showed higher expression levels than hB202, a finding consistent with Fig. 2B. The protein amount and fractionation procedure were analyzed by staining with anti-tubulin antibody (bottom panel).
Stable expression of truncated BARD1 results in MMC hypersensitivity.
Brca1+/+ cells which stably express the truncated hBARD1 peptide were generated to assess other phenotypes associated with defects in HDR. The hB202 expression cassette was introduced into a pim1 locus gene targeting vector containing a linked hygromycin resistance gene. Both targeted and nontargeted hygromycin-resistant clones (data not shown) were analyzed by Western blotting to confirm protein expression. All stable clones analyzed by Western blot analysis showed similar low expression levels (Fig. 4A, lanes 3 and 4, and data not shown), compared to hB202 expression achieved after transient transfection (Fig. 4A, lane 2). This might suggest that high stable expression of the dominant-negative hBARD1 peptide is toxic to cells, although this was not observed with transient transfections, or it may reflect the particularly robust level of expression achieved by transient transfection. Similar low levels of expression were observed for cells stably expressing the truncated mBard1 peptide, mB301 (data not shown).
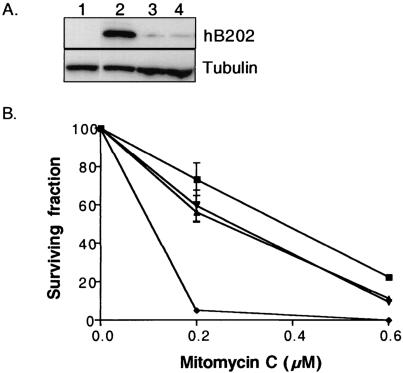
FIG. 4.
Stable expression of a truncated hBARD1 results in MMC hypersensitivity. (A) Comparison of hB202 expression in Brca1+/+ cells after transient transfection with the empty vector pCAGGs (lane 1), hB202 (lane 2), and cells selected for stable hB202 expression (lane 3, clone 6B, and lane 4, clone 9B). Multiple hygromycin-resistant cell lines that contained a stable integration of the hB202 construct were analyzed for expression of the hB202 peptide by Western analysis. None of the isolated cell lines exhibited strong expression of the hB202 peptide (representative clones, lanes 3 and 4), as was typically observed for transiently transfected cells (lane 2). (B) Clonogenic survival of parental control cells (▪), two independently isolated cell lines stably expressing hB202 (6B [▴] and 9B [▾]) and Brca1−/− cells (⧫) were assessed after treatment with two doses of MMC. At the higher 0.6 μM MMC concentration, the cells that stably express low levels of hB202 were significantly more sensitive than wild-type cells (twofold; P = 0.01). At the lower 0.2 μM MMC concentration, the difference in MMC sensitivity between the cells stably expressing low levels of the hB202 compared to wild-type cell was not statistically significant (P = 0.08). The Brca1−/− cells were highly sensitive to both doses of MMC compared to the parental cells and cells that stably express low levels of hB202. The error bars represent the standard error.
Vertebrate cells deficient in HDR are markedly hypersensitive to cross-linking agents, as previously demonstrated for the Brca1−/− cells (33). To assess cross-linking hypersensitivity in the stable hB202 expressing clones, a targeted (6B) and a randomly integrated clone (9B) were tested for clonogenic survival after exposure to MMC along with parental cells and Brca1−/− cells. At the higher dose of 0.6 μM MMC, the two hB202-expressing cell lines were significantly more sensitive than the parental cell line (P = 0.01) (Fig. 4B) but much less sensitive than the Brca1−/− cells. There was no statistically significant difference in survival between the parental control cells and the two hB202 cell lines when exposed to 0.2 μM MMC (P = 0.08). The hB202 cell lines had a similar decrease in survival percentage compared to the control cells; however, the variance in the data at 0.2 μM was greater, thereby increasing the P value. Similar results were obtained in cells with stable expression of dominant-negative mB301 (data not shown). The relatively mild degree of MMC hypersensitivity observed in the hB202-expressing cells may be explained by the low expression of the truncated BARD1 peptide in the presence of endogenous wild-type Bard1.
Analysis of BARD1 mutations in HDR: Cys83Gly and Gln564His.
