The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus (original) (raw)
Abstract
Tightly balanced antagonism between the Polycomb group (PcG) and the Trithorax group (TrxG) complexes maintain Hox expression patterns in Drosophila and murine model systems. Factors belonging to the PcG/TrxG complexes control various processes in plants as well but whether they participate in mechanisms that antagonize, balance or maintain each other's effects at a particular gene locus is unknown. CURLY LEAF (CLF), an Arabidopsis homolog of enhancer of zeste (EZ) and the ARABIDOPSIS HOMOLOG OF TRITHORAX (ATX1) control the expression of the flower homeotic gene AGAMOUS (AG). Disrupted ATX1 or CLF function results in misexpression of AG, recognizable phenotypes and loss of H3K4me3 or H3K27me3 histone H3-tail marks, respectively. A novel idea suggested by our results here, is that PcG and TrxG complexes function as a specific pair generating bivalent chromatin marks at the silent AG locus. Simultaneous loss of ATX1 and CLF restored AG repression and normalized leaf phenotypes. At the molecular level, disrupted ATX1 and CLF functions did not lead to erasure of the CLF- and ATX1-generated epigenetic marks, as expected: instead, in the double mutants, H3K27me3 and H3K4me3 tags were partially restored. We demonstrate that ATX1 and CLF physically interact linking mechanistically the observed effects.
INTRODUCTION
Polycomb group (PcG) complexes maintain the silencing of Hox genes, whereas TrxG maintain their expression (1–3). Two major classes of PcG repressor complexes (PRCs) have been recognized in animal systems: PRC2, containing EZ responsible for the histone H3 lysine 27 methylation, and PRC1, mediating the formation of transcription-resistant chromatin structure at target genes (4,5). Complexes containing the Trithorax histone H3 lysine 4-methyl transferase activity are COMPASS, in yeast, and TAC1, in Drosophila (6,7). PcG and TrxG factors regulate plant genes as well but plants have developed epigenetic mechanisms that are related, although not identical, with those of animals or yeast (8–11). For example, the PRC2 complex is conserved both structurally and functionally between animals and plants (9,12) but PRC1 homologs have not been identified. Arabidopsis components of PcG/TrxG complexes play repressing and activating roles for a number of plant genes (13–21) but whether PcG and TrxG act as antagonists at the same gene locus in plants is unknown. The Arabidopsis CLF gene encodes an EZ homolog, while the ATX1 gene encodes a Trx-homolog, acting as a repressor and activator of the homeotic AGAMOUS, AG, gene, respectively. Disruption of either function causes recognizable phenotypes (13,14). Here, we report that nucleosomes at silent AG loci carry both the activating, H3K4me3, and the repressing, H3K27me3, marks. Recent findings have suggested that simultaneously present H3K4me3 and H3K27me3 marks establish bivalent chromatin states of silent genes poised for transcription later in life (22). Furthermore, simultaneous loss of both ATX1 and CLF functions caused a remarkable shift towards the wild type. Analyzing the molecular mechanism behind this event we found that, contrary to expectations, loss of both functions resulted in partial restorations of the K4 and K27 marks on the AG nucleosomes in the double mutants. The results suggest that the antagonistic PcG- and TrxG-complexes form specific pairs to generate bivalent chromatin marks. The most unexpected result was the partial restoration of the methylation patterns and phenotypes in the double mutants. It suggested that in the absence of both ATX1 and CLF their roles could be undertaken by a different pair of antagonists. Restored patterns, however, were not identical with the initial patterns, an observation that could account for the variability and instability of phenotypes often seen in epigenetic mutants. Lastly, we demonstrate that ATX1 and CLF physically interact mechanistically linking the observed effects.
MATERIALS AND METHODS
Plant material
Homozygous atx1, clf-1, atx1−/−clf−/− and control wild type Arabidopsis thaliana (Ws), plants were grown under long day light conditions (14 h light/10 h darkness) at 24°C and similarly handled. atx1 plants were crossed with clf-1 both as the female and the male parent with no change of phenotypes or methylation profiles. Homozygocity of atx1−/−clf−/− lines was verified by PCR, RT-PCR and genetically after backcrossing to single mutant atx1−/− and _clf−/−_lines (Supplementary Table 1).
