Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development (original) (raw)
Abstract
Conservation of the vertebrate body plan has been attributed to the evolutionary stability of gene-regulatory networks (GRNs). We describe a regulatory circuit made up of Gata2, Fli1, and Scl/Tal1 and their enhancers, Gata2-3, Fli1+12, and Scl+19, that operates during specification of hematopoiesis in the mouse embryo. We show that the Fli1+12 enhancer, like the Gata2-3 and Scl+19 enhancers, targets hematopoietic stem cells (HSCs) and relies on a combination of Ets, Gata, and E-Box motifs. We show that the Gata2-3 enhancer also uses a similar cluster of motifs and that Gata2, Fli1, and Scl are expressed in embryonic day-11.5 dorsal aorta where HSCs originate and in fetal liver where they multiply. The three HSC enhancers in these tissues and in ES cell-derived hemangioblast equivalents are bound by each of these transcription factors (TFs) and form a fully connected triad that constitutes a previously undescribed example of both this network motif in mammalian development and a GRN kernel operating during the specification of a mammalian stem cell.
Keywords: hemangioblast, hematopoiesis, hematopoietic stem cell, network motif, transcription factor network
Hematopoietic stem cells (HSCs) represent the best-characterized adult multipotent stem cell population, with the availability of a plethora of markers and biological (clonogenic and transplantation) assays to identify long-term repopulating HSCs and distinguish them from multipotent progenitors with only limited self-renewal potential (1). Transcriptional regulation is a key mechanism controlling the formation and subsequent behavior of HSCs (2). Both gain and loss of function studies have identified a number of transcription factors (TFs), including Scl/Tal 1 (3, 4), Gata2 (5) and Runx1 (6, 7), as critical regulators of HSC development. Significantly, disruption of many of these TFs contributes to the pathogenesis of hematological malignancies, emphasising the importance of transcriptional regulation in both normal and leukaemic stem cells.
Biological complexity does not correlate with gene number but rather with the intricacy of gene regulation (8). Enhancers and other cis-regulatory elements play a central role in the coordinated expression of genes and a number of tissue-specific regulatory elements of key HSC TFs, such as Scl and Gata2, have been identified (9–13). However, it is clear from studies in nonvertebrate model organisms, such as Drosophila and the sea urchin, that to comprehend developmental processes, it is necessary to move beyond the study of individual genes and determine how regulatory genes interact to form functional gene regulatory-networks (GRNs) (reviewed in ref. 14).
Comprehensive analysis of the sea urchin endomesoderm GRN showed that they consist of assemblies of subcircuits made up of TF genes and their target cis-regulatory modules, with each subcircuit performing a distinct regulatory function during development (15). The linkages of these subcircuits are highly recursive with each cis-regulatory module receiving inputs from multiple TFs that make up the subcircuit. Subcircuits that perform essential functions in building body parts have been termed the kernels of the GRN (16). Disruption of TFs that make up the kernel often results in loss of the body part, and hence the basic architecture of kernels is highly conserved through evolution (16).
Scl, Gata2, and the Ets factor Fli1 are required for normal hematopoiesis in mice (reviewed in ref. 17) and are expressed in avian hematopoietic clusters (18) and hematopoietic mesodermal precursors in frog and zebrafish embryos (19, 20). In this article we describe a GRN kernel composed of Gata2, Fli1, and Scl, and their respective cis-regulatory modules. Using transgenic mice and in vivo ChIP assays of embryonic hematopoietic tissues, we demonstrate that this GRN kernel operates during key stages of mouse HSC specification in the aorta–gonad-mesonephros (AGM) region and in the midgestation fetal liver (FL).
Results
The Fli1+12 Hematopoietic Enhancer Targets Long-Term Repopulating Blood Stem Cells in the Embryo.
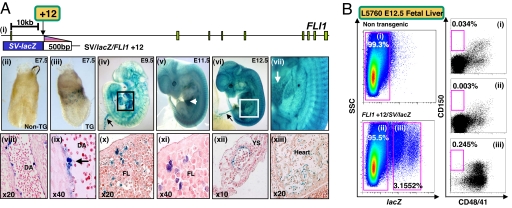
We have previously established that the Fli1+12 enhancer directs lacZ expression to blood and endothelium in F0 transgenic embryos (21). However, it was not known whether the Fli1+12 enhancer targeted expression to HSCs per se. To evaluate the in vivo activity of the Fli1+12 enhancer in detail, we established stable transgenic lines (L5760 and L5754) with founders generated using a 500-bp fragment of the human FLI1+12 locus cloned downstream of the SV minimal promoter and lacZ reporter (SV/lacZ/FLI1 + 12 in Fig. 1_A_i). Whole-mount analysis of gastrulating embryonic day-7.5 (E7.5) embryos showed expression of the transgene in the region of extraembryonic mesoderm that characterizes primitive hematopoiesis in the early yolk sac (compare Fig. 1_A_iii with Fig. 1_A_ii). This finding is consistent with expression in hematopoietic tissues and the vasculature during later stages of development (Fig. 1A iv–xiii).
Fig. 1.
