The structure of FtsZ filaments in vivo suggests a force-generating role in cell division (original) (raw)
Abstract
In prokaryotes, FtsZ (the _f_ilamentous _t_emperature _s_ensitive protein Z) is a nearly ubiquitous GTPase that localizes in a ring at the leading edge of constricting plasma membranes during cell division. Here we report electron cryotomographic reconstructions of dividing Caulobacter crescentus cells wherein individual arc-like filaments were resolved just underneath the inner membrane at constriction sites. The filaments' position, orientation, time of appearance, and resistance to A22 all suggested that they were FtsZ. Predictable changes in the number, length, and distribution of filaments in cells where the expression levels and stability of FtsZ were altered supported that conclusion. In contrast to the thick, closed-ring-like structure suggested by fluorescence light microscopy, throughout the constriction process the Z-ring was seen here to consist of just a few short (∼100 nm) filaments spaced erratically near the division site. Additional densities connecting filaments to the cell wall, occasional straight segments, and abrupt kinks were also seen. An ‘iterative pinching' model is proposed wherein FtsZ itself generates the force that constricts the membrane in a GTP-hydrolysis-driven cycle of polymerization, membrane attachment, conformational change, depolymerization, and nucleotide exchange.
Keywords: bacterial cytoskeleton, cell division, cryoelectron microscopy, FtsZ, tomography
Introduction
All life depends on the ability of cells to divide. While many aspects of this complex process are already understood in eukaryotic cells, our knowledge of the mechanisms driving what would appear to be the more simple process of ‘binary fission' in prokaryotes is surprisingly poor. At least 13 different proteins function cooperatively at the division site in Escherichia coli (Goehring and Beckwith, 2005; Margolin, 2005; Rothfield et al, 2005). FtsZ, the _f_ilamentous _t_emperature _s_ensitive protein Z, is the first to localize and assembles into a ring-like structure called the FtsZ-ring or Z-ring near the cytoplasmic membrane at midcell (Begg and Donachie, 1985; Taschner et al, 1988; Bi and Lutkenhaus, 1991). While the full role of the Z-ring is still unclear, it is required for the recruitment of the rest of the cell division machinery and might help to provide the force required to constrict the membrane. As such a vital molecule, FtsZ is present in virtually all prokaryotes and in some chloroplasts and mitochondria as well (Michie and Löwe, 2006).
FtsZ has been identified as a homolog of eukaryotic tubulin based on its possession of a tubulin signature motif and its remarkable similarity to tubulin in both structure and biochemical properties (Löwe and Amos, 1998; Nogales et al, 1998a, 1998b). While FtsZ polymerizes into protofilaments, sheets, ribbons, minirings, and tubes under different in vitro experimental conditions (Mukherjee and Lutkenhaus, 1994; Erickson et al, 1996; Gonzalez et al, 2003), which of these structures are physiologically relevant is still unclear. Like tubulin, FtsZ is a GTPase. GTP-tubulin polymerizes into straight microtubules, where the GTP at the dimer interfaces is quickly hydrolyzed. Because the GDP-bound state favors a curved protofilament conformation, microtubules must be stabilized by ‘caps' of GTP-bound dimers at their ends, and they exhibit dynamic instability: when the GTP cap is hydrolyzed, individual protofilaments quickly peel away from each other to produce curved rings (Nogales, 2001). How FtsZ assembles into polymers and the role of GTP hydrolysis is still unknown. As for tubulin, the GTP- and GDP-bound states of FtsZ seem to favor straight and curved conformations, respectively, but not exclusively (Lu et al, 2000). In vitro evidence suggests that FtsZ polymerizes in a cooperative manner (Caplan and Erickson, 2003; Chen et al, 2005), but an isodesmic mechanism that includes the cyclization of long single-stranded fibrils has also been proposed (Gonzalez et al, 2005).
Our knowledge of the in vivo structure of FtsZ has advanced incrementally with improvements in imaging technology. The Z-ring was first identified by gold-labeled antibodies that were seen by electron microscopy to localize to the leading edges of the midcell invagination (Bi and Lutkenhaus, 1991). Soon afterwards, the application of fluorescent light microscopic (fLM) techniques to bacterial cells (Margolin, 1998) revealed the Z-ring's donut-like structure in 3-D (Levin and Losick, 1996; Sun and Margolin, 1998). More recently, fLM techniques of living cells have revealed dynamics: FtsZ appears to assemble into arc-like and spiral structures that move and collapse during the cell cycle (Addinall and Lutkenhaus, 1996; Ben-Yehuda and Losick, 2002; Thanedar and Margolin, 2004). Fluorescence recovery after photobleaching (FRAP) experiments show that FtsZ molecules rapidly exchange between the Z-ring and the cytoplasmic pool with a half time of ∼10 s (Anderson et al, 2004). Here, we have used the emerging technique electron cryotomography (ECT) to further advance our understanding of the structure and function of FtsZ. ECT can reveal the three-dimensional ultrastructure of small, intact cells in a life-like, ‘frozen-hydrated' state (Lucic et al, 2005; Jensen and Briegel, 2007). ECT has already been used to visualize several bacterial cytoskeletal filaments directly in situ (Kurner et al, 2005; Komeili et al, 2006; Scheffel et al, 2006), and we recently used it to visualize a number of novel filament bundles in the widely studied model bacterium Caulobacter crescentus (Briegel et al, 2006).
C. crescentus is a favorite model organism for studying bacterial cell development because its characteristic asymmetrical division yields two morphologically different progeny: a sessile, stalked cell and a motile, flagellated cell. The stalked cell can immediately induce another round of cell division by replicating chromosomal DNA and expressing cell division proteins. In contrast, the flagellated ‘swarmer' cell must differentiate into a stalked cell by shedding its flagellum and assembling a stalk and holdfast at the newly vacated pole before it becomes replication competent. In accordance with these dramatic morphological changes, a large number of proteins experience cell cycle-dependent regulation on both the transcriptional and translational levels (Laub et al, 2000; Grunenfelder et al, 2001). The concentration of FtsZ begins at a very low number in swarmer cells, increases sharply before cell division, and then drops rapidly after division through proteolysis (Quardokus et al, 1996; Kelly et al, 1998). A process involving the protein MipZ positions the Z-ring at the midcell (Thanbichler and Shapiro, 2006).
By imaging the division sites of C. crescentus at high magnification with an electron cryomicroscope, we consistently observed arc-like filaments lying underneath the inner membrane at midcell. The filaments' position, configuration, time of appearance, and resistance to A22 all suggested that they were FtsZ. The filaments' absence in an FtsZ depletion strain, higher abundance in strains overexpressing FtsZ, and dramatically higher abundance and longer lengths in a strain expressing a more stable FtsZ mutant further indicated that the filaments were in fact FtsZ. Through analysis of these filaments, the architecture of the Z-ring in C. crescentus is described and an existing model for the role of FtsZ filaments in cell division is elaborated.
Results
Section one: Identification of FtsZ filaments by ECT
Observation of midcell arc-like filaments in predivisional wild-type C. crescentus. Predivisional NA1000 C. crescentus cells were plunge-frozen and observed in an electron cryomicroscope. Series of images were recorded of individual cells while the sample was tilted incrementally. Three-dimensional reconstructions of the cells (tomograms) were then calculated which clearly showed outer membranes, peptidoglycan layers, inner membranes, and crowded cytoplasms full of ribosome-like particles (Figure 1). Because good images of samples tilted to angles greater than ∼65° cannot yet be obtained (Iancu et al, 2005), the resolution of the reconstructions was anisotropic and the boundaries of objects were blurred in the direction parallel to the beam. Thus the membranes at the top and bottom of the cell were not resolved, and any filaments perpendicular to the beam would also have been difficult to discern.