RT-PCR analysis
Total RNA was isolated using Invisorb Spin Plant RNA Mini Kit (Invitek, Berlin, Germany) according to the manufacturer's instruction. First-strand cDNA synthesis was performed on 500 ng of RNA using M-MLV system for RT-PCR (Invitrogen) followed by PCR amplification with Taq DNA Polymerase (Invitrogen) according to the manufacturer's recommendation. PCR primers used for both RT-PCR and ChIP assays are shown in Supplementary Table 2. All PCR amplification reactions were carried out for 38 cycles (95° for 3 min, 95° for 30 min; 50° for 30 min; 72° for 10 min).
ChIP assays were performed following protocols as described (23). Anti-methylated histone antibodies obtained from different sources (Upstate) and (Abcam) were used to confirm reproducibility of patterns. Antibodies specific against ATX1 were raised in rabbits (CoCalico). Band intensities were quantified using ImageQuant™. Intensities were normalized versus the input sample, representing 15% of the DNA used as template (23). Each immunoprecipitation experiment was independently performed, as biological replicates, 3–5 times over a period of several months.
Protein–protein interactions
For testing protein interactions in yeast, ATX1 and CLF (cloned both as bait and pray in the pGBKT7 and pGADT7 vectors, respectively) were tested in the YEASTMAKER Yeast Transformation System’ kit (Clontech). Transformed yeast cells were grown on selective, high stringency, SD media lacking leucine, trypotphan, histidine and adenine. Growth was monitored over 15-day period. Growth of serially diluted transformed cells is shown in Supplementary Data (Figure 1). For the BiFC approach, CLF (At2g23380) and ATX1 (At2g31650) genes were amplified from cDNA templates using specific primers (GATEWAY®, Invitrogen GmbH, Karlsruhe, Germany). The PCR products were subsequently recombined into BiFC vectors (pE-SPYNE, pE-SPYCE) and YFP vector (pENSG-YFP), respectively (24) by attL × attR (LR) recombination at the GATEWAY™ recombination sites. BiFC vectors were kindly provided by C.S. Mayer and W. Dröge-Laser, Georg August University of Goettingen, Germany). The pDONR entry vectors were recombined with the BiFC destination vectors (pE-SPYNE and pE-SPYCE) in an LR reaction which placed the genes in frame with and downstream of the coding sequence of the YFP halves. The resulting expression vector drives expression of the fusion protein under the control of the 35S promoter. The control vectors pE-SPYNE(-)and pE-SPYCE(-) contained the YFP halves with the gateway cassettes replaced by ‘inert filler’ sequences. The generated ATX-YN/CLF-YC, ATX-YC/CLF-YN and control ATX-YN/(-)YC, CLF-YC/(-)NC constructs were delivered into onion epidermal cells and observed 24 h later. Coding sequences of two cytoplasmic proteins (EF1a and MYO1) were cloned in each of the vectors and used as negative controls for the complementation of ATX1.
Figure 1.
Phenotypes and expression of the homeotic gene AG in wild type, in single, and in double mutant plants. (a) Fourth rosette leaf from single (clf or atx1), from double (atx1/clf) mutant, and from wild type (wt) plants. (b) Same age plants are at different stages of development: atx1 and clf mutants are flowering, while atx1/clf and wt plants are still at vegetative stages and (c) expression of the CLF, ATX1 and AG genes in the different genetic backgrounds; ACTIN7, amplified from the respective templates under the same number of cycles is shown as a loading control. All samples were amplified from the same template.
Transient transformation of onion epidermal cells was carried out using the Helium Biolistic gene transformation system (BioRad, Hercules, CA, USA) tissue using a standard procedure with modifications (25,26). Co-bombardments were carried out with a total of 20 µg of DNA used per shot. Ten microgram of each plasmid was used in the combinations: pE-SPYNE –ATX/pE-SPYCE-CLF and pE-SPYNE –CLF/pE-SPYCE-ATX. Bombardments were repeated at least three times for each construct. Subcellular localization of fusion proteins was determined by Olympus FV500 Confocal Microscopy using an excitation wavelength of 488 nm. Emission was captured with a 532 nm band pass emission filter.
