The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor α (original) (raw)
Abstract
The Cbl protooncogene product has emerged as a negative regulator of receptor and nonreceptor tyrosine kinases. We recently demonstrated that oncogenic Cbl mutants upregulate the endogenous tyrosine kinase signaling machinery when expressed in the NIH 3T3 cells, and identified the platelet-derived growth factor receptor-α (PDGFRα) as one of the tyrosine kinases targeted by these oncogenes. These findings suggested a role for the normal Cbl protein in negative regulation of the PDGFRα. However, the mechanism of such negative regulation remained to be determined. Here we show that overexpression of the wild-type Cbl enhances the ligand-induced ubiquitination of the PDGFRα. Concomitantly, the PDGFRα in Cbl-overexpressing cells undergoes a faster ligand-induced degradation compared with that in the control cells. These results identify a role for Cbl in the regulation of ligand-induced ubiquitination and degradation of receptor tyrosine kinases and suggest one potential mechanism for evolutionarily conserved negative regulatory influence of Cbl on tyrosine kinases.
Tyrosine kinases provide a universal mode of signal transmission in response to extracellular cues that regulate cell proliferation and differentiation. Uncontrolled activation of tyrosine kinases is implicated in proliferation of cancerous cells, and their deficiencies result in pathological conditions such as developmental abnormalities and immunodeficiencies (1). Tight regulation of tyrosine kinase cascades is therefore critical to elicit an appropriate type and level of response to external stimuli.
Negative regulation of tyrosine kinase-mediated signaling is achieved through a number of distinct biochemical mechanisms. Cellular phosphotyrosine phosphatases provide one mechanism to reverse tyrosine kinase signals by removal of the phosphate moiety from phosphotyrosine (2). Another mechanism to regulate tyrosine kinase signaling is through the removal of receptors, such as epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor (PDGFR), from the cell surface via ligand-induced clathrin coat-mediated endocytosis (3, 4). A potent and evolutionarily conserved mechanism to regulate enzyme functions is through direct interactions with regulatory proteins. However, tyrosine kinase regulatory proteins have not been previously identified. Recent biochemical studies in mammalian cells (5–8) and independent genetic studies in Caenorhabditis elegans and Drosophila have identified the protooncogene product Cbl as a negative regulator of tyrosine kinases (9, 10).
Cbl was first identified as the cellular homologue of a viral oncogene v-cbl, which induces pre-B lymphomas and myeloid leukemias in mice (11). A large number of studies have now established that Cbl, a cytoplasmic 120 kDa polypeptide, is rapidly phosphorylated on tyrosine upon stimulation of cells through a number of receptor tyrosine kinases as well as receptors linked to nonreceptor tyrosine kinases (5–8). Notably, Cbl is either constitutively (e.g., Src-family kinases) or inducibly (e.g., EGFR, PDGFRα, or Syk/ZAP70 family) associated with tyrosine kinases themselves (5–8, 12–16). A critical determinant of the inducible association of Cbl with tyrosine kinases was identified recently. A phosphotyrosine-binding (PTB) domain within the N-terminal transforming region of Cbl, corresponding to sequences retained in the v-cbl oncogene, interacts directly with a specific phosphopeptide motif on tyrosine kinases to mediate a phosphorylation-dependent association between Cbl and protein tyrosine kinases (14, 15). In certain cases, such as the EGFR and PDGFRα, a second indirect mechanism of association is provided by the Grb2 adaptor protein, which binds to the proline-rich region of Cbl via its SH3 domains and to autophosphorylated receptors via its SH2 domain (10, 17, 18). Notably, the N-terminal PTB domain-containing region (Cbl-N) and an adjacent RING finger domain are the only two elements of Cbl highly conserved during evolution (5–8). Indeed, Drosophila Cbl is exclusively composed of these two regions, but nevertheless interacts with the EGFR (10).
The first evidence for the role of Cbl in negative regulation of tyrosine kinase signaling pathways was provided by studies of vulval development in C. elegans, a process that requires LET-23 receptor tyrosine kinase, a homologue of the mammalian EGFR (9). A gene identified as a negative regulator of signaling downstream of the LET-23 receptor, suppressor of lineage defect 1 (sli-1), encoded a Cbl homologue, SLI-1. Importantly, loss-of-function mutations identified in SLI-1 corresponded to either premature termination or a missense mutation in the conserved Cbl-N homology region. In the Drosophila eye, a functional EGFR homologue is critical for the development of the R7 photoreceptor cell and functions through a Ras/mitogen-activated protein kinase pathway analogous to the mammalian EGFR. The recently cloned Drosophila Cbl homologue (D-Cbl) was shown to bind to Drosophila and mammalian EGFRs and, when expressed as a transgene under control of a sevenless enhancer, induced the severe reduction in the development of the R7 photoreceptor, providing further evidence for a negative regulatory role of Cbl in receptor tyrosine kinase signaling (10).