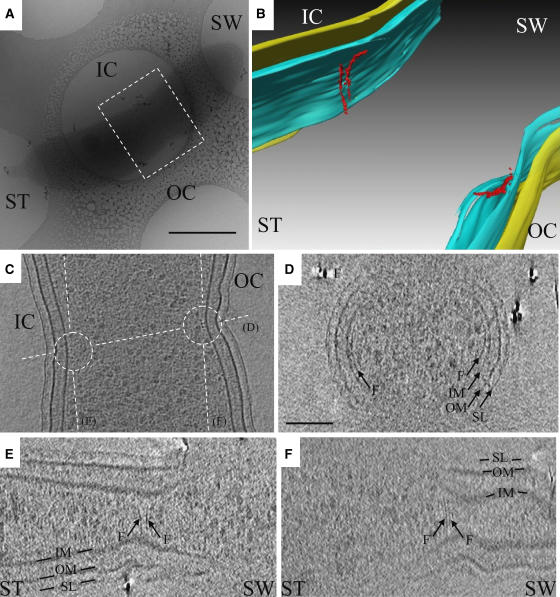
Figure 1.
Arc-like filaments at the division site in C. crescentus NA1000. (A) Low magnification cryo-EM view of a C. crescentus NA1000 cell plunge-frozen on an EM grid after acquisition of the tilt-series. The darker gray elongated object is the cell, suspended in vitreous ice across a circular hole in the carbon support film. The dashed box shows the region segmented in (B). ST: stalked pole; SW: pole opposite the stalked pole; IC: inner curvature; OC: outer curvature. Scale bar 500 nm. (B) 3-D segmentation of the inner membrane (blue), outer membrane (yellow), and arc-like filaments (red) in the reconstruction from the same cell. Because the image contains 3-D perspective effects, no scale bar is included. (C) 8 nm slice through the reconstruction parallel to the grid, showing the filaments in cross-section (small dark dots near the center of the circles) next to the membrane as well as the orientations of the other slices shown in panels (D–F) (dashed lines). (D–F) 5.4 nm slices through the reconstruction, showing that while filaments and other small proteins all appeared as small dots in cross-section, filaments were recognized by their continuous, elongated shape in ‘side' views such as these. Unfortunately, space does not allow all relevant slices to be shown. Thus here and in other figures, the locations and extents of all visible filaments were manually traced and presented as colorized, segmented models (like panel B), which are necessarily interpretations but do allow 3-D structures to be shown in 2-D figures. The colors and labels are consistent within all the figures: IM: inner membrane; OM: outer membrane; SL: surface layer; F: arc-like (FtsZ) filaments. The scale bar in (D) is 100 nm and serves for panels (C-F).
Close examination of the 3-D tomograms of predivisional cells revealed arc-like filaments lying just inside the inner membranes on one or both sides of the midcell constriction approximately perpendicular to the long axis of the cell (Figure 1B–F). Such filaments were seen in seven reconstructed predivisional cells, always just inside the constriction point (two out of seven cells had filaments just on one side, the other five cells had filaments on both sides). In the particular cell shown in Figure 1, the two filaments seen along the inner curvature of the cell were not strictly parallel (Figure 1E), but two other filaments on the outer curvature were (Figure 1F). The filaments were between 80 and 160 nm long, covered arc-lengths of 0.45–0.85 radians, and were ∼5 nm in diameter.
Appearance, configuration, and disassembly of the filaments during cell division. In ECT, the cells being imaged are frozen and cannot grow, and the high-energy electrons used to produce the images gradually destroy biomolecular structures, so it is not possible to gather data from the same cell repeatedly as it progresses through division. Instead, pseudo-time-courses must be built up by arranging reconstructions of different cells imaged at various stages of the process in relative order. During the course of these experiments, a total of 15 stalked NA1000 cells were reconstructed. Morphological features like the presence or absence of a flagellum, the length-to-width ratio of the cell body, and the degree of membrane constriction were used to place these cells in relative order approximating the progression of C. crescentus through cell division (Figure 2). Of the four stalked cells that were not yet visibly constricted, only the one with the longest length-to-width ratio (5.6) had midcell arc-like filaments (Figure 2B), consistent with the known timing of FtsZ localization to the midcell (Quardokus et al, 2001).
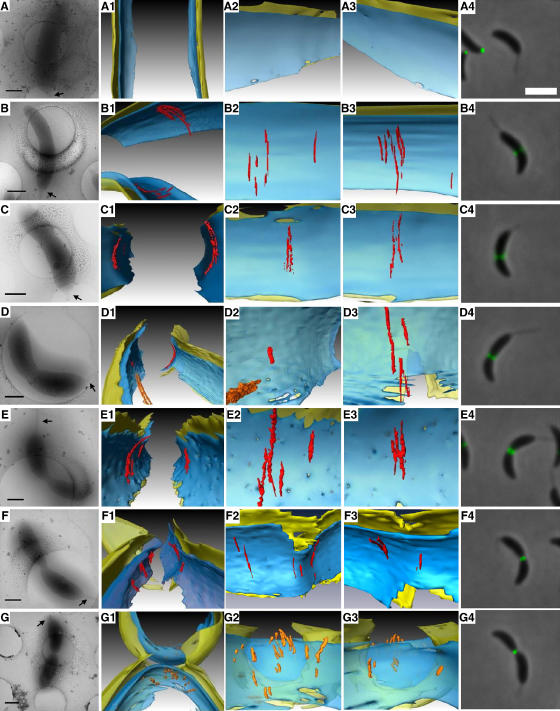
Figure 2.
Filament dynamics during cell division. Each row represents a different stage of cell division, arranged in approximate sequence from an unconstricted stalked state (A) until after fission of the inner membrane has formed two separate cytoplasmic compartments (G). Within each row, the first four columns show different depictions of a single cell reconstructed by ECT and the last column shows an fLM image of a different cell taken at an apparently analogous stage. (A–G) Low magnification cryo-EM images. Stalked poles are shown with arrows. Scale bars 500 nm. (A1–G1) 3-D segmentations of the division sites. (A2–G2) ‘Face-on' views from the cytoplasm of the ‘left' side of the cell wall. (A3–G3) ‘Face-on' views of the ‘right' side of the cell wall, again from the cytoplasm. (A4–G4) fLM images of NA1000 cells expressing FtsZ-YFP. Scale bar 2 μm. In D1–D2, an additional cytoplasmic filament bundle (Briegel et al, 2006) was surface-rendered in orange. In G1–G3, no arc-like filaments are observed; instead, the most filamentous densities are surface-rendered in orange for comparison. Note that the cells shown in rows C and E are the same as those shown in Figures 1 and 5AB, respectively, and that supporting unsegmented tomographic slices are shown in Supplementary Figures S1–S6.