RESULTS
Loss of both ATX1 and CLF functions rescue single-mutant phenotypes
Derepression of AG is partly responsible for the curly phenotype of Arabidopsis leaves and for the early flowering of clf mutants (14). The rosette leaves of atx1 mutants are not curled but are smaller and slightly serrated; atx1 plants also bolt earlier than wild type (Figure 1a and b). After introducing atx1−/− in the clf background, however, the phenotypes of homozygous double mutant plants shifted towards wild type: leaf-phenotype and flowering time of atx1/clf mutants were remarkably different from the single atx1 and clf mutants and phenocopied the wild type (Figure 1a and b); accordingly, AG was not detectably expressed in double mutant leaves (Figure 1c).
Rescue of clf phenotypes by the loss of ATX1 (and vice versa) implied antagonistic interactions. To determine epistasis, we tested the expression of ATX1 in the clf background and of CLF in the atx1 background. Lack of significant effects (Figure 1c) indicated that ATX1 was not involved in CLF expression and CLF did not affect ATX1 suggesting that, most likely, ATX1 and CLF do not function in the same genetic pathway.
Methylation patterns of histone H3 lysine 27 and lysine 4 of AG nucleosomes at the silent loci
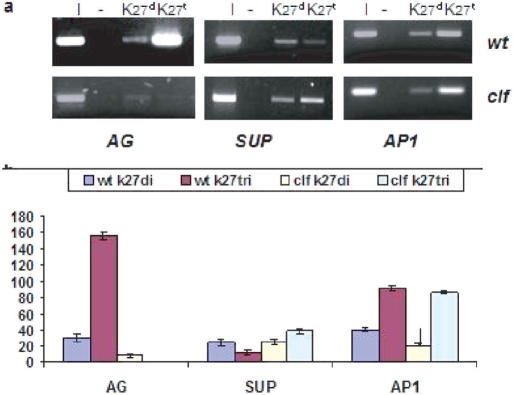
To reveal the molecular mechanism behind these effects, we analyzed the methylated profiles of histone H3 tails of AG nucleosomes in young (10-day) seedling and in leaf-chromatins where the AG gene is not transcribed (27). ATX1 carries HMT activity trimethylating lysine 4 (H3K4me3) (23), while CLF trimethylates K27/H3 of specific Arabidopsis genes (28–31). Immunoprecipitated wild type and clf seedling chromatins (with antibodies distinguishing di- and the tri-methylated K27/H3 isoforms) showed loss of H3K27me3 from _AG_-clf nucleosomes (Figure 2a and b) implicating CLF in trimethylating _AG_-K27/H3. The activity is specific: SUPERMAN (SUP) and APETALA1 (AP1) nucleosomes did not loose their K27/H3 marks in the clf background (Figure 2a and b) consistent with reported ability of CLF to regulate AG but not SUP and AP (14,30).
Figure 2.
H3K27me2 and H3K27me3 profiles of three Arabidopsis genes. (a) ChIP assays with antibodies specific against di- and tri-methylated H3K27; superscripts ‘t’ and ‘d’ stand for the tri-, or di-methylated isoforms, respectively. (I)-input sample is 15% of the immunoprecipitated DNA; (-) is negative control without antibody. Quantified bands from three independently performed ChIP assays are shown as graphs. Changes in H3K27me3 levels of wild type and clf SUP and AP1 nucleosomes are insignificant (p > 0.5).
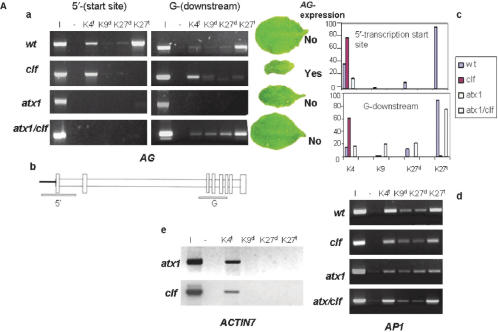
H3K27me3 is the predominant modification of wild type AG nucleosomes at both the 5′-transcription start site and at downstream gene (G)-regions (Figure 3a and c). H3K9me2 and H3K27me2 signals were low and did not change significantly in either atx1 or clf backgrounds (P > 0.05). Interestingly, nucleosomes from the silent AG gene carried also trimethylated K4 (Figure 3a and b). However, H3K4me3 is not sufficient to override the _AG_-non-transcribed state imposed by the presence of H3K27me3. Simultaneously present H3K27me3 and H3K4me3 marks in young seedling chromatin reflect chromatin states, referred to as bivalent, shown recently to silence genes poised for expression later in development (22). Accumulation of H3K4me3 at the _AG_-transcription start site correlates with derepression in the absence of H3K27me3 but with low expression when present together (Figures 1c and 3a), similar to the animal model (22). In contrast to H3K27me3, H3K4me3 is concentrated at the 5′-end _AG_-nucleosomes.