The FLI1+12 hematopoietic enhancer targets blood stem cells. (A) The FLI1+12 enhancer directs reporter activity to blood and blood vessels. (i) Schematic diagram of the human FLI1 locus. A fragment of DNA corresponding to the FLI1+12 region was used to generate transgenic mice. (ii–vii) E7.5–E12.5 X-Gal-stained whole-mount WT and L5760 SV/lacZ/FLI1+12 embryos. (ii) E7.5 WT embryo with no staining. (iii) E7.5 transgenic embryo showing staining within the extra-embryonic hematopoietic region. (iv) E9.5 transgenic embryo showing staining of the heart chambers (boxed) and vitelline vessels (arrow). (v) E11.5 transgenic embryo showing staining within the FL (arrow) and blood vessels. (vi) E12.5 transgenic embryo showing staining in the vitelline vessels (arrow). (vii) Magnified view of the boxed area in vi showing FL staining (arrow). (viii–xiii) Paraffin sections of a E11.5 SV/lacZ/FLI1+12 embryo. (viii) Sagittal section of the DA showing staining in the endothelium. (ix) High-power view of the DA showing staining in a hematopoietic cluster (arrow). (x) Section through the FL showing staining in hematopoietic cells. (xi) High-power view of x. (xii) Section through the yolk sac showing staining in the wall of a blood vessel. (xiii) Section through the heart showing endocardial staining. (B) Flow cytometry of FDG treated nontransgenic and transgenic E12.5 FLs from a litter of SV/lacZ/FLI1+12 (L5760) × WT crosses. CD150+/CD48−/CD41− cells are enriched in the population targeted by the transgene. YS, yolk sac.
To establish whether the Fli1+12 enhancer-targeted expression to FL HSCs, we first characterized the surface phenotype of FL cells targeted by the SV/lacZ/FLI1+12 transgene. One of every 5.7 CD150+/CD48−/CD41− FL cells gives long-term multilineage reconstitution in irradiated mice (22). FL cell suspensions were prepared from E12.5 embryos and stained with the fluorescent β-galactosidase substrate, fluoro-deoxy-d-glucose (FDG). As shown in Fig. 1B, ≈3% of FL cells from SV/lacZ/FLI1+12 transgenic embryos express the transgene. Approximately 0.034% of E12.5 FL cells have the CD 150+/CD48−/CD41− surface phenotype (Fig. 1_B_i) and cells targeted by the FLI1+12 enhancer are enriched for this phenotype (compare Fig. 1_B_i with Fig. 1_B_iii). Moreover, cells targeted by the transgene show a relative paucity of lineage-committed CD150−/CD48−/CD41− cells (compare Fig. 1_B_ii with Fig. 1_B_iii). We next performed in vitro colony assays to establish multilineage potential of _lacZ_-negative and _lacZ_-positive cells. As shown in supporting information (SI) Table 1, _lacZ_-positive cells were enriched for myeloid and erythroid colony forming cells. However colony assays do not identify true HSCs. We therefore performed long term reconstitution experiments by using irradiated adult mice as recipients.
E12.5 FL cell suspensions from SV/lacZ/FLI1+12 transgenic embryos were stained with FDG, and two sorting regions were set according to the relative levels of lacZ expression. Varying numbers of sorted FDG+ and FDG− cells were infused into irradiated recipient mice with 2 × 105 nontransgenic splenocytes to provide short-term radioprotection. Donor cell engraftment was examined 6 months after transplantation by analyzing recipient peripheral blood by PCR for the donor lacZ transgene. The cumulative results from two experiments demonstrated that the lacZ transgene was detected more than 6 months after transplant in ≈1/3 of recipient mice (5 of 15) transplanted with 104 FDG+ cells (SI Table 2). By contrast, the donor lacZ transgene was detected in zero of eight recipients transplanted with 1 × 105 FDG− cells and in one of five recipients transplanted with 5 × 105 FDG− cells. To determine whether transplantation of FDG+ FL cells resulted in multilineage hematopoietic engraftment, hematopoietic tissues and purified cells from all reconstituted recipients were analyzed by semiquantitative PCR for the lacZ transgene. The hematopoietic tissues of the reconstituted recipients contained 10–100% donor derived cells (data not shown). Taken together, these data demonstrate that the SV/lacZ/Fli1+12 construct targets the majority of FL HSCs capable of long-term multilineage reconstitution.
Ets and Gata Binding Motifs in the Fli1+12 HSC Enhancer Are Required for Its Activity.
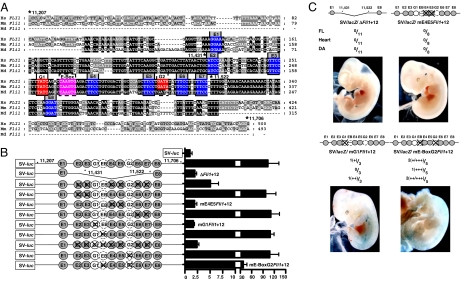
We have shown previously that clusters of highly conserved Ets and Gata sites are critical for the activity of early hematopoietic regulatory elements of the Scl and Lyl1 genes (17, 23). Clustering of Ets and Gata sites [two Ets and one Gata site with defined spacing and orientation constraints constitutes the Ets/Ets/Gata (E/E/G) signature] can be exploited in genome-wide computational screens to identify new hematopoietic stem/progenitor elements (21, 24). The Fli1+12 enhancer (Fig. 2A) has, in addition to the E/E/G (E5/E4/G1) signature, another six highly conserved Ets sites, an additional partially conserved Gata site, and an E-box (the consensus binding site for the Scl). We have previously shown that deletion of an 89-bp core (+11,430/+11,521) that includes the E/E/G signature abolished enhancer activity (21). To establish the relative contribution of the various conserved Ets, Gata, and E-box motifs, mutations were introduced individually and in combination, and the activity of the enhancer was tested in 416B hematopoietic progenitor cells (Fig. 2B). The activity of the WT and mutant constructs was expressed relative to that of the pGL2promoter vector containing the SV minimal promoter.