BRCA1 missense mutations at key cysteine residues residing in the RING motif are associated with human tumors (7, 12). Although these zinc-binding residues do not directly mediate the BRCA1-BARD1 interaction (5), mutations in these residues have been shown to interfere with the interaction of BRCA1 with RING interacting proteins (20, 59) and to abolish the formation of supramolecular assemblies of BRCA1 and BARD1 (23). These mutations also disturb BRCA1 function for both radiation resistance (42, 45) and ubiquitin ligase activity (15, 42).
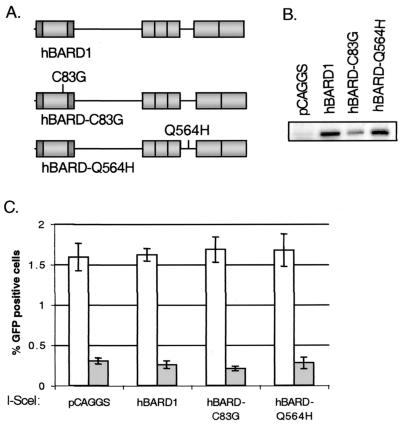
To begin to determine whether the zinc-binding cysteine residues in the BARD1 RING domain are important for its role in HDR, the C83G point mutation was introduced into the full-length human BARD1 (hBARD1-C83G) and into the truncated BARD1 (hB202-C83G) (Fig. 5A and 2A). As shown in Fig. 2E, hybrid polypeptides containing the first 304 amino acids of BRCA1 bind equally well to the hB202 and hB202-C83G peptides in the mammalian two-hybrid assay. However, in the context of our standard expression system, we found that both the hBARD-C83G and hB202-C83G peptides were underexpressed relative to the cognate hBARD1 and hB202 peptides, respectively (Fig. 2B and 5B). Cotransfection of I-_Sce_I and the hBARD1-C83G expression vectors in the Brca1+/+ and Brca1−/− cells showed no significant decrease in the number of GFP-positive cells, indicating that HDR was not significantly affected by the mutated full-length hBARD1, at least when expressed at this level (Fig. 5C). However, when the truncated hB202-C83G was coexpressed with I-_Sce_I in Brca1+/+ and Brca1−/− cells we observed an intermediate defect in HDR compared to expression of the hB202 peptide. The defect in HDR in Brca1+/+ cells was 1.8-fold (P = 0.004) and in Brca1−/− cells was 3.3-fold (P = 0.006) compared to control transfections in the respective cell lines with full-length hBARD1 (Fig. 5C). This indicates that the truncated mutant BARD1 is still able to interfere with HDR, albeit less well, probably as a result of its lower expression level.
FIG. 5.
Overexpression of mutant full-length hBARD1 does not effect HDR. (A) Schematic drawing of hBARD1, hBARD1 with a RING domain mutation (hBARD1-C83G) and the hBARD1 tumor-associated point mutation (hBARD1-Q564H). The Q564H mutation is located between the ankyrin and BRCT repeats. (B) As with the hB202-C83G peptide, hBARD1-C83G was expressed at lower levels compared to hBARD1 and hBARD1-Q564H as depicted in this representative Western analysis with an anti-FLAG antibody of Brca1+/+ cell lysates after transient transfection of hBARD1 expression plasmids. (C) HDR as deduced from the number of GFP-positive cells after transient cotransfection with I-_Sce_I and the constructs described above. The hBARD1-C83G and h BARD1-Q564H mutations did not decrease HDR in Brca1+/+ cells (open bars) or the Brca1−/− (gray bars). The error bars represent the standard error.
A germ line missense mutation occurring in BARD1 was found in a patient with late onset cancer of the ovary, breast, and endometrial tissues, with loss of the wild-type allele confirmed in the ovarian tumor (52). We examined whether a full-length hBARD1 protein containing this missense mutation, Q564H, would interfere with HDR. An expression plasmid containing BARD1 mutated at Q564H (hBARD1-Q564H) was cotransfected with the I-_Sce_I expression vector (Fig. 5A). We found that the hBARD1-Q564H was expressed at levels comparable to those for the wild-type protein (Fig. 5B). No significant decrease in the number of GFP-positive cells was observed in either the Brca1+/+ (P = 0.6) or the Brca1−/− (P = 1) cells (Fig. 5C) compared to hBARD1 expression. This indicates that, when transiently expressed, the BARD1 Q564H mutation does not interfere with gene conversion repair of an induced DSB.