There were 10 stalked cells (the seven mentioned earlier, plus three additional A22-treated cells that will be discussed later) that were visibly constricted to different degrees but whose inner membrane had not yet separated. Midcell arc-like filaments were seen in every case, but there was no discernable evolution in their number, position, or configuration (Figure 2C–F). Most of the filaments were between 40 and 120 nm, and the longest was ∼160 nm. Although the ‘missing wedge' in ECT smears details in the direction of the electron beam, the arc-like filaments were nevertheless clearly short chords and arcs rather than complete rings. In this respect, the two membranes served as omnipresent internal controls: they are consistently blurred at the same positions around the top and the bottom of the cell, but the arc-like filaments frequently ended nearer the center, before the missing wedge would have obscured them (see also Figures 7B, G and 10G). The numbers of filaments on each side were not consistent or equal. In the cell shown in Figure 2D, for example, there were four filaments of different length on the right-hand side but only one very short one on the left, but in the cell shown in Figure 2C, there were two filaments on each side. More pronounced asymmetries were also seen: in one case, there were nine filaments on one side and none on the other (data not shown). No regular bundling pattern, spacing, or relationship was observed between neighboring filaments. In one cell, an additional long filament bundle parallel to the long axis of the cell (orange filaments in Figure 2D) was observed in the cytoplasm near the midcell arc-like filaments that was similar to the ‘cytoplasmic bundle' identified in earlier work (Briegel et al, 2006).
In the last (15th) cell of the sequence, the outer membrane was deeply constricted and the inner membrane had separated to form two distinct cytoplasmic compartments (Figure 2G), as has been observed previously (Judd et al, 2005). The diameter across the constriction in the outer membrane was 154 nm. In this cell, no clearly arc-like filaments were observed at the division site. While there were a number of elongated densities (orange in Figure 2G), they were significantly shorter than the midcell arc-like filaments and their distances from the membrane were more variable.
fLM images of cells at apparently similar stages of division were collected for comparison (right column of Figure 2), showing that the midcell arc-like filaments assemble at the same time as Z-rings localize to the midcell (in very late phase unconstricted stalked cells), and both structures persist throughout constriction. Following separation of the inner membranes, FtsZ-YFP remained localized to the new poles (Thanbichler and Shapiro, 2006), but the midcell arc-like filaments were no longer identifiable.
Filament abundance and length in mutants with different levels of FtsZ expression and activity. Because the midcell arc-like filaments' position, orientation, and dynamics all suggested that they were FtsZ, the following further experiments were carried out to test that hypothesis. Since effective labeling methods for ECT of intact cells have not yet been developed, instead we looked at cells expressing different levels and forms of FtsZ to see if the number and/or length of the putative FtsZ filaments changed correspondingly. First, NA1000 cells overexpressing FtsZ were imaged (Quardokus et al, 2001). Presumably because the stoichiometry of FtsZ to other components of the divisome is critical, overexpression of FtsZ blocks cell division and produces extended constricted sites containing multiple FtsZ foci (Figure 3A). Tomograms of these cells exhibited arc-like filaments with the same diameter, curvature, distance from the membrane, and orientation with respect to the long axis of the cell as seen previously in dividing NA1000 cells, but the filaments were more numerous (14 arc-like filaments in ∼1.3 × 0.6 × 0.3 μm3) and were collected in bunches at the most constricted points (Figure 3C–E), just as would be expected for FtsZ in this overexpression strain.
Figure 3.
Increase in the abundance of arc-like filaments in cells overexpressing FtsZ. (A) Differential interference contrast (DIC) and fLM overlay image of cells overexpressing (wild-type) FtsZ, showing that FtsZ-YFP accumulates in the most constricted points. Scale bar 4 μm. (B) Low magnification cryo-EM view of a similar cell plunge-frozen in vitreous ice across an EM grid after acquisition of the tilt-series. The gray box shows the region segmented in panel (E). For scale, the circular hole in the carbon support film on the EM grid has a diameter of 2 μm. (C) 8 nm slice through the tomogram parallel to the grid. Scale bar 100 nm (for panels C and D). (D) 5 nm slice through the tomogram along the dash line in (C). The white arrow points to the arc-like filaments. (E) 3-D segmentation.
Next, cells overexpressing a hyperstable mutant form of FtsZ (FtsZG109S) were imaged. This mutation is located close to the nucleotide-binding pocket and corresponds to the classical mutant FtsZ84 in E. coli, which has been shown to slow both GTP-binding and hydrolysis activity. These changes lead to an ∼3-fold longer half-time of fluorescence recovery in FRAP experiments, indicating that the filaments it forms are less dynamic/more stable (Anderson et al, 2004). In C. crescentus, overexpression of FtsZG109S blocks cell division and produces long, deeply constricted regions containing large amounts of FtsZ, including extended patches (Wang et al, 2001) (Figure 4A). Tomograms of such deeply constricted regions revealed large numbers of tightly packed arc-like filaments (Figure 4B–E). In the ∼400 × 400 × 900 nm3 volume shown in Figure 4E, for instance, ∼50 arc-like filaments can be seen on both sides of the cell. Again these filaments were indistinguishable from those seen earlier, except that some filaments were much longer than any seen previously and they were much more numerous so that entire patches of the cell wall were covered and neighboring filaments were frequently seen aggregated into bundles of 4–5 filaments. Since many of these filaments extended across the top or bottom of the cells where they were difficult to resolve, their full lengths could not be determined. Nevertheless, because the extended division sites were thinner (∼300 nm diameter) than those in NA1000 cells, better resolutions were achieved in the reconstructions and at least at the thinnest point (boxed region in Figure 4E), it was almost possible to follow these longer mutant filaments around the entire circumference of the cell. Here the filaments appeared to form helices that wrapped all the way around the inner membrane (Figure 9C).
Figure 4.
Dramatic increase in the abundance and length of filaments in a mutant overexpressing FtsZG109S. (A) fLM image of cells overexpressing FtsZG109S, showing that FtsZ-YFP accumulates in the extended, constricted division sites. Scale bar 4 μm. (B) Low magnification cryo-EM image of a cell overexpressing FtsZG109S. Scale bar 500 nm. (C) 8 nm slice through the 3-D reconstruction parallel to the grid, showing tens of filaments in cross-section as small black dots close to the membrane. The dashed lines show the position of the slice shown in panel (D). Scale bar 100 nm (for panels C and D). (D) 5.4 nm ‘glancing' slice through the 3-D reconstruction showing tens of filaments just underneath the inner membrane. (E) 3-D segmentation with another cytoplasmic filament bundle colored orange. The box identifies a region referred to in Figure 9C.
As a final, ‘negative' control, strain YB1585 was imaged, wherein FtsZ can be conditionally depleted (Wang et al, 2001). When FtsZ is depleted, these cells elongate without visible constrictions (Supplementary Figure S8). While it is unclear where to look for filaments in these cells, tomograms of random samples throughout the cell body were collected, and no arc-like filaments were seen (Supplementary Figure S9).
Persistence of midcell arc-like filaments in the presence of the MreB-inhibitor A22. The correlation in the length, number, and distribution patterns of the filaments with the known FtsZ expression levels, stability, and fluorescence localization patterns in these three mutant strains strongly supported the proposition that the filaments were in fact FtsZ, rather than for instance some other, unknown filament that might also be found at the division site. Nevertheless there is at least one other filament, composed of the actin-like MreB, that is thought to be present in a ring at the midcell of predivisional cells. By fLM, MreB usually appears to form helical structures along the cell wall that span most of the long axis of the cell body, but these have been observed to condense into a ring-like structure when C. crescentus divides (Figge et al, 2004; Gitai et al, 2004). A22 was identified as a small molecule that diffuses rapidly through the cell membrane and depolymerizes MreB (Gitai et al, 2005). To rule out the possibility that the midcell arc-like filaments were MreB, two types of cells that exhibited these filaments (NA1000 cells and cells overexpressing FtsZG109S) were treated with A22 and imaged by ECT as before. Three A22-treated dividing NA1000 cells were reconstructed, and all three possessed arc-like filaments that were indistinguishable from those seen in the nontreated cells (Figure 5A and B). Similarly, two A22-treated cells overexpressing FtsZG109S were reconstructed which also both still exhibited large numbers of densely packed, unusually long filaments (Figure 5C and D). Supporting fLM studies were carried out in both cases to confirm that the A22 treatment did in fact delocalize the MreB foci as expected but had no perceptible effect on FtsZ (Supplementary Figure S10).