Fig. 2.
Conserved Ets and Gata TF binding sites in the FLI1+12 enhancer are required for its activity in hematopoietic cells. (A) Nucleotide sequence alignment of the FLI1 +12 enhancer with conserved Gata, Ets, and E-box sites marked in red, blue, and pink, respectively. Nucleotide numbering is from the translation start site. mm, mouse; hs, human; md, opossum. (B) (Left) Reporter constructs of human WT and mutated genomic fragments corresponding to the FLI1+12 enhancer. The conserved Ets, Gata, and E-box sites are represented as circles (dashed margins for partial conservation), numbered E1–8, G1–2, and EB, respectively; they are crossed out where mutated. (Right) Results of stable transfection assays in 416B cells corresponding to each construct. The luciferase activities are given as fold-increase over the activity of the pGL2-promoter vector (SV-luc) alone. (C) Summary of F0 transgenic embryos generated with various SV/lacZ/mutant FLI1+12 constructs. Representative X-Gal-stained whole-mount E11.5 embryos are also shown. (+) to (++++) indicates from week/rare to strong X-Gal staining.
Interestingly, mutating E4/E5 (Ets sites that are part of the Scl+19 signature) abolished enhancer activity, whereas mutating E2/E3 or E6/E7 which are equally conserved had no effect. Likewise, mutating the highly conserved GATA site G1, abolished enhancer activity, whereas mutating G2 alone had no effect. Significantly, although mutation of the partially conserved GATA site G2 and the E-box individually had no effect, loss of both reduced activity of the enhancer by ≈1/3.
Transgenic reporter analysis (Fig. 2C) of mutant FLI1+12 elements confirmed that binding sites within the Δ_FLI1_+12 region are also required for in vivo activity of the enhancer. Mutating E4/E5 resulted in complete absence of blood/endothelial/endocardial activity. Possibly owing to a degree of compensation, mutating G1 or G2/E-Box sites resulted in mild/moderate staining of the dorsal aorta (DA) and FL in a proportion of embryos. The staining, however, was considerably less than in WT embryos (compare sections in Fig. 1A with SI Fig 6).
Genome-Wide Computational Analysis Groups the Gata2-3 Region Together with the Fli1+12 E/E/G Regulatory Element.
Using the number of conserved Ets, Gata, and E-box motifs within the in vivo validated Fli1+12 hematopoietic enhancer as a gauge, we performed a series of genome-wide computational screens for related elements (SI Tables 3 and 4). Interestingly, all four screens returned a region 3 kb upstream of the Gata2 IS promoter as a potential match. This Gata2-3 region contains the 5H region of the 3.1-kb Gata2-EHRD promoter fragment previously shown to be sufficient and necessary for Gata2 expression in the hemogenic DA (9). Activity of this element has been solely attributed to conserved Gata sites, but its grouping with the Fli1+12 enhancer in screens for elements with multiple Ets, Gata, and E-Box motifs suggested that these other conserved sites could also be functionally important.
Ets Binding Motifs Are Indispensable for Gata2-3 Enhancer Activity.
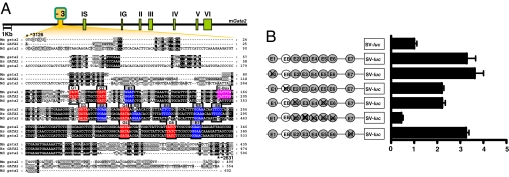
The 5H region (−3,097 to −2,762) of the Gata2-3 enhancer (Fig. 3A) has previously been shown to target reporter gene activity to the caudal DA, the site of origin of HSCs in the embryo, and this activity depends on the Gata binding motifs G1, G2, G4, G5, and G6 (9). Interspersed with these conserved Gata motifs are a number of conserved Ets and E-box motifs (Fig. 3A). This configuration is similar to that of the Scl+19, Fli1+12, and Hhex+1 enhancers, which are regulated by a combination of Ets factors and Gata2 in hematopoietic progenitors in vivo (17, 21). To examine the relative importance of these Ets and E-box motifs, a fragment corresponding to the murine −3,126/−2,631 enhancer was PCR amplified from human DNA and subcloned upstream of the SV minimal promoter in the luciferase reporter, pGL2p. Mutations were introduced individually and in combination and tested in stable transfections of 416B hematopoietic progenitor cells (Fig. 3B). Site-directed mutagenesis of individual Ets sites E1 and E7 had little effect on the overall activity of the enhancer, whereas mutation of E2 and E5 and of the E-box reduced activity by ≈1/3. Mutation of E2–E6 within the core of the enhancer was sufficient to abolish enhancer activity.