DISCUSSION
These results demonstrate that expression of truncated Bard1 peptides which disrupt the endogenous Brca1-Bard1 heterodimer can significantly decrease HDR of a chromosome DSB. That this effect is specific to the Brca1-Bard1 heterodimer is supported by results with a mutated peptide hB202-L107P, which is disrupted at the interface for interaction with BRCA1. As predicted, the hB202-L107P mutation completely disrupted the ability to interact with BRCA1 and did not affect repair by gene conversion of an induced DSB, despite being highly expressed and localized to the nucleus. From the HDR results, along with the MMC hypersensitivity of cells that stably express the truncated Bard1 peptide, we conclude that BARD1 participates with BRCA1 in the HDR of an induced chromosome break through its RING-RING interaction with BRCA1. Thus, these results implicate Bard1 sequences downstream from the RING domain for proper HDR by the complex.
The methods used to assess repair in mammalian cells typically involve the delivery of genotoxic damage through radiation or drugs, followed by cell survival assays or immunocytochemical detection of protein relocalization. However, these assays do not provide the molecular details required to assess specific repair pathways. Many DNA damage response proteins colocalize with BRCA1 to sites of damage (51, 57). The function of many of these colocalizing proteins includes protein activation and recruitment to the damage site, whereas others are involved in repair of the damage. The proteins involved in HDR, as demonstrated by repair assays that can assess the specific repair defects, include Rad51, BRCA1, BRCA2 (32, 34, 49, 50, 55), and now BARD1.
Truncated BARD1 peptides similar in size to our dominant-negative peptides have been shown to augment the E3 ubiquitin ligase activity of BRCA1 (8, 15, 26). Thus, although truncated BARD1 peptides appear to be sufficient for the intrinsic enzymatic function of the BRCA1-BARD1 heterodimer, the HDR function of the heterodimer appears to require full-length BARD1. The E3 ubiquitin ligase activity may therefore be separable from the HDR function of the heterodimer or, conversely, specific enzymatic substrates may require the full-length BRCA1-BARD1 heterodimer. Few in vivo substrates have yet been defined that undergo BRCA1-BARD1-mediated ubiquitination; however, autoubiquitination of the heterodimer or another protein associated with the complex has been noted (8). More importantly, the downstream effects of proteins that are ubiquitinated by the BRCA1-BARD1 heterodimer remain undefined.
The Brca1 mutant ES cell line we examined, like other BRCA1 mutant cells, is hypersensitive to cisplatin and MMC and incurs spontaneous chromosome instability (3, 33). As noted above, the Brca1−/− ES cells, as well as murine embryonic fibroblasts with a similar exon 11-deleted isoform, have diminished but detectable damage-induced Rad51 foci (3, 16). Because expression of the truncated Bard1 peptides in the Brca1-deficient cells further diminished the repair of induced DSBs by HDR, the Brca1Δ11 peptide appears to be a hypomorph for HDR function, implying that Brca1-null cells, if viable, would be severely reduced for HDR. Interestingly, the Brca1-deficient murine embryonic fibroblasts did not appear to undergo phosphorylation of the Brca1Δ11 peptide after DNA damage (16), indicating that damage-induced phosphorylation may not be necessary for the residual HDR activity.
These results have several implications. In the Brca1-conditional mammary tumor model, in which a proportion of the mammary epithelial cells express a Brca1Δ11 product, mammary tumors arose at an incidence of 25% (4). Given the hypomorphic HDR function of the Brca1Δ11 product, a greater reduction in cellular repair capability may be expected to have more profound effects, by increasing the mammary tumor incidence in the Brca1Δ11-conditional mouse model or, alternatively, leading to a rapid and overwhelming accumulation of chromosome damage and cell death, a result more akin to the phenotype observed in Brca1-deficient embryonic tissue (9). Notably, RAD51, the central protein of HDR, has not been found to be mutated or lost in human tumors (39). These results also raise the issue of whether the endogenously occurring BRCA1 spliced isoforms in humans that retain the RING and BRCT domains act to preserve genetic stability through residual repair function or are tumor promoting in the absence of full-length BRCA1 by allowing the propagation of chromosomally damaged cells. Since the variation in cancer incidence observed in clinical populations may be related to specific mutations that harbor residual function, it is important to further decipher these structure-function relationships.