In mammalian systems, Cbl overexpression in NIH 3T3 cells correlated with decreased autophosphorylation of the EGFR and lower JAK-STAT phosphorylation; conversely, higher EGFR autophosphorylation was observed in cells expressing lower levels of Cbl as a consequence of antisense Cbl transfection (19). Cbl overexpression in the rat basophilic leukemia cell line RBL-2H3 was shown to decrease the autophosphorylation and kinase activity of Syk that is induced on FcɛR1 stimulation, and mast cell degranulation was severely impaired (20). The in vivo negative regulatory phosphorylation site pY292 was identified as the Cbl PTB domain-binding site on ZAP-70 (16). NIH 3T3 cells expressing oncogenic forms of Cbl revealed a hyperphosphorylation of the PDGF receptor α (PDGFRα) and an up-regulation of signaling downstream of this receptor (14). Similarly, NIH 3T3 cells expressing oncogenic forms of Cbl showed increased autophosphorylation and kinase activity of a transfected human EGFR, both under serum-starved and EGF-stimulated conditions (21). Apparently, the effect of oncogenic mutants of Cbl on PDGFRα and EGFR reflects a reversal of the negative regulatory role of wild-type Cbl.
Together, these studies strongly support a role for Cbl as a negative regulator of diverse tyrosine kinases. However, the mechanism whereby Cbl negatively regulates tyrosine kinases has not been elucidated. Here we demonstrate that Cbl promotes ubiquitination and ligand-induced degradation of the receptor tyrosine kinase PDGFRα, suggesting one mechanism for the negative regulatory action of Cbl on tyrosine kinases.
MATERIALS AND METHODS
Cell Lines and Culture.
NIH 3T3 cells overexpressing hemagglutinin (HA) epitope-tagged human Cbl (3T3-HA-Cbl.8, here referred to as 3T3-Cbl.8) have been described (14). Additional clones (3T3-HA-Cbl.9 and 3T3-HA-Cbl.11) were derived by retroviral transfection as described (14). Cells were cultured in α-minimal essential medium (α-MEM) containing 10% fetal calf serum (FCS; HyClone) and 500 μg/ml of G418 (GIBCO/BRL).
Antibodies.
Murine monoclonal antibodies used were: 4G10 (antiphosphotyrosine, anti-pY) (22); 12CA5 (anti-HA epitope tag) (23); and OKT8 (anti-CD8, used as a nonbinding negative control; American Type Culture Collection). Polyclonal rabbit antibodies used were: anti-PDGFRα (sc-431, Santa Cruz Biotechnology); anti-ubiquitin (NCL-UBIQ, NovoCastra, Newcastle, U.K.), and normal rabbit serum (negative control) obtained from unimmunized rabbits.
PDGF Stimulation.
For stimulation experiments, cells were cultured for 24 hr in α-MEM medium containing 0.5% FCS (prestarvation), and then stimulated in α-MEM containing 0.5% FCS supplemented with 20 ng/ml recombinant human PDGF-AA or PDGF-BB (Upstate Biotechnology, Lake Placid, NY). At the indicated times after adding PDGF, the medium was aspirated and cells were lysed in cold lysis buffer [0.5% Triton X-100 (Fluka)/50 mM Tris, pH 7.5/150 mM sodium chloride/1 mM phenylmethylsulfonyl fluoride/1 mM sodium orthovanadate/10 mM sodium fluoride/1 μg/ml each of leupeptin, pepstatin, chymostatin, antipain, and aprotinin (Sigma)].
Immunoprecipitation and Immunoblotting.
Antibodies were added to aliquots of lysates equalized for protein content by the Bradford assay (Bio-Rad; using the BSA standard). After 1–2 hr of rocking at 4°C, 20 μl of protein A-Sepharose 4B beads (Pharmacia) were added and incubation was continued for 45 min. Beads were washed six times in lysis buffer, and bound proteins were solubilized in Laemmli sample buffer with 2-mercaptoethanol (2-ME) and resolved by SDS/PAGE. Polypeptides were transfered to polyvinylidene difluoride membranes (Immobilon-P, Millipore) and immunoblotted by using horseradish peroxidase (HRP)-conjugated protein A (Cappel-Organon Technika, Durham, NC), followed by enhanced chemiluminescence (DuPont/NEN). Membranes were stripped and reprobed as described (14).
Biotinylation of Cell Surface PDGFRα and Time Course of Degradation.