Fig. 3.
Conserved Ets TF binding sites in the Gata2-3 enhancer are required for its activity in hematopoietic cells. (A) Schematic diagram of the mouse Gata2 locus and nucleotide sequence alignment with conserved Gata, Ets, and E-box sites marked in red, blue, and pink respectively. The −3,126/−2,631 m_Gata2-3_ region includes the 5H region (−3,097/−2,762) of the 3.1-kb Gata2-EHRD (9). Nucleotide numbering is from the mouse IS exon. mm, mouse; hs, human; md, opossum. (B) (Left) Reporter constructs of human WT and mutated Gata2-3 genomic fragments. The conserved Ets sites and E-box are represented as circles numbered E1–E7 and EB respectively; they are crossed out where mutated. (Right) Results of stable transfection assays in 416B cells corresponding to each construct. The luciferase activities are given as the fold-increase over the activity of the pGL2-promoter vector (SV-luc) alone.
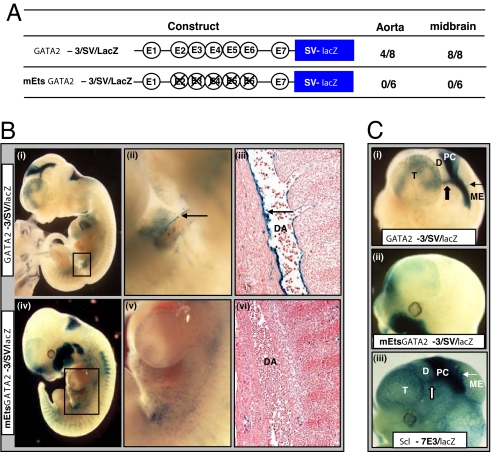
To evaluate the role of the Ets binding motifs in vivo, the Gata2-3 enhancer fragment was cloned upstream of the SV minimal promoter (Gata2-3/SV/lacZ in Fig. 4A) in a lacZ reporter vector and tested in F0 transgenics. Transgenic expression in the caudal DA was seen in four of eight transgenic embryos (Fig. 4B i–iii). In contrast, zero of six transgenic embryos generated with a GATA2 fragment with targeted mutations in E2–E6 Ets binding motifs (mEts Gata2-3/SV/lacZ) showed reporter activity in the DA (Fig. 4 A and B iv–vi).
Fig. 4.
The Gata2-3 enhancer targets the hemogenic aortic endothelium and requires Ets sites for its in vivo activity. (A) Transgenic lacZ reporter constructs with WT enhancer (Gata2-3) or enhancer with mutant Ets sites (mEts Gata2-3). The number of embryos showing staining in the DA (AGM region) or midbrain out of the total number of E10.5 and E11.5 F0 transgenic embryos generated with each construct is listed alongside. (B) Representative F0 embryos stained with X-Gal for lacZ expression. (i) Gata2-3/SV/lacZ E10.5 whole-mount showing staining in the DA (boxed). (ii) Magnified view of the boxed area in i showing concentration of the reporter along the ventral surface of the DA (arrow). (iii) Histological section corresponding to region in (ii) showing prominent endothelial staining (arrow). (iv) mEts Gata2-3/SV/lacZ E11.5 whole-mount embryo showing nonspecific staining in the region of the DA (boxed). (v) A magnified view of the boxed area in iv. (vi) Section corresponding to region in v showing absence of endothelial staining. (C) Magnified lateral view of heads of X-Gal stained F0 embryos in B. (i) Gata2-3/SV/lacZ whole-mount embryo showing prominent lacZ expression in the posterior commissure (block arrow) and tectal neurons into the mesencephalon (arrow). (ii) mEts Gata2-3/SV/lacZ whole-mount embryo showing loss of _l_acZ staining. The ectopic staining in the maxillary region was seen in only one of six F0 mutant transgenic embryos. (iii) _Scl_-7E3/lacZ wholemount embryo showing staining similar to that seen with the Gata2-3/SV/lacZ transgenics. D, diencephalon; ME, mesencephalon; PC, posterior commissure; T, telencephalon.
The full-length 3.1-kb Gata2-EHRD promoter fragment directs reporter activity to neural and hematopoietic tissues in the embryo (9). The 496-bp Gata2-3 enhancer fragment, together with a heterologous promoter (SV minimal promoter), also directs reporter activity to neural tissues (Gata2-3/SV/lacZ in Fig. 4 A and _C_i). In contrast, transgenic embryos generated with the mutant transgene that lacks E2–E6 Ets sites (mEts Gata2-3/SV/lacZ in Fig. 4A) show no activity in the midbrain (Fig. 4_C_ii). Therefore the Ets sites within the 5H region are required for both hematopoietic and midbrain activity of the Gata2-3 enhancer. Furthermore, the neural activity of this enhancer is remarkably similar to that of the Scl promoter proximal enhancers 1a and 1b, which are also regulated by Ets and Gata TFs (Fig. 4_C_iii) (11).
In Vivo Expression Analysis and Enhancer Occupancy Are Consistent with a Recursively Wired Regulatory Network.