Figure 3.
Methylation profiles of histone H3-tail lysines of _AG_-nucleosomes. (a) ChIP assays with antibodies specific against tri-methylated H3K4 di- and tri-methylated H3K27, and di-methylated H3K9. Amplified regions are from the 5′-transcription start site and from downstream gene (G)-regions (see b). The characteristic leaf phenotypes are shown in parallel with the methylation patterns at 5′-end and downstream nucleosomes, and corresponding expression of AG is indicated; (b) schematic representation of the AG gene: empty vertical boxes represent exons, horizontal boxes represent introns. Bars below the line show regions amplified in the PCR-assay; 5′-represents the region upstream of the transcription start site; G (gene) stands for downstream coding sequences; (c) quantified bands showing the relative change in methylation modifications at the 5′-end and the downstream (G)-nucleosomes; results are from three independently performed ChIP assays; (d) the _AP1_-methylation profiles are shown as controls illustrating the quality of the templates; the same amounts of template DNA were used in the respective amplification experiments and (e) ACTIN7, a constitutively expressed gene carries only ‘activating’ m3K4/H3 tags.
To check for possible presence of ATX1 at the AG locus, we performed ChIP assays with ATX1-specific antibodies. The results suggested binding of ATX1 to _AG_- nucleosomes at the transcription start site but absent from downstream (G)-nucleosomes (Figure 4). Thereby, ATX1 and H3K4me3 distribution domains overlap, supporting a role of ATX1 in _AG_-K4-trimethylation. Interestingly, ATX1 was bound to the 5′-end AG nucleosomes even when transcription was off. We note also that ATX1 was not bound to AP1 nucleosomes, despite the ATX1-activation of AP1 transcription (13). The results suggest that ATX1 controls AP1 indirectly.
Figure 4.
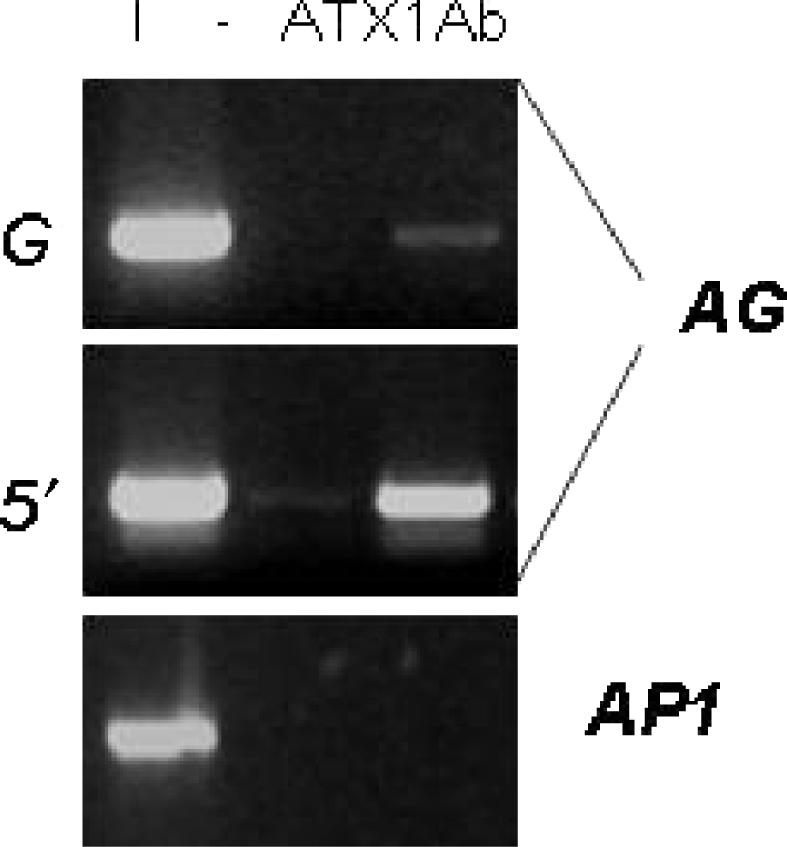
ChIP assays with antiATX1 antibodies. Bands corresponding to the 5′-end _AG_-region amplified after a ChIP with antiATX1 antibodies; downstream region nucleosomes were only weakly represented in the precipitated fraction, while AP1 nucleosomes were not found in the ATX1-bound fraction.