Cell monolayers were washed with ice-cold PBS containing 20 mM Hepes buffer solution (pH 7.35) and then incubated in the same buffer with sulfo-n-hydroxysulfosuccinimide (NHS)-biotin (Pierce) for 15 min at 4°C. After washing, the cells were incubated in α-MEM containing 1 mg/ml BSA (tissue culture grade; Sigma) with or without PDGF for 15 min at 4°C. The cells were washed, incubated at 37°C in α-MEM for the indicated times, and lysates were prepared as described above. Anti-PDGFRα immunoprecipitates of these lysates were blotted with avidin-HRP conjugate (Vector Laboratories) followed by enhanced chemiluminescence, as described above. Densitometry was carried out by using the Hewlett–Packard ScanJet 4C and Scion Images for Windows software. Densitometric data were expressed as arbitrary units.
RESULTS AND DISCUSSION
Cbl Enhances the Ligand-Induced Ubiquitination of PDGFRα.
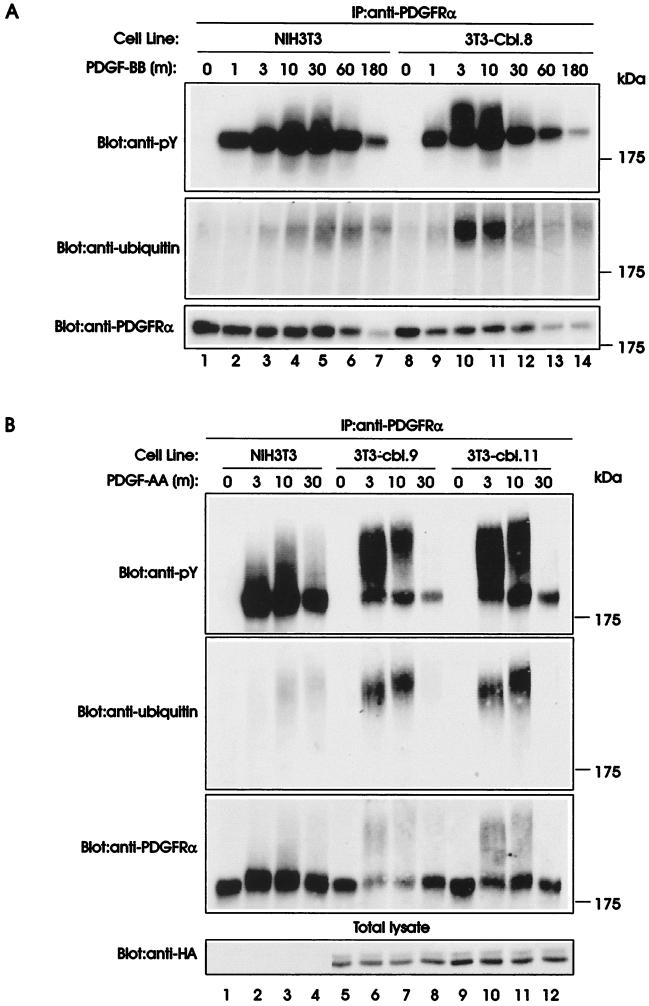
Our earlier studies demonstrated that the PDGFRα was hyperactive in NIH 3T3 cells expressing various oncogenic mutants of Cbl when compared with cells expressing wild-type Cbl, and led us to suggest that normal Cbl functions as a negative regulator of PDGFRα signaling (14). In these earlier studies, we noted that the kinetics of the PDGFRα autophosphorylation in response to PDGF-AA was slower in mutant Cbl-transfected cells compared with wild-type Cbl-transfected cells (14). In addition, a more prominent upward mobility shift (seen as smearing) of the PDGFRα was observed in anti-pY immunoblots of the wild-type Cbl transfectants. Because the PDGFRα and the closely related PDGFRβ are known to undergo ubiquitination upon ligand stimulation (24, 25), we considered the possibility that the observed mobility shift in PDGFRα may be because of ubiquitination, and that this process may be altered by Cbl. To test this possibility, serum-starved parental NIH 3T3 cells and Cbl-transfected NIH 3T3 cells (3T3-Cbl.8) were stimulated with PDGF-BB at 37°C for various times. PDGFRα was immunoprecipitated from lysates of unstimulated and PDGF-stimulated cells, resolved by SDS/PAGE, and visualized by anti-pY immunoblotting (Fig. 1A Top).
Figure 1.
Ubiquitination of the PDGFRα upon PDGF stimulation of parental and Cbl-transfected NIH 3T3 cells. (A) Serum-starved parental (NIH 3T3) and Cbl-overexpressing NIH 3T3 cells (3T3-Cbl.8) were stimulated with 20 ng/ml human recombinant PDGF-BB and lysed at the indicated times (m, minutes). Immunoprecipitations (IP) were performed with an anti-PDGFRα antibody and 1 mg aliquots of each lysate, and were analyzed by anti-pY immunoblotting (Top). The membrane was stripped and serially reprobed with anti-ubiquitin (Middle) and anti-PDGFRα antibodies (Bottom). (B) Serum-starved parental (NIH 3T3) and Cbl-overexpressing NIH 3T3 cells (3T3-Cbl.9 and 3T3-Cbl.11) were stimulated with 20 ng/ml human recombinant PDGF-AA and lysed at the indicated times (m, minutes). Immunoprecipitation and immunoblotting with anti-pY (Top), anti-ubiquitin (Upper Middle) and anti-PDGFRα antibodies (Lower Middle) were performed as in A. Total lysates collected at each time point were immunoblotted with an anti-HA antibody (Bottom).