During mouse development, the first long-term reconstituting HSCs are thought to be generated from the floor of the DA in the AGM region at E10.5. These putative HSCs are recognized as blood clusters and are thought to originate from the aortic endothelium or the underlying mesenchyme. HSC numbers are subsequently amplified in the FL and other sites (reviewed in ref. 25). Using in situ hybridization, we demonstrate that Scl, Gata2, and Fli transcripts are all present in hematopoietic intra-aortic clusters located within the floor of the DA, in the aortic endothelium, and in FL cells at E11.5, consistent with their essential roles in hematopoiesis (Fig. 5A). As expected, the expression patterns of Gata2, Fli1, and Scl were not identical. For example, only Scl was expressed in the majority of circulating blood cells in the DA and FL and corresponds with Scl (but not Gata2 or Fli1) expression in erythroid progenitors, which constitute the dominant blood cell type at E11.5. However, coexpression in DA endothelium and blood clusters was consistent with the cross-regulation suggested by our transgenic analysis of mutant enhancer constructs described above.
Fig. 5.
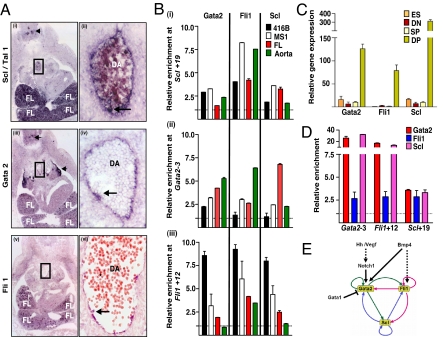
Gata2, Fli1 and Scl bind Gata2-3, Fli1+12 and Scl+19 in vivo. (A) (i and ii) In situ hybridization for Scl expression. (i) Transverse cryosection through a E11.5 embryo at the level of the AGM showing expression in FL, DA (boxed) and neural tissue (arrow). (ii) Magnified view of the boxed area showing expression in the endothelium and in a blood cluster (arrow). (iii and iv) In situ hybridization for Gata2 expression. (iii) Transverse cryosection through the AGM region of a E11.5 embryo showing Gata2 expression in the FL and DA (boxed). Prominent Gata2 expression is also seen in neural tissue (arrows). (iv) Magnified view of the boxed area in iii showing Gata2 expression in the endothelium and in a blood cluster (arrow). (v and vi) In situ hybridization for Fli1 expression. (v) Transverse cryosection through the AGM region of a E11.5 embryo showing Fli1 expression in the FL. (vi) Transverse paraffin section through the AGM region showing expression in the endothelium and blood clusters (arrow). (B) (i–iii) ChIP assays of blood progenitor (416B) and endothelial (MS1) cell lines and primary hematopoietic tissues (abdominal aorta and FL) from E11.5 embryos. Levels of enrichment were normalized to that obtained with a control rabbit antibody and are plotted as a fold-increase over that measured at a control region (Scl+21). (C) Variation in Gata2, Fli1, and Scl gene expression during in vitro differentiation of GFP-Bry ES cells to EBs. (D) Quantitative ChIP assay on in vitro differentiated, unsorted GFP-Bry EBs, of which ≈49% of cells were GFP+Flk1+ (DP) at day 3 (SI Fig 7). (E) A fully connected triad of haematopoietic TFs. The Gata2/Fli1/Scl triad with putative initiators. Direct binding interactions are represented by solid lines.
To correlate transgenic in vivo enhancer activity with in vivo TF occupancy, ChIP experiments were performed with well characterized antibodies (see SI Materials and Methods for details) by using dissected midgestation DA and FL, as well as with hematopoietic progenitor (416B) and endothelial cell lines (MS1) (Fig. 5B). In the dissected hematopoietic tissues, Gata2, Fli1, and Scl were enriched 1- to 8-fold at the Gata2-3, Fli1+12, and Scl+19 enhancers relative to Scl +21 (a region that has no known enhancer activity). All three TFs were enriched at least 2-fold at each of the three HSC enhancers in either AGM region or FL cells. It is important to note that although HSCs are enriched at these sites, not all cells within the dissected tissues are HSCs. TF enrichments at Gata2-3, Fli1+12, and Scl+19 in clonal blood progenitor (416B) and endothelial (MS1) cell populations are shown for comparison.
Following differentiation, ES cells in culture form colonies known as embryoid bodies (EBs) that contain mesodermal (Brachyury+) progenitors that display both blood and endothelial potential [blast colony-forming cell (BL-CFC)] (26). This transient population (day 2.5–4 of differentiation) represents the in vitro equivalent of the yolk-sac hemangioblast. The ES/EB model system has been applied to an ES cell line with GFP targeted to the Brachyury locus (GFP-Bry ES) (27). By sorting day 3–3.5 EBs based on GFP and Flk1 expression, it is possible to identify three distinct cell populations, GFP−Flk1− (DN), GFP+Flk1− (SP), and GFP+Flk1+ (DP) that represent a developmental progression ranging from premesoderm (DN) to prehemangioblast mesoderm (SP) to the hemangioblast (DP). The expression of Gata2, Fli1 and Scl is dramatically up-regulated in cells with hemangioblast potential (Fig. 5C). To assess TF binding at the HSC enhancers, ChIP assays were performed on day-3 EBs. All three TFs were enriched at least 2.5-fold at each of the three enhancers relative to a control region, Scl +21 (Fig. 5D).