Tumor-associated missense mutations in BRCA1 include cysteine residues involved in zinc binding, although thus far the zinc-binding residues in BARD1 have not been found to be mutated in human tumors (13, 52). Although the tumor associated C61G mutation in BRCA1 abolishes the ubiquitin ligase activity of the BRCA1-BARD1 heterodimeric RING (15, 42), the corresponding mutation in BARD1, C83G, results in a slight reduction or no reduction in ubiquitin ligase activity (15, 26, 60). In addition, the corresponding Xenopus BARD1 C77G mutation did not abolish the xBRCA1-xBARD1 interaction in frog embryos (22). The intermediate reduction in gene conversion repair we observed with the hB202-C83G peptide and the lack of a repair defect with full-length hBARD1-C83G could potentially be explained by decreased protein expression observed by Western blot analysis. As the hB202 peptide itself reduces gene conversion repair, the reduced level of gene conversion with the hB202-C83G peptide may reflect the dominant-negative effect of the truncation rather than an effect due to the point mutation. The reason for the observed lower expression levels of this mutant protein in both the full-length and truncated BARD1 constructs is not evident, especially in light of the strong interaction with the BRCA1 peptide observed in the mammalian two-hybrid experiments. However, a mutation in the corresponding cysteine residue in BRCA1 is substantially more sensitive to proteolytic degradation in vitro, presumably due to compromised zinc binding (5); thus, it is possible that the BARD1 C83G mutation could similarly affect protein stability in vivo.
A search for BARD1 mutations in both familial and sporadic cancers has yielded few data to support BARD1 as a commonly mutated tumor suppressor in human tumors (13, 52). A germ line missense mutation occurring in BARD1 was found in a patient with late-onset cancer of the ovary, breast, and endometrial tissues, with loss of the wild-type allele confirmed in the ovarian tumor (52). This mutation, Q564H, is located between the ankyrin and the BRCT repeats at a residue that is conserved between mouse and human BARD1. Functional analysis has shown that this mutation does not interfere with binding of BARD1 to BRCA1; however, it exhibits decreased binding to CstF-50 and disrupts the ability of BARD1 to inhibit 3′-cleavage of mRNA precursors (24). In our assays, overexpression of BARD1-Q564H did not diminish gene conversion repair of an induced DSB; therefore, this mutant BARD1 does not exhibit dominant-negative activity in the presence of wild-type BARD1. This result does not rule out the possibility that the BARD1 Q564H mutation could have an effect on HDR in the absence of wild-type BARD1 or a tumor-promoting effect through impaired regulation of mRNA processing. However, the mutation appears to be a rare event, and its role in tumorigenesis needs confirmation. Since Bard1-deficient cells have not been identified and the Bard1 knockout results in early embryonic lethality (28), hypomorphic alleles that allow for cellular viability may be necessary to further elucidate the effect of specific BARD1 mutations.
It is hypothesized that defects in HDR lead to unrepaired and misrepaired chromosomes during the normal course of DNA replication, thereby leading toward increased chromosome instability and an increased propensity for tumorigenesis. This spontaneous chromosome instability has been observed in cells deficient for BRCA1 and BRCA2 and mouse models that are either hypomorphic or have tissue-restricted deletions of either gene develop cancer (19, 31). However, chromosome instability is also observed in cells and mouse models with a spectrum of deficiencies, many of which involve the cellular response to DNA damage (11). The importance of identifying at the molecular level how this instability arises and how each protein functions within the DNA damage response pathway that leads to genomic instability becomes increasingly important since not all of these deficiencies have been documented to predispose to tumorigenesis.
Acknowledgments
We thank Shridar Ganesan and David Livingston (Dana-Farber) for the generous gift of the mouse Brca1 GH118 antibody.