Figure 5.
Arc-like filaments persist in the presence of A22. Low magnification cryo-EM views (A, C) and corresponding segmented 3-D reconstructions (B, D) of a NA1000 cell (A, B) and a cell overexpressing FtsZG109S (C, D), both after treatment with A22, again showing arc-like filaments. Scale bar in (A) 500 nm. For scale in (C), the circular hole in the carbon support film on the EM grid has a diameter of 1.3 μm. See also supporting fLM studies in Supplementary Figure S10.
Taken together, these results provide compelling evidence that the arc-like filaments observed at the division site were in fact FtsZ: (1) they appeared at the leading edge of the constriction where FtsZ is known to localize; (2) their width, position inside the membrane, and orientation were as expected for FtsZ filaments; (3) the timing of their appearance and their persistence during cell division followed FtsZ's known dynamic pattern; (4) they persisted in the presence of A22, which depolymerizes the other filament (MreB) thought to exist at midcell in dividing cells; (5) their abundance correlated with increases and decreases in the expression of FtsZ; (6) their numbers and length were both dramatically increased in a strain overexpressing a more stable, mutant form of FtsZ; and (7) the details of their distribution pattern (bunches versus extended patches) in two different overexpression strains matched what had been observed for FtsZ by fLM.
Section two: Characterization and measurement of FtsZ filaments
In total, 248 putative FtsZ filaments were seen (36 filaments in 11 NA1000 cells, 30 filaments in two cells overexpressing wild-type FtsZ, and 182 filaments in three cells overexpressing FtsZG109S).
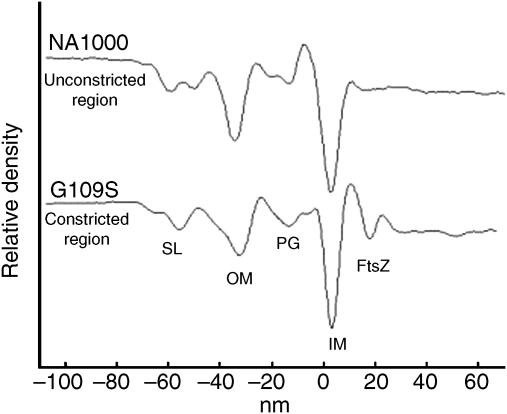
Distance from and connections to the inner membrane. FtsZ filaments were close to but not lying directly against the inner membrane. To measure their average distance from the membrane, a ‘profile' of all the densities seen as a function of distance both above and below the inner membrane was calculated in one of the constricted regions of a cell overexpressing FtsZG109S (Figure 6). Outside the inner membrane, the expected peaks for the peptidoglycan layer, outer membrane, and surface layer were observed. Inside the inner membrane, an additional peak was resolved 16 nm (peak to peak) into the cytoplasm corresponding to the accumulated FtsZ filaments. As a control, a similar calculation on an unconstricted region of an NA1000 cell where no filaments were observed showed all the same peaks except the cytoplasmic peak attributed to FtsZ.
Figure 6.
FtsZ filaments are on average 16 nm away from the membrane. Density profiles through the cell wall of a cell overexpressing FtsZG109S in the region of an extended division site containing FtsZ filaments (lower curve) and a random, unconstricted region of one NA1000 cell body not containing FtsZ as a control (top curve). The horizontal axis is the distance either outside (negative numbers) or inside (positive numbers) the inner membrane. The surface layer (SL), outer membrane (OM), peptidoglycan (PG), inner membrane (IM), and FtsZ filament layers can be resolved.
Most of the FtsZ filaments showed no apparent connection to the inner membrane (one example from a cell overexpressing FtsZG109S is shown in Figure 7A). In a minority of cases (7, 4, and 23 filaments analyzed in NA1000 cells, cells overexpressing wild-type FtsZ, and cells overexpressing FtsZG109S, respectively), however, mid-filament ‘bridge-like' densities connected the filaments to the inner membrane (Figure 7B and C) and/or the peptidoglycan (Figure 7D). In seven additional cases (all from cells overexpressing FtsZG109S), the filaments appeared to originate (or terminate) in the inner membrane (Figure 7E and F). In some rare instances, the inner membrane and the filament seemed to deviate from smooth arcs together, as if their separation was rigidly controlled (Figure 7G), but in some other rare cases, filaments were seen unusually close to the membrane (Figure 7H). Unfortunately, the identities of the bridging macromolecules remain unclear, some of them may have been present for reasons other than to fulfill a specific bridging function, and it is possible that additional bridges existed which were too small to resolve by these methods.
Figure 7.
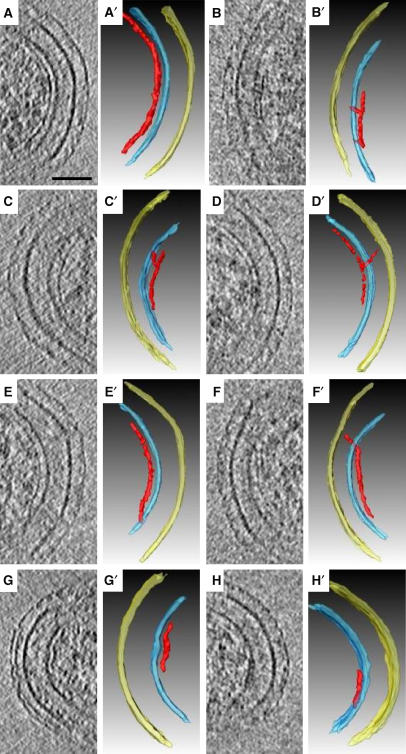
Connections between FtsZ filaments and the cell wall. (A–H) 6.7 nm slices containing individual FtsZ filaments. (A′–H′) Corresponding interpretive 3-D segmentations. The different panels show a smoothly curved filament without apparent connections to the cell wall (A), filaments connected to the inner membrane in their midsections (B and C), a filament connected to the peptidoglycan (D), filaments connected to the membrane at their termini (E and F), a filament tightly following the curvature of an indentation in the membrane (G), and a filament unusually close to the membrane with multiple connections (H). Panels B (B′) and G (G′) are NA1000 cells, the other panels are cells overexpressing FtsZG109S. Scale bar 50 nm.