Taken together, these data are consistent with a recursively wired gene regulatory subcircuit that operates during the specification of HSCs in the embryo (Fig. 5E).
Discussion
The basic body plan has undergone little evolutionary change since the early Cambrian (≈540 million years ago) (16). This remarkable conservation has been attributed to the evolutionary stability of GRN kernels that specify various body parts. The heart–field specification kernel that is used in both Drosophila and vertebrates (28) [protostomes and deuterostomes split ≈700 million years ago (29)] and the endomesoderm specification kernel common to sea urchin and starfish (who last shared an ancestor >500 million years ago) (15), display conservation of core subcircuits surrounded by network linkages that are not conserved (16). On the basis of sequence conservation of the cis-regulatory modules and requirement for the TFs in specifying definitive hematopoiesis, the GRN kernel that we describe in this manuscript has probably existed since the divergence of Actinopterygii (zebrafish) and Mammalia ≈400 million years ago (29) and is possibly older, because several key components that regulate hematopoiesis have been conserved between mammals and Drosophila through ≈700 million years of evolution. Drosophila Gata factor, Serpent, plays a central role in committing mesodermal cells to a hemocyte fate and is the functional homologue of murine hematopoietic Gata factors, Gata1, -2, and -3 (30). Drosophila pointed regulates the number of circulating hemocytes in larvae (31) and is the prototype of the pointed domain-containing subfamily of Ets proteins of which Fli1 is a member. A Drosophila Scl homologue (HLHB3) exists, but its role in hematopoiesis has not as yet been rigorously evaluated (32).
The development of HSCs from the hemogenic endothelium is a Notch 1-regulated event and is impaired in Notch 1-deficient mouse embryos (33). Notch 1 and Gata2 are coexpressed in endothelial cells lining the floor of the aorta at E10.5, and Notch 1 binds the Gata2 promoter and acts as an upstream regulator of Gata2 expression during the onset of definitive hematopoiesis in the embryo (33, 34). Therefore, as a functionally relevant upstream regulator of Gata2 expression, Notch 1 is a potential initiator of the circuit (Fig. 5E). In zebrafish embryos, Notch 1 is a component of a signaling cascade that involves Hedgehog and Vegf and is required to specify the DA and blood stem cells (35, 36). Bmp4 transcripts are also concentrated along the ventral aspect of the DA in E11.5 embryos (24). The Gata2 promoter responds to Bmp4 signaling (37), which also initiates _Fli1_- expression in Xenopus, and as such Bmp4 is another candidate initiator of the circuit (Fig. 5E). Lineage-labeling studies in Xenopus have demonstrated that the dorsal lateral plate (DLP) mesoderm gives rise to both the DA and clusters of cells attached to the floor of the DA that probably represent HSCs and express Gata2, Scl and _X_fli1. Bone morphogenetic protein (BMP) signaling is required for dorsal lateral plate (DLP) formation, and disruption of BMP signaling results in loss of _X_fli1 and Scl expression (38).
We have previously reported that Gata2 and Fli1 regulate Scl and Fli1 expression during fetal hematopoiesis by binding the Scl+19 and Fli1+12 enhancers respectively (17, 21). We now show that Scl also binds and regulates both enhancers. Gata2 was known to regulate the Gata2-3 enhancer during hematopoiesis (9). Our data show that Fli1 and Scl also bind the Gata2-3 enhancer and regulate its activity. The recently identified Gata2 intronic endothelial enhancer (39) has a cluster of Gata, Ets, and E-Box motifs and consistently grouped together with the Gata2-3 haematopoietic enhancer in our genome-wide computational analysis (SI Table 4). Pending investigation of the Gata sites, full activity of the Gata2 intronic endothelial enhancer was shown to rely on Ets and E-Box motifs with binding of a Scl-E12 heterodimer to the E-Box motif in vitro (39). The Scl+19 element does not have an E-Box motif, but Scl is enriched at this element in FL cells and in blast colony-forming cells (BL-CFCs). It is pertinent to note that protein complexes, which include Scl, Lmo2, and Gata3, have been shown to bind DNA through a GATA site alone (40) and that the E-Box motif in the Gata–E-Box composite element within the −3.7-kb HS1 Gata1 enhancer is dispensable for in vivo binding of the Gata1/Scl/Lmo2/Ldb1/E2A complex (41). Taken together, it is likely that Notch and/or Bmp initiate Gata2 and Fli1 expression, which then autoregulate and combine to initiate Scl. These three factors can then maintain expression of each other in HSCs after Bmp and Notch signaling ceases.