An expected time-dependent PDGF-induced tyrosine phosphorylation of the PDGFRα was observed in parental NIH 3T3 cells and this phosphorylation was maximal by 10 min. Additionally, a proportion of the phosphorylated PDGFRα immunoprecipitated from lysates of parental NIH 3T3 cells migrated with a slower mobility and was apparent as a smear above the main PDGFRα species. The mobility shift was observed by 3 min of PDGF treatment (Fig. 1A, lane 3), peaked by 10–30 min (Fig. 1A, lanes 4 and 5), and progressively decreased thereafter (Fig. 1A, lanes 6 and 7). In contrast to parental NIH 3T3 cells, Cbl-overexpressing 3T3-Cbl.8 cells yielded a remarkably different pattern of PDGF-induced smearing of the tyrosine phosphorylated receptor. First, the smearing was already visible after 1 min of PDGF stimulation (Fig. 1A, lane 9), and reached a maximal intensity after 3 min (Fig. 1A, lane 10) with a substantial decline by 30 min (Fig. 1A, lane 12). Second, the proportion of the receptor undergoing an upward mobility shift was markedly greater than that seen in the parental cells (Fig. 1A, compare lanes 10 and 11 with lanes 4 and 5). This altered pattern of ligand-induced PDGFRα phosphorylation was confirmed in two additional Cbl-transfected NIH 3T3 clones stimulated with PDGFRα-selective ligand PDGF-AA (26) (Fig. 1B, Top).
To directly determine whether the ligand-induced smearing of the PDGFRα reflected its ubiquitination, we reprobed the membrane with an anti-ubiquitin antibody. The slower-migrating PDGFRα species indeed selectively reacted with anti-ubiquitin antibody (Fig. 1 A and B, Upper Middle, seen as a smear), whereas the major faster-migrating PDGFRα species was nonreactive. Importantly, the anti-ubiquitin antibody blot directly confirmed the enhanced and faster ubiquitination of the PDGFRα upon PDGF stimulation of the Cbl-transfected cells. Anti-PDGFRα immunoblotting showed comparable levels of receptor expression on both the parental and Cbl-expressing NIH 3T3 cells (Fig. 1A, Bottom). The antibody, however, reacts poorly with the slower-migrating ubiquitinated species of the PDGFRα, accounting for the lack of this species in anti-PDGFRα blot in the exposure shown. Prolonged exposures confirmed the presence of smeared PDGFRα species in appropriate lanes (Fig. 1B, Lower Middle and data not shown).
To quantify the relative levels of PDGFRα ubiquitination in parental and Cbl-overexpressing NIH 3T3 cells, densitometric quantification of the upward-shifted and unshifted PDGFRα signals shown in anti-pY blot in Fig. 1B was performed. The ratio of unshifted to upward-shifted PDGFR signals in lane 3 (NIH 3T3), which showed the maximum smearing for this cell line, was 1:0.87. In contrast, the ratios for 3T3-Cbl.9 (lane 6) and 3T3-Cbl.11 (lane 10) cell lines, at 3-min time points after PDGF addition were 1:2.9 and 1:2.8, respectively. Thus, an approximately 3-fold higher level of ubiquitination of the PDGFRα was seen in Cbl-overexpressing compared with parental NIH 3T3 cells. Altogether, these findings demonstrate that the overexpression of Cbl protein leads to enhanced and more rapid ligand-induced ubiquitination of the PDGFRα.
Cbl Associates with Ubiquitinated PDGFRα.