This work was supported by the Breast Cancer Research Foundation and NIH grant PO1 CA94060. U.K.W. was supported by the Swedish Foundation for International Cooperation in Research and Higher Education (STINT) and the Swedish Cancer Society. R.B. was supported by NIH grant PO1 CA97403.
REFERENCES
- 1.Ayi, T. C., J. T. Tsan, L. Y. Hwang, A. M. Bowcock, and R. Baer. 1998. Conservation of function and primary structure in the Brca1-associated ring domain (Bard1) protein. Oncogene 17**:**2143-2148. [DOI] [PubMed] [Google Scholar]
- 2.Baer, R., and T. Ludwig. 2002. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12**:**86-91. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya, A., U. S. Ear, B. H. Koller, R. R. Weichselbaum, and D. K. Bishop. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275**:**23899-23903. [DOI] [PubMed] [Google Scholar]
- 4.Brodie, S. G., X. Xu, W. Qiao, W. M. Li, L. Cao, and C. X. Deng. 2001. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene 20**:**7514-7523. [DOI] [PubMed] [Google Scholar]
- 5.Brzovic, P. S., J. E. Meza, M. C. King, and R. E. Klevit. 2001. BRCA1 RING domain cancer-predisposing mutations. Structural consequences and effects on protein-protein interactions. J. Biol. Chem. 276**:**41399-41406. [DOI] [PubMed] [Google Scholar]
- 6.Brzovic, P. S., P. Rajagopal, D. W. Hoyt, M. C. King, and R. E. Klevit. 2001. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8**:**833-837. [DOI] [PubMed] [Google Scholar]
- 7.Castilla, L. H., F. J. Couch, M. R. Erdos, K. F. Hoskins, K. Calzone, J. E. Garber, J. Boyd, M. B. Lubin, M. L. Deshano, L. C. Brody, et al. 1994. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nat. Genet. 8**:**387-391. [DOI] [PubMed] [Google Scholar]
- 8.Chen, A., F. E. Kleiman, J. L. Manley, T. Ouchi, and Z. Q. Pan. 2002. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J. Biol. Chem. 277**:**22085-22092. [DOI] [PubMed] [Google Scholar]
- 9.Deng, C. X., and S. G. Brodie. 2001. Knockout mouse models and mammary tumorigenesis. Semin. Cancer Biol. 11**:**387-394. [DOI] [PubMed] [Google Scholar]
- 10.Fabbro, M., J. A. Rodriguez, R. Baer, and B. R. Henderson. 2002. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J. Biol. Chem. 277**:**21315-21324. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg, E. C., and L. B. Meira. 2003. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage, version 5. DNA Repair 2**:**501-530. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, L. S., E. A. Ostermeyer, C. I. Szabo, P. Dowd, E. D. Lynch, S. E. Rowell, and M. C. King. 1994. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat. Genet. 8**:**399-404. [DOI] [PubMed] [Google Scholar]
- 13.Ghimenti, C., E. Sensi, S. Presciuttini, I. M. Brunetti, P. Conte, G. Bevilacqua, and M. A. Caligo. 2002. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes Cancer 33**:**235-242. [DOI] [PubMed] [Google Scholar]
- 14.Gowen, L. C., B. L. Johnson, A. M. Latour, K. K. Sulik, and B. H. Koller. 1996. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 12**:**191-194. [DOI] [PubMed] [Google Scholar]
- 15.Hashizume, R., M. Fukuda, I. Maeda, H. Nishikawa, D. Oyake, Y. Yabuki, H. Ogata, and T. Ohta. 2001. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276**:**14537-14540. [DOI] [PubMed] [Google Scholar]
- 16.Huber, L. J., T. W. Yang, C. J. Sarkisian, S. R. Master, C. X. Deng, and L. A. Chodosh. 2001. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol. Cell. Biol. 21**:**4005-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irminger-Finger, I., W. C. Leung, J. Li, M. Dubois-Dauphin, J. Harb, A. Feki, C. E. Jefford, J. V. Soriano, M. Jaconi, R. Montesano, and K. H. Krause. 2001. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 8**:**1255-1266. [DOI] [PubMed] [Google Scholar]