Large integrated transcriptional networks are made up of recurring patterns of smaller recognizable network motifs and the structure of these motifs have been described in Escherichia coli and Saccharomyces cerevisiae (42). The GRN kernel in blood/endothelial progenitors conforms best to a fully connected triad (also known as a clique) that is rare in lower organisms. Furthermore, if, as our data show, all of these interactions are positive, then the Gata2/Fli1/Scl triad would show a tendency to be locked into an ON state (Fig. 5E). As such, the fully connected triad would have the ability to endow a newly specified HSC with “memory” so that it retains pluripotency when it leaves the AGM niche (where it likely receives Notch/Bmp signals that initiate the circuit) and transits to the FL and eventually the bone marrow niche. Steady-state kinetics of individual TFs, threshold concentrations at which they trigger expression, and the requirement for combinatorial binding could all be factors in this circuit reaching equilibrium in HSCs. Interestingly, Gata1 has been shown to disrupt Gata2-positive autoregulation by binding precisely to the Gata2-3 region and mediating domain-wide chromatin remodeling (43). This Gata switch has been proposed as a mechanism of abrogating Gata2 expression in HSCs and inducing differentiation. This and other mechanisms probably operate in HSCs to unlock the Gata2/Fli1/Scl triad when necessary during cell differentiation.
It is salient to note that, although the fully connected triad is very rare in prokaryotic networks (where these motifs have been systematically studied), it may be used in metazoan transcriptional networks to “hardwire stemness” into stem cells. The TFs Oct4, Sox2, and Nanog play key roles in maintaining pluripotency of ES cells, and the promoters of these genes in ES cells are bound by all three factors in vivo (44). These genes and their promoters potentially represent another example of a fully connected triad, but all of the regulatory inputs would first need to be validated using appropriate transfection and transgenic assays.
Switching on the Gata2/Fli1/Scl triad could be a prerequisite for specifying HSCs. Once established, the circuit could maintain expression of its component TFs, which in turn would then be at hand to regulate other genes that are required for hematopoiesis and to interact with gene differentiation batteries that specify the various hematopoietic lineages.
Materials and Methods
In Silico Identification of Clusters of Conserved Binding Sites.
Clusters of conserved TF binding sites were identified using TFBScluster as described in ref. 21.
Reporter Constructs.
Candidate enhancer sequences were PCR amplified from human genomic DNA. Mutations were generated by PCR using oligonucleotides with mismatches and verified by DNA sequencing (SI Materials and Methods and ref. 13).
Transgenic Analysis and Histology.
F0 transgenic mouse embryos were generated by pronuclear injection of lacZ reporter fragments (13). In situ hybridizations were performed using digoxigenin-labeled riboprobes and detected with an alkaline phosphatase-conjugated antidigoxigenin antibody (45).
Cell Transfection and Analysis.
For 416B stable transfections, 10μg of linearized plasmid DNA and 1 μg of linearized pGK neo were co-electroporated. Transfected cells were selected at 24 h by adding 500 μg/ml G418 and were assayed 2 weeks later (17).
FACS.
Single-cell suspensions of E12.5 FLs (L5760 × WT) were stained and analyzed as in SI Materials and Methods and ref. 13. FDG+ and FDG− live cell populations were sorted for in vitro colony and transplantation assays (10, 13).
In Vitro Colony Assays.
Sorted E12.5 FL cells were counted and cultured in cytokine supplemented Methocult GF-3434 for erythroid and myeloid colony formation (10). See SI Materials and Methods for details.
Transplantation Assays.
Sorted E12.5 FL cells were counted, suspended in PBS, and injected into the tail vein of 3- to 5-month-old irradiated (CBA × C57Bl/6) F1 mice along with 2 × 105 nontransgenic splenic cells for radioprotection (10). See SI Materials and Methods for details.
ES Cell Culture and Analysis.
Bry-GFP ES cells were maintained and differentiated as described in ref. 27. See SI Materials and Methods for details.
ChIP Assays.
See SI Materials and Methods for details.
Supplementary Material
Supporting Information
Acknowledgments
We thank Aimee Parker for help with embryo dissections; Paula Braker, Sandie Piltz, and Michelle Hammett for generating transgenic mice and help with transplants; Rachael Walker for help with FACS; Martin Gering, Aldo Ciau-Uitz, Uri Alon, Diego Miranda-Saavedra, and Brian Huntly for helpful discussions; and Catherine Porcher (Oxford University, Oxford, U.K.) for the anti-Scl antibody used in the ChIP. This work was supported by grants from the Leukaemia Research Fund, the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and by fellowships from the National Health and Medical Research Council (Australia) (to J.E.P.), the Kay Kendall Leukaemia Fund (to K.O.), and the Canadian Institutes for Health Research (to J.R.L.).