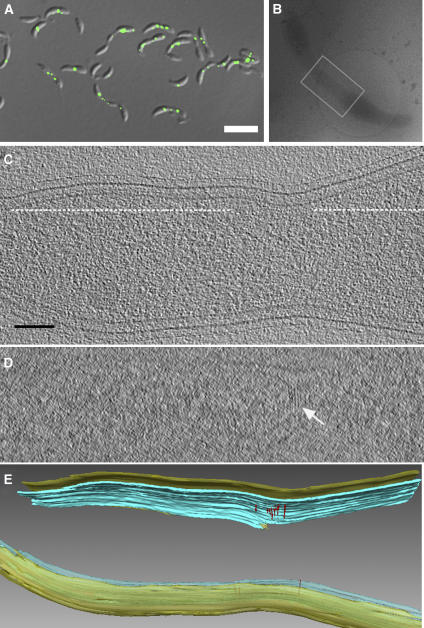
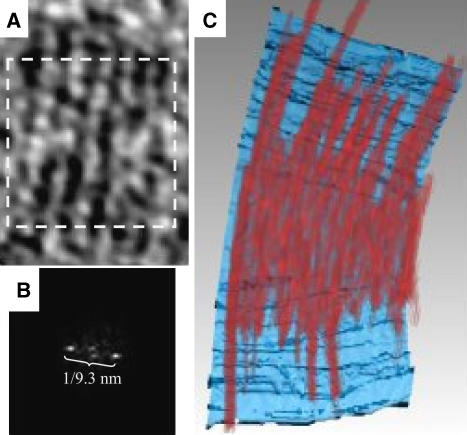
Lateral spacing in the cells overexpressing FtsZG109S. Extensive lateral interactions between tubulin molecules play a fundamental role in microtubule assembly (Nogales et al, 1998b). In vitro, FtsZ can form sheets, tubes, or even 2-D crystals, suggesting that FtsZ too can become ordered through stabilizing lateral interactions (Löwe and Amos, 1999). In the cells overexpressing FtsZG109S, a regular spacing between neighboring arc-like filaments was observed. The power spectrum of a 2-D slice through several filaments revealed this spacing to be 9.3 nm on average (Figure 8). This spacing is different, however, from that seen in calcium-induced 2-D crystals of FtsZ (7.2 nm), where ‘thick' filaments interacted through FtsZ C-termini (Löwe and Amos, 1999). No discernable regularity of spacing was seen between the sparser filaments present in NA1000 or cells overexpressing wild-type FtsZ.
Figure 8.
Lateral spacing of FtsZ filaments in cells overexpressing FtsZG109S. (A) 6.7 nm ‘glancing' slice just inside the inner membrane of the cell overexpressing FtsZG109S depicted in Figure 5C showing a flat ribbon of six adjacent filaments. (B) Power spectrum of the boxed area in panel (A). The single pair of prominent peaks reveals the average distance between filaments as 9.3 nm. (C) Volume-rendering of the same ribbon of filaments in red, allowing the relative density of all the voxels of the 3-D structure to be perceived without interpretive segmentation. In the background, the inner membrane is surface-rendered in blue.
Orientation with respect to the cell body. The FtsZ filaments lay in planes approximately perpendicular to the long axis of the cell body. Because the rod-shaped cells lay flat on the specimen grid parallel to one face of the reconstruction, the pitch, or angle, of each filament with respect to the long axis of the cell could be measured quite accurately by noting the angular rotation required to cause each filament in the reconstruction to appear most completely within a single thin tomographic slice (Figure 9A). In the NA1000 cells, cells overexpressing wild-type FtsZ, and cells overexpressing FtsZG109S, the mean pitch of the filaments was 8±6° (_n_=36, median 5°), 8±7° (_n_=30, median 6°), and 11±11° (_n_=182, median 8°), respectively (Figure 9B). Because C. crescentus has two distinct cell poles and an inner curvature, these landmarks were used to define a unique 3-D coordinate system within each cell. Each filament was then classified as spiraling either ‘right-' or ‘left-' handed, but no patterns that would have potentially related cell polarity and the filament pitch sense were detected. As mentioned above, some regions of the cells overexpressing FtsZG109S were thin enough that individual filaments could almost be traced across the top and bottom of the cell in the reconstructions. In one case, three adjacent single filaments that all nearly circled the cell were seen to be interlaced helices rather than closed rings (Figure 9C).
Figure 9.
Orientation of FtsZ filaments. (A) 3-D segmentation superimposed on a ‘glancing' slice through the reconstruction of the extended division site of a cell overexpressing FtsZG109S. Pitch (θ) was defined as the angle of the plane of each filament with respect to the long axis of the cell. (B) Pitches of observed filaments from three cells overexpressing FtsZG109S (182 filaments), 11 NA1000 cells (36 filaments), and two cells overexpressing (wild-type) FtsZ (30 filaments). (C) Two orthogonal views of a segmentation of the volume boxed in Figure 4E, showing that FtsZ filaments did not form closed rings, but rather interlaced spirals. Here each filament is surface-rendered in a different color.
Straight FtsZ filaments and conformational changes. While most of the filaments were smoothly curved, many straight segments were also seen (Figure 10). In the three cells overexpressing FtsZG109S, for instance, 28 out of 182 filaments analyzed (15%) were either completely straight or had a clearly straight segment (Figure 10A–F). Straight segments were also observed in NA1000 (eight out of 36 filaments, Figure 10G and H) and cells overexpressing wild-type FtsZ (10 out of 30 filaments, Supplementary Figure S11). Where straight segments approached the membrane, they either terminated in the membrane (white arrowheads in Figure 10A and B), exhibited distinct kinks (black arrows in Figure 10), or transitioned into smoothly curved conformations following the contours of the membrane (for example, Figure 10F).
Figure 10.
Straight and curved segments of FtsZ filaments. 6.7 nm slices containing example straight and curved segments of FtsZ filaments. Abrupt ‘kinks' were sometimes seen (black arrows) as well as direct connections of straight filaments to the membrane (white arrowheads). Panels (G) and (H) are NA1000 cells, the other panels (A–F) are cells overexpressing FtsZ G109. Scale bar 50 nm.
Discussion
Although the localization of FtsZ had been visualized previously by immuno-electron microscopy, the actual filaments themselves were not resolved probably because their structures were lost during the fixation, dehydration, plastic-embedment, and staining required for conventional EM. It now appears that cryo-EM techniques that preserve cells in more native, frozen-hydrated states will make it possible to resolve the architecture of the bacterial cytoskeleton. Filament-like structures were recently found at the constriction ring of Enterococcus gallinarum septum by cryo-EM of vitreous sections and suspected to be FtsZ (Zuber et al, 2006). In recent work on _C. crescentu_s, four types of large filament bundles were observed in the tomograms of NA1000 cells in different developmental stages (Briegel et al, 2006). One of the bundles (the so-called ‘ring-like' bundle) was speculated to be FtsZ, and shared many of the same characteristics as the midcell arc-like filaments described here (short arcs, perpendicular to long axis of cell, and following the contours of the inner membrane), but the filaments were spaced differently (11 versus 9.3 nm here) and appeared in a stacked bundle. As reported in the previous paper, the ring-like bundle was seen only once, and we have not yet seen it again in the ∼100 additional NA1000 cells that have since been imaged. Whether the ring-like bundle was in fact a very short-lived configuration of FtsZ or something else remains to be seen. In the current study, we imaged the division sites of various strains of C. crescentus at higher magnification. Under these conditions, arc-like filaments were consistently resolved at the leading edges of constricting sites (where filaments had not previously been seen at lower magnifications; Judd et al, 2005; Briegel et al, 2006). In addition to their position, orientation, time of appearance, and resistance to A22, the most compelling evidence that these filaments were in fact FtsZ was that their abundance correlated with the expression levels of FtsZ in depletion and overexpression strains and both their number and length dramatically increased in cells overexpressing FtsZG109S, a mutant form which is known to produce hyperstable FtsZ filaments in E. coli (Anderson et al, 2004).