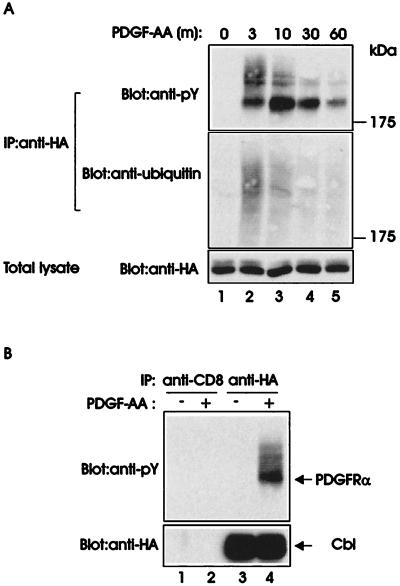
In previous work, we demonstrated that Cbl associates with PDGFRα upon PDGF-AA stimulation (14). To assess whether Cbl associates with the ubiquitinated PDGFRα, we carried out anti-HA immunoprecipitations from lysates of 3T3-Cbl.8 cells that were either left unstimulated or were stimulated for various times with PDGF-AA. Cbl-associated PDGFRα was visualized by immunoblotting with anti-pY antibody (Fig. 2A, Top). As seen in Fig. 2A, a substantial level of slower-migrating smear, corresponding to ubiquitinated PDGFRα, was coimmunoprecipitated with Cbl in addition to the expected 185 kDa tyrosine phosphorylated PDGFRα. The association of Cbl with the slower-migrating PDGFRα species was maximal at 3 min of PDGF stimulation and gradually decreased thereafter, correlating with the time kinetics of ligand-induced ubiquitination of the PDGFRα (Fig. 1). Reprobing of the same membrane with an anti-ubiquitin antibody confirmed that the slower-migrating Cbl-associated PDGFRα species were ubiquitinated (Fig. 2A, Middle). Anti-HA immunoblotting demonstrated comparable levels of HA-tagged Cbl in all lysates (Fig. 2A, Bottom). Control immunoprecipitations with an anti-CD8 antibody, using the lysates of unstimulated 3T3-Cbl.8 cells or cells stimulated with PDGF-AA for 3 min, revealed that coimmunoprecipitation of the ubiquitinated PDGFRα with anti-HA antibody was specific (Fig. 2B, compare lane 4 with lane 2). These results demonstrate that Cbl forms a complex with the ubiquitinated PDGFRα upon PDGF stimulation.
Figure 2.
Association of Cbl with the ubiquitinated PDGFRα. (A) Serum-starved Cbl-expressing cells were stimulated with 20 ng/ml human recombinant PDGF-AA and lysed at the indicated times (m, minutes). Immunoprecipitations (IP) were carried out by using an anti-HA antibody (12CA5) and 1 mg of each lysate, followed by immunoblotting with an anti-pY antibody (Top). The membrane was stripped and reprobed with an anti-ubiquitin antibody (Middle). Total lysates collected at each time point were immunoblotted with anti-HA antibody (Bottom). (B) Lysates of unstimulated 3T3-Cbl.8 cells (−) and stimulated with 20 ng/ml PDGF-AA for 3 min (+) were immunoprecipitated with an anti-CD8 control antibody or anti-HA antibody followed by anti-pY immunoblotting (Upper). The membrane was stripped and reprobed with an anti-HA antibody (Lower).
Cbl Facilitates Ligand-Induced Degradation of the PDGFRα.
Given the well-documented role of ubiquitination in targeting cellular proteins for degradation (27, 28), we considered the possibility that enhanced ubiquitination of the PDGFRα in Cbl-overexpressing cells may result in increased PDGFRα degradation. Consistent with this possibility, previous work has demonstrated that a ubiquitination-defective mutant of the PDGFRβ undergoes reduced ligand-dependent turnover (25). To test the effect of Cbl overexpression on the PDGFRα turnover, we quantified the PDGFRα levels in parental NIH 3T3 cells and Cbl-transfected NIH 3T3 cells at various times after stimulation with PDGF-AA. To selectively analyze the ligand-responsive cell surface-expressed PDGFRα, serum-starved cells were surface labeled with biotin for 15 min at 4°C, followed by 15 min incubation with 20 ng/ml PDGF-AA at 4°C. After washing, the cells were shifted to 37°C to initiate PDGF-AA-induced turnover and incubation was continued at 37°C for the indicated times. Lysates were prepared at each time point and immunoprecipitated with an anti-PDGFRα antibody, and blotted with avidin-HRP to quantify the level of surface-labeled PDGFRα.
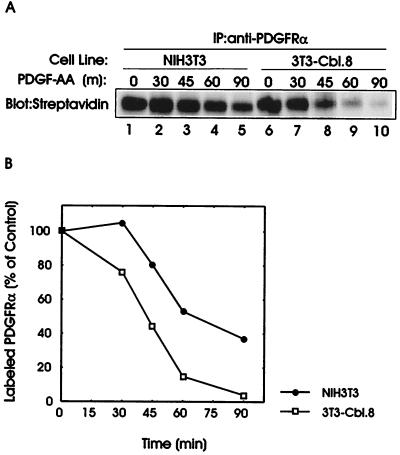
As shown in Fig. 3A, a time-dependent decrease in the intensity of the receptor band was observed following ligand stimulation of both the parental NIH 3T3 cells and Cbl-transfected 3T3-Cbl.8 cells. However, although significant biotinylated PDGFRα was still detected in parental cells at 90 min after PDGF stimulation (Fig. 3A, lane 5), relatively little PDGFRα was detected in Cbl-transfected cells at 60–90 min after PDGF stimulation (Fig. 3A, lanes 9 and 10).