In clf chromatin, the H3K27me3 signal disappeared but the H3K4me3 remained. This profile paralleled AG derepression, as well as the appearance of the ‘curly’ phenotype (Figure 3a). Unexpectedly, loss of H3K4me3 in atx1 chromatin concurred with loss of H3K27me3 implicating ATX1 in the trimethylation of K27. The effects are _AG_-specific: AP1 sequences were amplified from the same chromatin preparations attesting to the quality of the template (Figure 3d). ACTIN7, used as additional control, showed H3K4me3-bands in atx1 chromatin, indicating that absent K4me3 signals were not due to immunoprecipitation artifacts or poor templates (Figure 3e).
Methylation patterns of histone H3 lysine 27 and lysine 4 in atx1/clf double mutants
Functional relevance of ATX1-CLF interaction was pursued further by determining the AG methylation patterns in atx1/clf double mutants. We predicted that no H3K27me3 or H3K4me3 marks would be present in the absence of both ATX1 and CLF. Contrary to the expectation, however, H3K27me3 and H3K4me3, in addition to low levels of H3K9me2 and H3K27me2, were revealed on downstream (G)-nucleosomes but not on 5′-end nucleosomes (Figure 3a). The patterns were reproducible and overall levels of H3K4me3 and H3K27me3 in the clf/atx1 background were comparable to wild type (p < 0.3 for both H3K4me3 and H3K27me3). A notable difference, however, was the presence of H3K4me3 and H3K27me3 tags on downstream-, but not promotor-associated, nucleosomes. Thereby, restored patterns were not identical with the wild type. In support, within homozygous atx1−/−clf−/− lines ‘normalized’ phenotypes were displayed by ∼95% of the plants. About 5% of progenies of wild-type looking double mutants (n = 333; mean value 5.5; SD: 1.36) reverted to smaller, precociously bolting, small rosette-leave (some curled) phenotypes. These results have an important implication for interpreting spontaneous reversals in epigenetic events, as discussed later.
Re-appearance of H3K27me3 and H3K4me3 indicated that HMT activities, different from ATX1 and CLF, could undertake _AG_-modifications but only when both ATX1 and CLF were absent. Restoration of H3K27me3 on (G)-nucleosomes is apparently sufficient to silence AG and to rescue phenotypes associated with loss of CLF and ATX1. The dependence of K27-trimethylation upon ATX1 suggests that ATX1 and CLF ‘communicate’ at _AG_-nucleosomes.
ATX1 and CLF physically interact
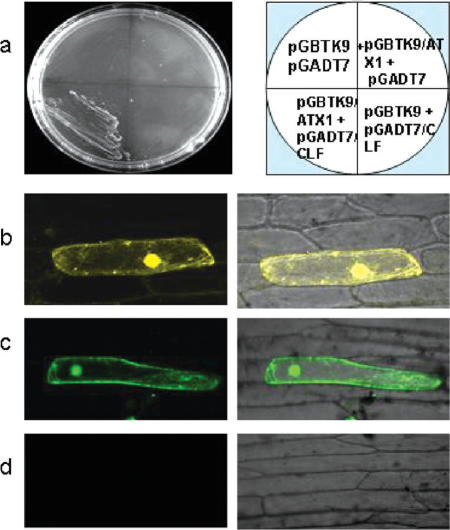
To get a clue about the molecular mechanism of the ATX1-CLF communication, we tested their ability to bind each other. In earlier studies, while using ATX1 as bait, we had isolated an ATX1-interacting clone from yeast and have identified it as CLF (unpublished data). At the time, we did not pursue this observation further but the current results prompted us to revisit this finding. Cloned reciprocally as ‘prey’ and ‘bait’ and tested under two different stringency conditions, CLF and ATX1 did show interaction in yeast (Figure 5a; Supplementary Figure 1). To establish whether ATX1 and CLF would bind in plant cells as well, we used a novel approach developed for the visualization of interactions between proteins in living cells: non-fluorescent fragments of YFP could reconstitute the fluorophore only when brought together by interactions between proteins covalently linked to each fragment (32). ATX1 and CLF were cloned as fusion proteins with halves of the YFP protein and transiently expressed in plant cells to be inspected by the bimolecular fluorescence complementation (BiFC) approach. Accordingly, the fluorescence seen in the nuclei of transformed onion epidermal cells (Figure 5b and c) demonstrate that ATX1 and CLF physically interact. Vectors expressing the YFP halves alone did not produce any signal; nor did the EF1α protein, cloned and tested as a complementary half for the ATX1-linked fluorophore. These experiments underscored the specificity of the ATX1-CLF interaction (Figure 5d). Moreover, loss of H3K27me3 in the atx1 background implicated ATX1 in recruiting CLF to _AG_-nucleosomes. Attempts to co-immunoprecipitate recombinantly produced in Escherichia coli ATX1 and CLF were unsuccessful suggesting that protein modifications might be involved and required in their interaction.