- 18.Irminger-Finger, I., J. V. Soriano, G. Vaudan, R. Montesano, and A. P. Sappino. 1998. In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J. Cell Biol. 143**:**1329-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jasin, M. 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21**:**8981-8993. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, D. E., M. Proctor, S. T. Marquis, H. P. Gardner, S. I. Ha, L. A. Chodosh, A. M. Ishov, N. Tommerup, H. Vissing, Y. Sekido, J. Minna, A. Borodovsky, D. C. Schultz, K. D. Wilkinson, G. G. Maul, N. Barlev, S. L. Berger, G. C. Prendergast, and F. J. Rauscher III. 1998. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16**:**1097-1112. [DOI] [PubMed] [Google Scholar]
- 21.Jin, Y., X. L. Xu, M. C. W. Yang, F. L. Wei, T. C. Ayi, A. M. Bowcock, and R. Baer. 1997. Cell cycle-dependent colocalization of Bard1 and Brca1 proteins in discrete nuclear domains. Proc. Natl. Acad. Sci. USA 94**:**12075-12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joukov, V., J. Chen, E. A. Fox, J. B. Green, and D. M. Livingston. 2001. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. USA 98**:**12078-12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kentsis, A., R. E. Gordon, and K. L. Borden. 2002. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. USA 99**:**15404-15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleiman, F. E., and J. L. Manley. 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285**:**1576-1579. [DOI] [PubMed] [Google Scholar]
- 25.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96**:**11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallery, D. L., C. J. Vandenberg, and K. Hiom. 2002. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 21**:**6755-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marians, K. J. 2000. Replication and recombination intersect. Curr. Opin. Genet. Dev. 10**:**151-156. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy, E. E., J. T. Celebi, R. Baer, and T. Ludwig. 2003. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol. Cell. Biol. 23**:**5056-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meza, J. E., P. S. Brzovic, M. C. King, and R. E. Klevit. 1999. Mapping the functional domains of BRCA1: interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem. 274**:**5659-5665. [DOI] [PubMed] [Google Scholar]
- 30.Miki, Y., J. Swensen, D. Shattuck-Eidens, P. A. Futreal, K. Harshman, S. Tavtigian, Q. Liu, C. Cochran, L. M. Bennett, W. Ding, R. Bell, J. Rosenthal, C. Hussey, T. Tran, M. McClure, C. Frye, T. Hattier, R. Phelps, D. Haugren-Strano, H. Katcher, K. Yakumo, Z. Gholami, D. Shaffer, S. Stone, S. Bayer, C. Wray, R. Bogden, P. Dayanath, J. Ward, P. Tonin, S. Narod, P. Bristow, F. Norris, L. Helvering, P. Morrison, P. Rostecl, M. Lai, C. Barrett, C. Lewis, S. Neuhasen, L. Cannon-Albright, D. Goldgar, R. Wiseman, A. Kamb, and M. Skolnick. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266**:**66-71. [DOI] [PubMed] [Google Scholar]
- 31.Moynahan, M. E. 2002. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene 21**:**8994-9007. [DOI] [PubMed] [Google Scholar]
- 32.Moynahan, M. E., J. W. Chiu, B. H. Koller, and M. Jasin. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4**:**511-518. [DOI] [PubMed] [Google Scholar]
- 33.Moynahan, M. E., T. Y. Cui, and M. Jasin. 2001. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61**:**4842-4850. [PubMed] [Google Scholar]
- 34.Moynahan, M. E., A. J. Pierce, and M. Jasin. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7**:**263-272. [DOI] [PubMed] [Google Scholar]
- 35.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108**:**193-199. [DOI] [PubMed] [Google Scholar]
- 36.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8**:**499-504. [DOI] [PubMed] [Google Scholar]
- 37.Pierce, A. J., P. Hu, M. Han, N. Ellis, and M. Jasin. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15**:**3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13**:**2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce, A. J., J. M. Stark, F. D. Araujo, M. E. Moynahan, M. Berwick, and M. Jasin. 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol. 11**:**S52-S59. [DOI] [PubMed] [Google Scholar]
- 40.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12**:**3831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez, J. A., and B. R. Henderson. 2000. Identification of a functional nuclear export sequence in BRCA1. J. Biol. Chem. 275**:**38589-38596. [DOI] [PubMed] [Google Scholar]