Figure 3.
Time course of the ligand-induced degradation of the surface-labeled PDGFRα in parental and Cbl-transfected NIH 3T3 cells. Serum-starved parental NIH 3T3 and Cbl-expressing cells were surface labeled with biotin. Labeled cells were incubated either without (0) or with PDGF-AA for 15 min at 4°C. Cells were washed, incubated at 37°C for the indicated times, and lysed. Anti-PDGFRα immunoprecipitates of the lysates (1 mg) were subjected to blotting with avidin-HRP followed by enhanced chemiluminescence detection. (A) A representative blot. (B) Densitometry was performed and the intensity of PDGFRα signal at various time points was expressed as a percentage of that in unstimulated cells at time zero. •, PDGF-AA-stimulated NIH 3T3 cells; □, PDGF-AA-stimulated 3T3-Cbl.8 cells.
The loss of cell surface PDGFRα in the parental and Cbl-transfected NIH 3T3 cells was quantified by densitometry of the avidin blot (Fig. 3B). The ligand-independent loss of PDGFRα measured at 45 and 90 min, was comparable in parental and Cbl-transfected cells (data not shown). In contrast, a substantially earlier and more pronounced loss of the surface-labeled PDGFRα was observed upon PDGF stimulation of the Cbl-overexpressing NIH 3T3 cells as compared with that in the parental cells. The half-life of the surface-labeled PDGFRα following PDGF-AA stimulation was 36% shorter in Cbl-transfected NIH 3T3 cells compared with that in parental cells (42 min vs. 66 min, respectively). Similar results were observed in three independent experiments (data not shown). Together these results show that enhancement of the PDGFRα ubiquitination as a result of Cbl overexpression is associated with a faster ligand-induced loss of the surface PDGFRα.
Genetic studies in the yeast Saccharomyces cerevisiae have revealed that ubiquitination of the Ste2p, an integral membrane protein and receptor for the α-factor pheromone, is required for its endocytosis (29, 30). Similar roles for ubiquitination have been described for Ste3p (31), Ste6p (32), and uracil permease in the yeast system (33), and for the growth hormone receptor in mammalian cells (34). In these systems, ubiquitination is thought to provide a signal for endocytosis and target these proteins for degradation in the vacuoles/lysosomes (30), although a recent study found that Ste6p degradation was impaired in mutants defective for proteasome degradation (35). Independently, ubiquitination has been implicated in the degradation of the PDGFRβ (36) and Met receptor tyrosine kinases (37). In these latter cases, it has been concluded that ubiquitination targets these receptors for proteasome-mediated degradation, although the effects of two potent proteasome inhibitors were modest and not substantially different from the effects of a lysosomotropic inhibitor (36). Thus, although it is clear that ubiquitination targets receptors for ligand-induced degradation, the cellular machinery responsible for degradation remains controversial. It will be of interest to determine whether Cbl-dependent regulation involves targeting of ubiquitinated receptors to lysosomal degradation, proteasome-mediated degradation, or both.
How Cbl may regulate the ubiquitination of tyrosine kinases is unclear. Given that Cbl overexpression led to a marked enhancement of the PDGFRα ubiquitination, it is possible that Cbl influences this proximal event. As showed earlier, an intact Cbl PTB domain is critical for the tyrosine kinase regulatory functions of Cbl. Notably, one of the two autophosphorylation sites in the PDGFRβ, which are critical for its ubiquitination, conforms to a consensus Cbl PTB domain-binding site (DNDY1021) (16). Cbl binding to an autophosphorylation site on the PDGFR may stabilize a conformation that favors ubiquitination. Alternatively, Cbl may recruit other proteins, such as components of the ubiquitination machinery, in a manner similar to the role of the F-box protein-mediated recruitment of ubiquitin-conjugating enzymes to phosphorylated substrates in the yeast system (38). Interestingly, Cbl has also emerged as a critical negative regulator of Syk/ZAP70 cytoplasmic tyrosine kinases associated with lymphocyte antigen receptors; the antigen receptors are also known to be ubiquitinated in a ligand-dependent and kinase-dependent manner (39, 40).