Figure 5.
Interaction of ATX1- and CLF-in yeast and in plant cells. (a) ATX1 and CLF cloned as bait and prey in the pGBKT9 and the pGADT7 vectors; yeast cells transformed with the constructs, as shown, and grown on selective SD media lacking leucine, trypotphan, histidine and adenine (high stringency); (b) BiFC assay of ATX1-YN/CLF-YC. The yellow signal indicates binding of ATX1 and CLF fusion proteins inside the nucleus; (c) BiFC assay of ATX1-YC and CLF-YN (ATX1 and CLF expressed in the opposite combinations of YFP-peptides). Fluorescence is shown in green, using a filter, to distinguish interactions of a different combination of complementing YFP-halves. (d) Cells transformed with the control vectors ATX1-YN/(-)YC and CLF-YN/(-)YC did not generate fluorescence. The images on the right represent merges of the left images with the DIC images.
DISCUSSION
Our findings offer novel insights into the workings of the Arabidopsis PcG and TrxG antagonists. An interaction between ATX1 and CLF for the control of the AG silent state is supported by three types of evidence: first, restoration of _AG_-repression in the double mutants and rescued leaf phenotypes associated with AG misexpression in single atx1 and clf mutants, provide compelling genetic evidence; second, bivalent chromatin marks established by the two methylases at the silent AG locus constitute epigenetic evidence. Lastly, the propensity of ATX1 and CLF to form specific complexes in vivo mechanistically links the observed effects. Restoration of AG repression upon loss of both ATX1 and CLF and loss of H3K27me3 in the atx1 background support the idea that the two methylases interact. Partial restoration of the H3K4me3 and H3K27me3 signals in the double atx1/clf mutants illustrated that _AG_-nucleosomes could be targeted by a different (surrogate) pair of methylases but only when both ATX1 and CLF were absent. Why methylations were restored on downstream nucleosomes but not at the 5′-nucleosomes is unclear but one plausible possibility is that a specific transcription factor directing ATX1 and CLF to the promotor does not interact with the surrogate pair.
Presence of both K4me3 and K27me3 tags at silent embryonic stem cell loci was interpreted as a bivalent mark on genes poised for transcription later in development (22). It is quite remarkable, then, that a similar bivalent mark was found at the flower homeotic gene, silent in young seedlings but expressed during flowering (33). ChIP assay, currently used for determining chromatin modification states, is not a feasible approach for analysis of the active AG profiles because flower chromatin is a mix of cells (whorls) expressing and non-expressing AG (33). Alternative approaches need to be developed to assess AG modification patterns in flowers.
It is interesting to note that the nucleosomes of another homeotic regulator, AP1, also carry the bivalent K4me3 and K27me3 marks in silent chromatins (Figures 2a and 3d). However, an apparently different antagonistic pair establishes the bivalent marks of AP1 nucleosomes because the K27me3 signal in clf chromatin is preserved as is the K4me3 signal in atx1 chromatin (Figures 1d and 2c). While CLF does not affect AP1 (14), consistent with preservation of K27me3 in clf chromatin, ATX1 activates AP1 transcription (13). ChIP analysis with antiATX1 antibodies did not reveal presence of ATX1 at _AP1_nucleosomes (Figure 4) consistent with a conclusion that ATX1 does not modify AP1 nucleosomes. Thereby, activation of AP1 transcription is, most likely, indirect.