Structure of the Z-‘ring'
In stark contrast to the thick, well-organized, complete ring-like structure suggested by fLM images, the Z-‘ring' of (wild-type) C. crescentus was seen here to consist of short (∼100 nm), independent filaments situated erratically near the division site in small numbers (just one or a few present at any given time). The plunge-freezing process is unlikely to have reduced the number, length, or positions of the filaments artefactually, since it happens so quickly that even the water in the buffer does not crystallize (Dubochet, 2007) (see also Supplementary Discussion). Their ∼5 nm diameter suggested that they were single protofilaments, which are known from crystal structures to be ∼4 nm thick (Löwe and Amos, 1998) and which have been shown by previous EM studies to form in vitro (Mukherjee and Lutkenhaus, 1994; Erickson et al, 1996). Nevertheless, our resolution was insufficient to rule out the possibility that they were actually doublets, such as the so-called ‘thick filaments' seen in 2-D crystals from Methanococcus jannaschii (Löwe and Amos, 1999). The filaments' short lengths suggested the same continuous fragmentating and reannealing behavior of FtsZ filaments observed by atomic force microscopy (Mingorance et al, 2005). Our results also agree with studies showing that only ∼30% of FtsZ molecules are assembled into filaments in Bacillus subtilis and Escherichia coli at any given moment, from which it was concluded that there are probably only 2–3 filaments in the Z-ring (Anderson et al, 2004). In our reconstructions of NA1000 cells, complete rings were not seen and the number of filaments on one side of the cell seemed unrelated to the number on the opposite side. Based on the Z-ring's 8–9 s turnover half-time in B. subtilis and E. coli, the longest single protofilament was estimated to contain ∼90 subunits and be ∼400 nm long (Anderson et al, 2004), which is less than even the diameter of predivisional cells and therefore much too short to form a complete ring. Even in cells overexpressing FtsZG109S, where the filaments were seen to be longer, the filaments did not form closed rings but rather interlaced helices. Unfortunately, the resolution of the reconstructions was insufficient to detect periodicity within a single filament, average individual subunits, or dock X-ray structures, so no information could be extracted about which side of FtsZ might face the membrane or mediate lateral contacts, or whether such characteristics are even consistent.
Existing models for the function of FtsZ
These structural results have important implications for models of the role of FtsZ in binary fission. The first model we will discuss arose through analogy to eukaryotic cytokinetic rings, where myosin molecules act on a band of antiparallel actin filaments to cause contraction. This ‘sliding' model predicts that some unidentified motor protein binds at least two different filaments of FtsZ and slides them past each other, gradually cinching the Z-ring. Our data fail to support this model since in wild-type cells filaments were short and rings were not seen, isolated filaments were common, and when multiple filaments were seen close to each other, there did not seem to be any regular spacing or bundling pattern that would suggest the presence of motor proteins connecting them. In addition, no myosin-like motor proteins that would act on cytoskeletal filaments have been identified in bacteria.
A different model, which we will call here the ‘scaffold' model, postulates that the function of FtsZ is simply to position peptidoglycan-synthetic machinery at the division plane. The construction of new, special, encroaching peptidoglycan would gradually displace the membrane inwards. In this model, the highly dynamic nature of FtsZ filaments seems only disadvantageous, except that it might facilitate remodeling in response to the shrinking aperture to maintain a consistent fit. While our structural data are compatible but not particularly supportive of this model, species of the Gram-negative genus Mycoplasma are problematic because they lack peptidoglycan entirely and have only a single cytoplasmic membrane (Wang and Lutkenhaus, 1996). Mycoplasma genomes generally lack most of the division proteins found in other prokaryotes, but often harbor a minimal set that includes FtsZ. Thus at least in the Mycoplasmas, FtsZ apparently plays a role other than positioning peptidoglycan synthetic machinery.
The last model we will discuss was based on the in vitro observation that FtsZ polymers adopt straight or bent conformations when bound to GTP or GDP, respectively (Lu et al, 2000). This ‘conformational change' model suggested that GTP hydrolysis by each FtsZ monomer could cause a transition from a straight to a curved conformation, which at the larger scale of a polymer would provide the force that constricts the membrane (Erickson, 1997). Although crystal structures of FtsZ failed to reveal notable differences between its GTP-bound form and GDP-bound form, it was suspected that the current GDP-bound FtsZ structure did not reflect the GDP state in the hydrolysis cycle (Oliva et al, 2004). The idea of tubulin homologs generating force is not without precedence either, since in the case of microtubules, the conformational change from straight to bent does appear to generate the forces that drive DamK rings to slide along the microtubule (Westermann et al, 2006). Our tomograms strongly support this ‘conformational change' model because both straight and bent conformations of FtsZ filaments were observed and because the model is compatible with the shortness, isolation, and erratic/asymmetric distribution of the filaments seen here.
An ‘iterative pinching' model for bacterial cell division
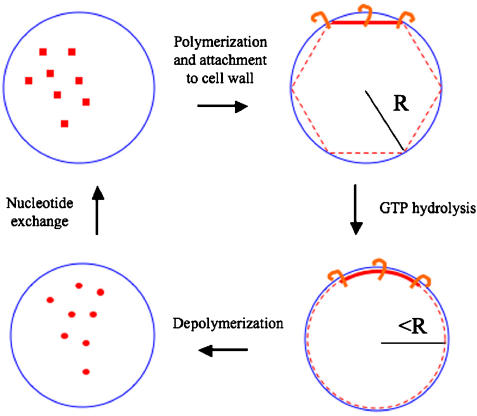
Elaborating on the ‘conformational change' model (Erickson, 1997, 2001), we propose an ‘iterative pinching' model to explain how FtsZ constricts the cell membrane. First, under a physiological (millimolar) concentration of GTP, FtsZ monomers polymerize into filaments in a ‘straight' conformation at or near the membrane at midcell. As or after FtsZ filaments form, other division proteins like FtsA bind the filament and anchor it to the membrane (Pichoff and Lutkenhaus, 2005). Then, when the nucleotide in each FtsZ molecule is hydrolyzed from GTP to GDP, the filament changes its conformation from straight to curved. This has the effect of drawing the ends closer together, which in the context of a membranous constriction pinches the aperture slightly. Another way to conceptualize this is in reference to Figure 11, where a straight filament is shown bound at its ends to the membrane. If a hypothetical closed polygon is circumscribed within a ‘reference' circle, and then maintains its total length but changes shape to form a new ‘constricted' circle, the diameter of the constricted circle is smaller than the reference circle (in the case of a hexagon, the diameter is reduced by 5%). After thus pinching the constriction, the FtsZ filament depolymerizes into FtsZ monomers that exchange their nucleotides in the cytoplasm for the next round of polymerization. The cycle repeats itself perhaps thousands of times in the vicinity of the division plane, so that binary fission proceeds through a large number of small pinching events. If FtsZ filaments fragment, loose subunits, and reanneal in vivo as they do in vitro (Mingorance et al, 2005), this could also potentially pinch the membrane. In bacteria like C. crescentus with peptidoglycan, FtsZ probably also serves as a scaffold to recruit synthetic enzymes, and the construction of new peptidoglycan behind constricting membrane preserves the progress made by each pinching event. In bacteria like Mycoplasmas without peptidoglycan, our model predicts that there must be some other mechanism to maintain progress between pinching events. One possibility would be that the FtsZ filaments are more abundant, so that by the time one filament releases from the membrane several others would have already anchored to stabilize and then advance that progress.
Figure 11.