Based on results presented here, we propose that Cbl is a regulator of the ligand-induced ubiquitination and degradation of receptor kinases. The precise step at which Cbl participates to enhance receptor ubiquitination and/or degradation remains conjectural at present. It is notable that whereas null mutations of either the C. elegans Cbl homologue, SLI-1, or another negative regulator of LET-23, UNC-101, have no overt phenotypes, combined mutation of these genes leads to a multivulva phenotype indicating a potential functional interaction between SLI-1 and UNC-101. UNC-101 has been identified as a homologue of the 47 kDa component of the clathrin coat-associated adaptin complex (41). Thus, Cbl may regulate a step in receptor endocytosis. Studies of the PDGFRβ show a requirement for binding of the PI3-kinase to effect a postendocytic step in ligand-induced internalization (42, 43). Given the prominent activation-induced association between phosphorylated Cbl and PI3 kinase, it is possible that Cbl-PI3 kinase complexes are involved in the Cbl-mediated effects observed here, although additional mechanisms cannot be ruled out at present.
A mutant PDGFRβ defective in ubiquitination was previously reported to have mediated an enhanced mitogenic signal (44). However, in a previous study (14), we were unable to detect a significant difference in the growth rates of 3T3-Cbl.8 and parental NIH 3T3 cells. A similar lack of significant effect on growth rate of Cbl-transfected NIH 3T3 cells was reported by another group (19). In recent experiments, we confirmed the lack of difference between parental NIH 3T3 cells and the late-passage 3T3-Cbl.8 cell clone comparable to those used in earlier studies. However, preliminary experiments reveal a substantial reduction in the PDGF-AA-induced proliferation of all three Cbl-overexpressing clones (3T3-Cbl.8, 3T3-Cbl.9, and 3T3-Cbl.11) when examined at relatively early passages compared with parental NIH 3T3 (data not shown). Further investigations are clearly warranted to clarify whether Cbl-mediated changes in PDGFR ubiquitination and turnover result in a change in PDGFRα-mediated mitogenesis. It will be equally important to determine the effects of Cbl overexpression on other biologic responses mediated by the PDGFRα, such as chemotaxis or specific gene induction (17).
In conclusion, our findings that overexpression of Cbl enhances the ligand-induced ubiquitination and degradation of a receptor tyrosine kinase, PDGFRα, provide a mechanism of the function of Cbl as a negative regulator of tyrosine kinases. Although we have only examined the effect of Cbl in the context of the PDGFRα, given the ability of Cbl to associate with and negatively regulate a number of other tyrosine kinases, and the conservation of ligand-induced loss of receptors as a mechanism to down-regulate signaling, it is likely that regulation of ubiquitination and protein degradation may represent a general mechanism of the function of Cbl.
Acknowledgments
We thank Dr. Nancy L. Lill for a critical review of the manuscript. This work was supported by Grant CA76118 from the National Institutes of Health and Grant 97–139-MGO from the American Cancer Society to H.B. S.M. was a recipient of a postdoctoral fellowship from the Uehara Memorial Foundation (Tokyo) and M.L.L. is a Breast Cancer Research Fellow of the Massachusetts Department of Public Health.
ABBREVIATIONS
PDGF
platelet-derived growth factor
PDGFR
platelet-derived growth factor receptor
EGFR
epidermal growth factor receptor
PTB
phosphotyrosine binding
HRP
horseradish peroxidase
HA
hemagglutinin
References
- 1.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 2.Tonks N K, Neel B G. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 3.van der Geer P, Hunter T, Lindberg R A. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 4.Seaman M N, Burd C G, Emr S D. Curr Opin Cell Biol. 1996;8:549–556. doi: 10.1016/s0955-0674(96)80034-2. [DOI] [PubMed] [Google Scholar]
- 5.Langdon W Y. Aust NZ J Med. 1995;25:859–864. doi: 10.1111/j.1445-5994.1995.tb02892.x. [DOI] [PubMed] [Google Scholar]
- 6.Miyake S, Lupher M L, Jr, Andoniou C E, Lill N L, Ota S, Doillard P, Rao N, Band H. Crit Rev Oncogen. 1998;8:189–218. doi: 10.1615/critrevoncog.v8.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 7.Lupher, M. L., Jr., Andoniou, C. E., Bonita, D., Miyake, S. & Band, H. (1998) Int. J. Biochem. Cell Biol., in press. [DOI] [PubMed]
- 8.Lupher, M. L., Jr., & Band, H. (1998) Immunol. Today, in press. [DOI] [PubMed]