De-repression of AG in clf mutants is associated with a strong curly leaf phenotype (14) but, interestingly, atx1 plants also show a leaf phenotype despite the fact that AG is not expressed in atx1 leaf chromatin (Figure 1a and c). Most likely, other ATX1-regulated genes contribute to leaf size and shape; in addition, both H3K4me3 and H3K27me3-tags might be needed to establish ‘normal’ silencing of AG. We note also that the AG transcription pattern as well as histone H3-K4, K9 and K27 methylation profiles of _AG_-nucleosomes did not depend on the genetic background of the plants as they were indistinguishable in chromatins isolated from Col, Ws and Ler background plants. Furthermore, the bivalent-state silencing ‘code’ may be gene-specific; for instance, Clarke Kent epialleles of SUP require simultaneously present H3K27me2 and H3K9me2 to maintain DNA methylation and to keep the gene silent (31).
There are three EZ and multiple _ATX1_-related and putative K4-trimethylase genes in the genome of Arabidopsis (16,34,35). At least three different PRC2-like complexes, containing different EZ homologs, have been proposed (9,29). Despite some partial redundancy, however, none of the CLF homologs (MEA or SWN) could complement clf mutants consistent with the idea that each one participates in a different complex (9,14). Likewise, the inability of any of the ATX1-related proteins to substitute for ATX1 in the atx1 background (Figure 1a and b; 14,23) suggest that K4 methyl transferases assemble, most likely, distinct complexes as well. Given that a functionally equivalent methylase cannot substitute a missing relative within a complex, our results take the specificity one step further, by suggesting that PcG- and TrxG-complexes form specific pairs establishing bivalent marks at targeted loci.
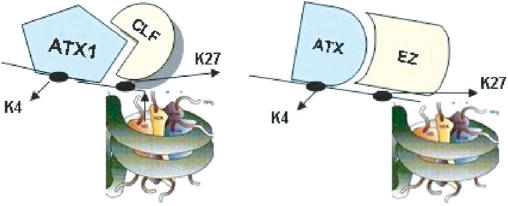
The idea of how two antagonistic epigenetic complexes might interact is illustrated in the proposed model (Figure 6). A central assumption is that each PRC2 and TrxG complex assembles a specific set of subunits around a particular EZ or TRX homolog. Only when both complexes are missing could their roles be undertaken by another pair. Such a model could explain partial rescue of single mutant atx1 and clf phenotypes in the absence of both ATX1 and CLF. Furthermore, restored patterns were not identical with the wild type, i.e. they did not occur at 5′-end nucleosomes, nor did they take place 100% of the times. Thereby, inability of surrogate complexes to fully rescue native patterns provides a basis for interpreting spontaneous reversals, variability and instability of phenotypes associated with epigenetic mutations.
Figure 6.
A model for the interaction of the ATX1-containing TrxG and CLF-containing PcG complexes at the AG locus. In wild type leaf chromatin, CLF and ATX1 counterbalance each other. At the molecular level, this is associated with non-transcribed AG and with presence of m3K4/H3 and m3K27/H3. Both ATX1 and CLF activities are required for the ‘normal’ repression of AG generating, a bivalent chromatin state. ATX1 and CLF participate in antagonistic complexes functioning as a specific pair (nucleosome on the left). Loss of either CLF or ATX1 cannot be substituted by a homologous activity within the pair. ATX1 at the _AG_- 5′-nucleosomes may recruit CLF but not vice versa. This may account for the m3K4 and m3K27 patterns in the single mutants (see text). Elimination of both ATX1 and CLF allows a different pair of antagonists to label AG (nucleosome on the right). However, they modify only downstream nucleosomes because the specific factor taking ATX1/CLF to the start site fails to recruit their homologs. Assembly of substitute complexes restoring the methylation tags might not be occurring 100% of the time accounting for the variability in rescued phenotypes.