- 42.Ruffner, H., C. A. Joazeiro, D. Hemmati, T. Hunter, and I. M. Verma. 2001. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA 98**:**5134-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21**:**208-214. [PubMed] [Google Scholar]
- 44.Scully, R., J. Chen, R. L. Ochs, K. Keegan, M. Hoekstra, J. Feunteun, and D. M. Livingston. 1997. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90**:**425-435. [DOI] [PubMed] [Google Scholar]
- 45.Scully, R., S. Ganesan, K. Vlasakova, J. Chen, M. Socolovsky, and D. M. Livingston. 1999. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell 4**:**1093-1099. [DOI] [PubMed] [Google Scholar]
- 46.Scully, R., and D. M. Livingston. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408**:**429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharan, S. K., M. Wims, and A. Bradley. 1995. Murine Brca1: sequence and significance for human missense mutations. Hum. Mol. Genet. 4**:**2275-2278. [DOI] [PubMed] [Google Scholar]
- 48.Snouwaert, J. N., L. C. Gowen, A. M. Latour, A. R. Mohn, A. Xiao, L. DiBiase, and B. H. Koller. 1999. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a Brca1 transgene. Oncogene 18**:**7900-7907. [DOI] [PubMed] [Google Scholar]
- 49.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19**:**5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stark, J. M., P. Hu, A. J. Pierce, M. E. Moynahan, N. Ellis, and M. Jasin. 2002. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 277**:**20185-20194. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, G. S., B. Wang, C. R. Bignell, A. M. Taylor, and S. J. Elledge. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421**:**961-966. [DOI] [PubMed] [Google Scholar]
- 52.Thai, T. H., F. H. Du, J. T. Tsan, Y. Jin, A. Phung, M. A. Spillman, H. F. Massa, C. Y. Muller, R. Ashfaq, J. M. Mathis, D. S. Miller, B. J. Trask, R. Baer, and A. M. Bowcock. 1998. Mutations in the Brca1-associated ring domain (Bard1) gene in primary breast, ovarian and uterine cancers. Hum. Mol. Genet. 7**:**195-202. [DOI] [PubMed] [Google Scholar]
- 53.Thakur, S., H. B. Zhang, Y. Peng, H. Le, B. Carroll, T. Ward, J. Yao, L. M. Farid, F. J. Couch, R. B. Wilson, and B. L. Weber. 1997. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol. Cell. Biol. 17**:**444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsan, J. T., Z. Wang, J. Y., L. Y. Hwang, R. O. Bash, and R. Baer. 1997. Mammalian cells as hosts for two-hybrid studies of protein-protein interaction, p. 217-232. In P. L. Bartel and S. Fields (ed.), The yeast two-hybrid system. Oxford University Press, Oxford, England.
- 55.Tutt, A., D. Bertwistle, J. Valentine, A. Gabriel, S. Swift, G. Ross, C. Griffin, J. Thacker, and A. Ashworth. 2001. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20**:**4704-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, H., N. Shao, Q. M. Ding, J. Cui, E. S. Reddy, and V. N. Rao. 1997. BRCA1 proteins are transported to the nucleus in the absence of serum and splice variants BRCA1a, BRCA1b are tyrosine phosphoproteins that associate with E2F, cyclins and cyclin dependent kinases. Oncogene 15**:**143-157. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14**:**927-939. [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, C. A., M. N. Payton, G. S. Elliott, F. W. Buaas, E. E. Cajulis, D. Grosshans, L. Ramos, D. M. Reese, D. J. Slamon, and F. J. Calzone. 1997. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-Δ11b. Oncogene 14**:**1-16. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L. C., Z. W. Wang, J. T. Tsan, M. A. Spillman, A. Phung, X. L. Xu, M. C. Yang, L. Y. Hwang, A. M. Bowcock, and R. Baer. 1996. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Genetics 14**:**430-440. [DOI] [PubMed] [Google Scholar]
- 60.Xia, Y., G. Pao, H. W. Chen, I. M. Verma, and T. Hunter. 2003. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 278**:**5255-5263. [DOI] [PubMed] [Google Scholar]
- 61.Yu, X., and R. Baer. 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275**:**18541-18549. [DOI] [PubMed] [Google Scholar]