An ‘iterative pinching' model for the role of FtsZ in cell division. Cytosolic GTP-bound FtsZ monomers (red squares, upper left) polymerize into straight filaments (red lines, upper right) and bind to the inner membrane (blue circle) through anchor proteins (orange hooks). GTP hydrolysis drives a conformational change to a curved filament (lower right), pinching the membrane inwards slightly in that region. GDP-bound FtsZ monomers depolymerize (red circles, lower left), and then exchange their nucleotide to complete the cycle. While FtsZ filaments are usually short and span only a chord within the circle (radius R) rather than a complete ring or inscribed polygon, the constricting effect of the conformational change can be appreciated by considering a hypothetical polygon and a corresponding circle of equal circumference (dashed purple lines with radius slightly less than R). The resulting small constrictions in each round could be stabilized before the filaments depolymerize by the construction of new peptidoglycan behind the advancing membrane.
The essential part of this model is that a large number of individual short FtsZ filaments generate small forces by themselves through GTP-hydrolysis-driven conformational changes. It offers explanations for (1) why FtsZ filaments are so dynamic; (2) why filament lengths can be so much smaller than the circumference of a cell (i.e., why complete rings are not necessary to induce invaginations; Addinall and Lutkenhaus, 1996); (3) how the small number of fairly isolated filaments seen at any given time in this study could be part of a process that is broad in both space and time; and (4) how a single mechanism could function throughout the entire range of constricting cell diameters required. The very last stages of constriction, when the cell diameter is only a few tens of nanometers, is particularly challenging to other models since it is difficult to imagine how an FtsZ filament in the ‘scaffold' or ‘slipping' models could remain stable after being bent so severely. Our model allows the possibility that straight FtsZ filaments even just a few monomers long could still anchor across and further pinch a constriction site through a conformational change, although it does not suggest anything about the molecular details of how the lipid bilayer would finally be severed. Our model also relaxes the precision to which the dynamic FtsZ filaments must be localized, since instead of being the conclusion of a single initiating ring-formation event, constriction is the sum of a large number of independent events that can be stochastically distributed over a region. This is particularly true in C. crescentus, where the constriction site is broad and gradual, as opposed to E. coli, where the division septum is much sharper. While unfortunately E. coli cells are too thick to obtain high-resolution ECT, our model would predict that to produce this sharp septum, the FtsZ filaments must be more consistently and tightly localized. It remains unclear, however, how the average 16 nm distance between the FtsZ filaments and the membrane is maintained, especially in curved segments which are presumably exerting tension on the membrane. It is possible that more numerous, thin connections are present which were not resolved here.
The information obtainable through fLM and ECT is complementary. While fLM allows particular proteins to be localized with fluorescent tags and followed through dynamic transitions in actively dividing cells, only ECT provides the resolution necessary to see individual filaments and their configurations. Our reconstructions of a few, short, isolated FtsZ filaments can be reconciled with the classic bright, continuous Z-ring seen by fLM by noting that labels attached to FtsZ monomers will fluoresce regardless of their polymerization state. The Z-rings seen by fLM prove that there is a detectably large concentration of FtsZ molecules near the membrane at midcell throughout cell division, but taken together all the available data suggest that this pool of FtsZ is rapidly polymerizing and depolymerizing and that only a few true filaments exist at any given time, as seen here by ECT. It is not clear, however, how to reconcile our findings with recent fLM images suggesting FtsZ filaments form continuous, dynamic helices that wrap around the cell multiple times with variable pitch, stretching almost to the cell poles and then collapsing on the midplane before cell division (Ben-Yehuda and Losick, 2002; Thanedar and Margolin, 2004; Peters et al, 2007).
Surprisingly, the FtsZ filaments were found 16 nm from the membrane. Our model depends on the existence of anchor proteins that link FtsZ to the membrane, and in species with peptidoglycan, other machinery that both stabilizes progress made and coordinately remodels the rest of the cell wall. In this way, our model echoes other recent findings that bacterial cell shape is governed by cytoskeletal filaments, but stabilized by construction of rigid peptidoglycan layers (Gitai et al, 2004; Cabeen and Jacobs-Wagner, 2005; Divakaruni et al, 2005; Dye et al, 2005). Densities connecting the filaments to the cell wall were seen in our reconstructions both at the ends of the filaments and in their middle, terminating sometimes in the membrane and sometimes in the peptidoglycan layer. As might have been expected, the ratio of these connections to FtsZ filaments was higher in the NA1000 cells than in the cells overexpressing FtsZ. Two division proteins, FtsA and ZipA, are known to bind directly to FtsZ in an early phase of Z-ring formation (Hale and de Boer, 1997; Din et al, 1998). While ZipA is not found in C. crescentus or its family, the α-proteobacteria, FtsA, binds both the C-terminal of FtsZ and the cytoplasmic membrane (Pichoff and Lutkenhaus, 2005). FtsA is therefore a likely constituent of the connections seen here. Later in division, the FtsQLB complex forms, and thus may also have been a part of some of the connections (Goehring and Beckwith, 2005), as could FtsI (PBP3), which is a membrane protein involved in peptidoglycan synthesis that attaches to FtsZ via FtsW (Wang et al, 1998; Mercer and Weiss, 2002). Effective labeling techniques for ECT are needed to locate all these components and dissect the rest of the division apparatus.
Materials and methods
Sample preparation
C. crescentus NA1000 cells were grown at 30°C in PYE until they reached log phase (OD600=0.2∼0.4). Swarmer cells were then isolated by the centrifugation method, resuspended in M2G minimal medium (Johnson and Ely, 1977), and allowed to progress through the cell cycle (Tsai and Alley, 2001). At different time points, 4 μl aliquots were removed, mixed with 10 nm colloidal gold and plunge-frozen onto glow-discharged Quantifoil R2/2, R2/1, or R1.2/1.3 grids (Quantifoil Micro Tools GmbH) using a Vitrobot (FEI) (Iancu et al, 2006). For the A22 experiments, A22 dissolved in methanol was added to log-phase cultures to a final concentration of 10 μg/ml (Gitai et al, 2005). In the case of NA1000 cells, the A22 was allowed to act for 10–20 min before samples were plunge-frozen as above. The FtsZG109S overexpression plasmid, pUJ142FtsZG109S (Wang et al, 2001), was conjugated from E. coli S17-1 into NA1000 to give YB1524. NA1000 containing the parent plasmid pUJ142 was examined as a control (Din et al, 1998). The C. crescentus strains overexpressing FtsZG109S or wild-type FtsZ were cultured as described previously (Quardokus et al, 2001; Wang et al, 2001) and induced with xylose for ∼3 h before plunge-freezing. When applicable, A22 was added to cells overexpressing FtsZG109S at the same time as the xylose. The FtsZ depletion strain (YB1585) was cultured as described previously (Wang et al, 2001) and deprived with xylose for ∼6 h before plunge-freezing.
ECT
Samples were stored in liquid nitrogen and maintained frozen throughout transfer into and imaging in an FEI G2 Polara transmission electron microscope operating at 300 keV. Energy-filtered ‘tilt-series' of images of individual cells were collected automatically from approximately −63° to +63° at 1° intervals using the UCSF predictive tomography software (Zheng et al, 2004) on either a 2k × 2k Gatan CCD camera or a 4k × 4k lens-coupled Gatan UltraCAM. The energy slit-width was 20 eV, the defocus was ∼12 μm (first contrast transfer function zero at ∼5 nm resolution), the total dose for each tilt-series was ∼150–180 e−/Å2, and the magnification was set such that each CCD pixel corresponded to 0.67 nm at the specimen level. The dose and defocus were chosen empirically to most clearly resolve the position and extent of the filaments, but under these experimental conditions individual monomers were not distinguishable.