- 9.Yoon C H, Lee J H, Jongeward G D, Sternberg P W. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 10.Meisner H, Daga A, Buxton J, Fernandez B, Chawla A, Banerjee U, Czeck M P. Mol Cell Biol. 1997;17:2217–2225. doi: 10.1128/mcb.17.4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langdon W Y, Hartley J W, Klinken S P, Ruscetti S K, Morse H C. Proc Natl Acad Sci USA. 1989;86:1168–1172. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukazawa T, Reedquist K A, Trub T, Soltoff S, Panchamoorthy G, Druker B, Cantley L, Shoelson S E, Band H. J Biol Chem. 1995;270:19141–19150. doi: 10.1074/jbc.270.32.19141. [DOI] [PubMed] [Google Scholar]
- 13.Fukazawa T, Miyake S, Band V, Band H. J Biol Chem. 1996;271:14554–14559. doi: 10.1074/jbc.271.24.14554. [DOI] [PubMed] [Google Scholar]
- 14.Bonita D, Miyake S, Lupher M L, Jr, Langdon W Y, Band H. Mol Cell Biol. 1997;17:4597–4610. doi: 10.1128/mcb.17.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupher M L, Jr, Reedquist K A, Miyake S, Langdon W L, Band H. J Biol Chem. 1996;271:24063–24068. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- 16.Lupher M L, Jr, Zhou S Y, Shoelson S E, Cantley L C, Band H. J Biol Chem. 1997;272:33140–33144. doi: 10.1074/jbc.272.52.33140. [DOI] [PubMed] [Google Scholar]
- 17.Kazlauskas A. Curr Opin Genet Dev. 1994;4:5–14. doi: 10.1016/0959-437x(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 18.Bazenet C E, Gelderloos J A, Kazlauskas A. Mol Cell Biol. 1996;16:6926–6936. doi: 10.1128/mcb.16.12.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno H, Sasaki K, Miyagawa K, Honda H, Mitani K, Yazaki Y, Hirai H. J Biol Chem. 1997;272:8739–8743. doi: 10.1074/jbc.272.13.8739. [DOI] [PubMed] [Google Scholar]
- 20.Ota Y, Samelson L E. Science. 1997;276:418–420. doi: 10.1126/science.276.5311.418. [DOI] [PubMed] [Google Scholar]
- 21.Thien C B F, Langdon W Y. Oncogene. 1997;15:2909–2919. doi: 10.1038/sj.onc.1201468. [DOI] [PubMed] [Google Scholar]
- 22.Druker B J, Mamon H J, Roberts T M. N Engl J Med. 1989;321:1383–1391. doi: 10.1056/NEJM198911163212007. [DOI] [PubMed] [Google Scholar]
- 23.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 24.Mori S, Claesson-Welsh L, Okuyama Y, Saito Y. Biochem Biophys Res Commun. 1995;213:21–29. doi: 10.1006/bbrc.1995.2094. [DOI] [PubMed] [Google Scholar]
- 25.Mori S, Heldin C-H, Claesson-Welsh L. J Biol Chem. 1992;267:6429–6434. [PubMed] [Google Scholar]
- 26.Claesson-Welsh L. J Biol Chem. 1994;269:32023–32026. [PubMed] [Google Scholar]
- 27.Weissman A M. Immnunol Today. 1997;18:189–198. doi: 10.1016/s0167-5699(97)84666-x. [DOI] [PubMed] [Google Scholar]
- 28.Hochstrasser M. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 29.Hicke L, Riezman H. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 30.Hicke L. FASEB J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 31.Roth A F, Davis N G. J Chem Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolling R, Hollenberg C P. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galan J M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 34.Strous G J, Kerkhof P, Govers R, Ciechanover A, Schwartz A L. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- 35.Loayza D, Michaelis S. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori S, Tanaka K, Omura S, Saito Y. J Biol Chem. 1995;271:29447–29452. doi: 10.1074/jbc.270.49.29447. [DOI] [PubMed] [Google Scholar]
- 37.Jeffers M, Taylor G A, Weidner M, Omura S, Vande Woude G F. Mol Cell Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowyra D, Craig K L, Tyers M, Elledge S J, Herper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 39.Cenciarelli C, Hou D, Hsu K-C, Rellahan B L, Wiest D L, Smith H T, Fried V A, Weissman A M. Science. 1992;257:795–797. doi: 10.1126/science.1323144. [DOI] [PubMed] [Google Scholar]
- 40.Cenciarelli C, Wilhelm K G, Guo A, Weissman A M. J Biol Chem. 1996;271:8709–8713. doi: 10.1074/jbc.271.15.8709. [DOI] [PubMed] [Google Scholar]
- 41.Lee J, Jongeward G D, Sternberg P W. Genes Dev. 1994;8:60–73. doi: 10.1101/gad.8.1.60. [DOI] [PubMed] [Google Scholar]
- 42.Joly M, Kazlauskas A, Fay F S, Corvera S. Science. 1994;263:684–687. doi: 10.1126/science.8303278. [DOI] [PubMed] [Google Scholar]
- 43.Joly M, Kazlauskas A, Fay F S, Corvera S. J Biol Chem. 1995;270:13225–13230. doi: 10.1074/jbc.270.22.13225. [DOI] [PubMed] [Google Scholar]
- 44.Mori S, Heldin C-H, Claesson-Welsh L. J Biol Chem. 1993;268:577–583. [PubMed] [Google Scholar]