Abbreviations
AGM
aorta–gonad-mesonephros
DA
dorsal aorta
E_n_
embryonic day n
EB
embryoid body
FDG
fluoro-deoxy-d-glucose
FL
fetal liver
GRN
gene-regulatory network
HSC
hematopoietic stem cell
TF
transcription factor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Weissman IL. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 2.Enver T, Greaves M. Cell. 1998;94:9–12. doi: 10.1016/s0092-8674(00)81215-5. [DOI] [PubMed] [Google Scholar]
- 3.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 4.Robb L, Elwood NJ, Elefanty AG, Kontgen F, Li R, Barnett LD, Begley CG. EMBO J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 6.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine M, Tjian R. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi-Osaki M, Ohneda O, Suzuki N, Minegishi N, Yokomizo T, Takahashi S, Lim KC, Engel JD, Yamamoto M. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez MJ, Bockamp EO, Miller J, Gambardella L, Green AR. Development (Cambridge, UK) 2001;128:4815–4827. doi: 10.1242/dev.128.23.4815. [DOI] [PubMed] [Google Scholar]
- 11.Sinclair AM, Gottgens B, Barton LM, Stanley ML, Pardanaud L, Klaine M, Gering M, Bahn S, Sanchez M, Bench AJ, et al. Dev Biol. 1999;209:128–142. doi: 10.1006/dbio.1999.9236. [DOI] [PubMed] [Google Scholar]
- 12.Wozniak RJ, Boyer ME, Grass JA, Lee Y, Bresnick EH. J Biol Chem. 2007;282:14665–14674. doi: 10.1074/jbc.M700792200. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez M, Gottgens B, Sinclair AM, Stanley M, Begley CG, Hunter S, Green AR. Development (Cambridge, UK) 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- 14.Levine M, Davidson EH. Proc Natl Acad Sci USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Proc Natl Acad Sci USA. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson EH, Erwin DH. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 17.Gottgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient R, Green AR. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffredo T, Gautier R, Eichmann A, Dieterlen-Lievre F. Development (Cambridge, UK) 1998;125:4575–4583. doi: 10.1242/dev.125.22.4575. [DOI] [PubMed] [Google Scholar]
- 19.Ciau-Uitz A, Walmsley M, Patient R. Cell. 2000;102:787–796. doi: 10.1016/s0092-8674(00)00067-2. [DOI] [PubMed] [Google Scholar]
- 20.Gering M, Rodaway AR, Gottgens B, Patient RK, Green AR. EMBO J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donaldson IJ, Chapman M, Kinston S, Landry JR, Knezevic K, Piltz S, Buckley N, Green AR, Gottgens B. Hum Mol Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. [DOI] [PubMed] [Google Scholar]
- 22.Yilmaz OH, Kiel MJ, Morrison SJ. Blood. 2006;107:924–930. doi: 10.1182/blood-2005-05-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan WY, Follows GA, Lacaud G, Pimanda JE, Landry JR, Kinston S, Knezevic K, Piltz S, Donaldson IJ, Gambardella L, et al. Blood. 2007;109:1908–1916. doi: 10.1182/blood-2006-05-023226. [DOI] [PubMed] [Google Scholar]
- 24.Pimanda JE, Donaldson IJ, de Bruijn MF, Kinston S, Knezevic K, Huckle L, Piltz S, Landry JR, Green AR, Tannahill D, et al. Proc Natl Acad Sci USA. 2007;104:840–845. doi: 10.1073/pnas.0607196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dzierzak E. Curr Opin Hematol. 2005;12:197–202. doi: 10.1097/01.moh.0000160736.44726.0e. [DOI] [PubMed] [Google Scholar]
- 26.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. Development (Cambridge, UK) 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 27.Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V. Development (Cambridge, UK) 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 28.Cripps RM, Olson EN. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 29.Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. Proc Natl Acad Sci USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal L, Banerjee U, Hartenstein V. Nat Genet. 2004;36:1019–1023. doi: 10.1038/ng1404. [DOI] [PubMed] [Google Scholar]
- 31.Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, Hultmark D. Proc Natl Acad Sci USA. 2004;101:14192–14197. doi: 10.1073/pnas.0403789101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varterasian M, Lipkowitz S, Karsch-Mizrachi I, Paterson B, Kirsch I. Cell Growth Differ. 1993;4:885–889. [PubMed] [Google Scholar]
- 33.Kumano K, Chiba S, Kunisato A, Sata M, Saito T, Nakagami-Yamaguchi E, Yamaguchi T, Masuda S, Shimizu K, Takahashi T, et al. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 34.Robert-Moreno A, Espinosa L, de la Pompa JL, Bigas A. Development (Cambridge, UK) 2005;132:1117–1126. doi: 10.1242/dev.01660. [DOI] [PubMed] [Google Scholar]
- 35.Gering M, Patient R. Dev Cell. 2005;8:389–400. doi: 10.1016/j.devcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Lawson ND, Vogel AM, Weinstein BM. Dev Cell. 2002;3:127–136. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 37.Oren T, Torregroza I, Evans T. Nucleic Acids Res. 2005;33:4357–4367. doi: 10.1093/nar/gki746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walmsley M, Ciau-Uitz A, Patient R. Development (Cambridge, UK) 2002;129:5683–5695. doi: 10.1242/dev.00169. [DOI] [PubMed] [Google Scholar]
- 39.Khandekar M, Brandt W, Zhou Y, Dagenais S, Glover TW, Suzuki N, Shimizu R, Yamamoto M, Lim KC, Engel JD. Development (Cambridge, UK) 2007;134:1703–1712. doi: 10.1242/dev.001297. [DOI] [PubMed] [Google Scholar]
- 40.Ono Y, Fukuhara N, Yoshie O. Mol Cell Biol. 1998;18:6939–6950. doi: 10.1128/mcb.18.12.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Development (Cambridge, UK) 1999;126:2799–2811. doi: 10.1242/dev.126.12.2799. [DOI] [PubMed] [Google Scholar]
- 42.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. London: Chapman & Hall; 2007. [Google Scholar]
- 43.Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. Proc Natl Acad Sci USA. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ottersbach K, Dzierzak E. Dev Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information