Image processing
Images were binned two-fold before tilt-series were reconstructed using the IMOD package (Kremer et al, 1996). All the reconstructions were denoised using nonlinear anisotropic diffusion (Frangakis and Hegerl, 2001) in Bsoft (Heymann and Belnap, 2007). Segmentation and 3-D visualization were carried out manually using the IMOD or Amira (Mercury Computer Systems) packages. To calculate density profiles near the inner membrane (Figure 6), the cytoplasmic membrane was first segmented manually to generate a triangle-mesh surface. Density values were then sampled and averaged along normals to each triangle using Amira modules developed ‘in-house.'
Supplementary Material
Supplementary Information
Supplementary Movie
Acknowledgments
We thank H Jane Ding and David J Rosenman for computational help. This work was supported in part by NIH grant R01 AI067548 to GJJ, R01 GM51986 to YVB, DOE grant DE-FG02-04ER63785 to GJJ, a Searle Scholar Award to GJJ, the Beckman Institute at Caltech, and gifts to Caltech from the Gordon and Betty Moore Foundation and Agouron Institute.
References
- Addinall SG, Lutkenhaus J (1996) FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol 22: 231–237 [DOI] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP (2004) Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol 186: 5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Donachie WD (1985) Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol 163: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R (2002) Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109: 257–266 [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ (2006) Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol 62: 5–14 [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C (2005) Bacterial cell shape. Nat Rev Microbiol 3: 601–610 [DOI] [PubMed] [Google Scholar]
- Caplan MR, Erickson HP (2003) Apparent cooperative assembly of the bacterial cell division protein FtsZ demonstrated by isothermal titration calorimetry. J Biol Chem 278: 13784–13788 [DOI] [PubMed] [Google Scholar]
- Chen Y, Bjornson K, Redick SD, Erickson HP (2005) A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys J 88: 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N, Quardokus EM, Sackett MJ, Brun YV (1998) Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol 27: 1051–1063 [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW (2005) The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc Natl Acad Sci USA 102: 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J (2007) The physics of rapid cooling and its implications for cryoimmobilization of cells. Method Cell Biol 79: 7–21 [DOI] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z (2005) Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci USA 102: 18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP (1997) FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol 7: 362–367 [DOI] [PubMed] [Google Scholar]
- Erickson HP (2001) The FtsZ protofilament and attachment of ZipA—structural constraints on the FtsZ power stroke. Curr Opin Cell Biol 13: 55–60 [DOI] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D (1996) Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA 93: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW (2004) MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R (2001) Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135: 239–250 [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L (2004) An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA 101: 8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L (2005) MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120: 329–341 [DOI] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J (2005) Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol 15: R514–R526 [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Jimenez M, Velez M, Mingorance J, Andreu JM, Vicente M, Rivas G (2003) Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J Biol Chem 278: 37664–37671 [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Velez M, Jimenez M, Alfonso C, Schuck P, Mingorance J, Vicente M, Minton AP, Rivas G (2005) Cooperative behavior of Escherichia coli cell-division protein FtsZ assembly involves the preferential cyclization of long single-stranded fibrils. Proc Natl Acad Sci USA 102: 1895–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunenfelder B, Rummel G, Vohradsky J, Roder D, Langen H, Jenal U (2001) Proteomic analysis of the bacterial cell cycle. Proc Natl Acad Sci USA 98: 4681–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA (1997) Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88: 175–185 [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM (2007) Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol 157: 3–18 [DOI] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, Gan L, He Y, Jensen GJ (2006) Electron cryotomography sample preparation using the Vitrobot. Nat Protocols 1: 2813–2819 [DOI] [PubMed] [Google Scholar]
- Iancu CV, Wright ER, Benjamin J, Tivol WF, Dias DP, Murphy GE, Morrison RC, Heymann JB, Jensen GJ (2005) A ‘flip-flop' rotation stage for routine dual-axis electron cryotomography. J Struct Biol 151: 288–297 [DOI] [PubMed] [Google Scholar]
- Jensen GJ, Briegel A (2007) How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol 17: 260–267 [DOI] [PubMed] [Google Scholar]
- Johnson RC, Ely B (1977) Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH (2005) Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol 187: 6874–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV (1998) Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev 12: 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ (2006) Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311: 242–245 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Kurner J, Frangakis AS, Baumeister W (2005) Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307: 436–438 [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148 [DOI] [PubMed] [Google Scholar]
- Levin PA, Losick R (1996) Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev 10: 478–488 [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature 391: 203–206 [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA (1999) Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J 18: 2364–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Reedy M, Erickson HP (2000) Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol 182: 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W (2005) Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem 74: 833–865 [DOI] [PubMed] [Google Scholar]
- Margolin W (1998) A green light for the bacterial cytoskeleton. Trends Microbiol 6: 233–238 [DOI] [PubMed] [Google Scholar]
- Margolin W (2005) FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6: 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KL, Weiss DS (2002) The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J Bacteriol 184: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Löwe J (2006) Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem 75: 467–492 [DOI] [PubMed] [Google Scholar]
- Mingorance J, Tadros M, Vicente M, Gonzalez JM, Rivas G, Velez M (2005) Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J Biol Chem 280: 20909–20914 [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J (1994) Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol 176: 2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E (2001) Structural insight into microtubule function. Annu Rev Biophys Biomol Struct 30: 397–420 [DOI] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Löwe J (1998a) Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5: 451–458 [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH (1998b) Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391: 199–203 [DOI] [PubMed] [Google Scholar]
- Oliva MA, Cordell SC, Löwe J (2004) Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 11: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Peters PC, Migocki MD, Thoni C, Harry EJ (2007) A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol Microbiol 64: 487–499 [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J (2005) Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55: 1722–1734 [DOI] [PubMed] [Google Scholar]
- Quardokus E, Din N, Brun YV (1996) Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci USA 93: 6314–6319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus EM, Din N, Brun YV (2001) Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol Microbiol 39: 949–959 [DOI] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL (2005) Spatial control of bacterial division-site placement. Nat Rev Microbiol 3: 959–968 [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D (2006) An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440: 110–114 [DOI] [PubMed] [Google Scholar]
- Sun Q, Margolin W (1998) FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol 180: 2050–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner PE, Huls PG, Pas E, Woldringh CL (1988) Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol 170: 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L (2006) MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162 [DOI] [PubMed] [Google Scholar]
- Thanedar S, Margolin W (2004) FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr Biol 14: 1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Alley MR (2001) Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol 183: 5001–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Khattar MK, Donachie WD, Lutkenhaus J (1998) FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol 180: 2810–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J (1996) Characterization of the ftsZ gene from Mycoplasma pulmonis, an organism lacking a cell wall. J Bacteriol 178: 2314–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jones BD, Brun YV (2001) A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol 40: 347–360 [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G (2006) The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440: 565–569 [DOI] [PubMed] [Google Scholar]
- Zheng QS, Braunfeld MB, Sedat JW, Agard DA (2004) An improved strategy for automated electron microscopic tomography. J Struct Biol 147: 91–101 [DOI] [PubMed] [Google Scholar]
- Zuber B, Haenni M, Ribeiro T, Minnig K, Lopes F, Moreillon P, Dubochet J (2006) Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J Bacteriol 188: 6652–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Movie