Partial normalization of axial–skeletal transformations in mice, observed when both Mll (a human homolog of trithorax) and _BMI_-1 (a PcG component) were deleted, led to the suggestion that direct interaction between MLL and a PcG-subunit was the molecular mechanism of phenotype rescue (36). Here, we demonstrated that the Trx and EZ homologs could, indeed, bind directly; interactions among other subunits of the complex pair are not excluded. It is interesting to consider our results in the context of two recent models of PcG-TrxG interactions in animal systems. At the _UBX_-PRC sites, the Trx protein is constitutively bound with PcG complexes compatible with a possibility that the two complexes might directly interact, although binding partners were not identified (37). It was suggested that Ash1 and Trx HMTases were not ‘coactivators’ for the activation of HOX genes but function as antirepressors of PcG function to prevent inappropriate silencing of HOX. Methylated K4/H3 was suggested to prevent binding of PcG (35). Tri-methylated K4 were confined to the first 1 kb, and tri-methylated K27 were found throughout the coding sequence (38) similar to the H3K27me3 and H3K4me3 distribution at the AG locus. However, the molecular mechanism keeping the homeotic AG gene silent is apparently different: presence of K4-methylation did not prevent binding of a CLF-containing repressive complex; to the contrary, ATX1 was required for methylations at K27 to occur. Our results agree well with the model suggesting that simultaneous presence of TRX and PcG (and the activating K4 and repressing K27 marks, respectively) establish bivalent chromatin states in embryonic stem cells (22). In a bivalent state, certain genes are silent but the chromatin structure is poised for activation later in development. It is tempting to suggest that the presence of K4- and K27-trimethyl marks at the non-expressing AG locus in young seedling chromatin reflects a bivalent chromatin state prepared for expression at a later developmental stage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
[Supplementary Material]
ACKNOWLEDGEMENTS
The authors are grateful to B. Bernstein and to J. Goodrich for critically reading the manuscript and helpful suggestions, to J. Goodrich for his gift of clf seeds, and to C.S. Mayer and W. Dröge-Laser for the gifts of BiFC vectors. This work partially supported by NSF, through grant MCB-0343934 (Z.A.). Funding to pay this Open Access publication charges for this article was provided by NSF, through grant award MCB-0343934.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 2.Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, van der Lugt NM, Shashikant CS, Ruddle FH, et al. Mammalian Trithorax and Polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl Acad. Sci. USA. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb group and thrithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 4.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a Polycomb group protein. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 5.Levine SS, King IFG, Kingston RE. Division of labor in Polycomb group repression. Trends Biochem. Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 8.Avramova Z. Heterochromatin in animals and plants; similarities and differences. Plant Phys. 2002;129:40–49. doi: 10.1104/pp.010981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 10.Loidl P. A plant dialect of the histone language. Trends Plant Sci. 2004;9:84–90. doi: 10.1016/j.tplants.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Meyerowitz EM. Plants compared to animals: the broadest comparative study of development. Science. 2002;295:1482–1485. doi: 10.1126/science.1066609. [DOI] [PubMed] [Google Scholar]
- 12.Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Curr. Opin. Plant. Biol. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX1, an Arabidopsis homolog of Trithorax has histone methylase activity and activates flower homeotic genes. Curr. Biol. 2003;13:627–634. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 14.Goodrich J, Puangsomiee P, Martin M, Long D, Meyerowitz E, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 15.Katz A, Oliva M, Mosquna A, Hakim O, Ohad N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 2004;37:707–719. doi: 10.1111/j.1365-313x.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim SY, He Y, Jacob Y, Noh YS, Michaels S, Amasino R. Establishment of the vernalization-responsive. Winter-annual habitat in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell. 2005;17:3301–3310. doi: 10.1105/tpc.105.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 19.Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 20.Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl Acad. Sci. USA. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S. EMBRYONIC FLOWER2, a novel Polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell. 2001;13:2471–2481. doi: 10.1105/tpc.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Venegas R, Avramova Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 2005;33:5199–5206. doi: 10.1093/nar/gki830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weltmeier F, Ehlert A, Mayer C, Dietrich K, Wang X, Schutze K, Alonso R, Harter K, Vicente-Carbajosa J, et al. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 2006;25:3133–3143. doi: 10.1038/sj.emboj.7601206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon-Kamm W, Spencer T, Mangano M, Adams T, Daines R, Start W, O'Brien J, Chambers S, Adams W, Jr, et al. Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell. 1990;2:603–618. doi: 10.1105/tpc.2.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleh A, Lumbreras V, Lopez C, Puigjaner E, Kizis D, Pagès M. Maize DBF1-interactor protein 1 containing an R3H domain is a potential regulator of DBF1 activity in stress responses. Plant J. 2006;46:747–757. doi: 10.1111/j.1365-313X.2006.02742.x. [DOI] [PubMed] [Google Scholar]
- 27.Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schonrock N, Bouveret R, Leroy O, Borghi L, Kohler C, Gruissem W, Hennig L. Polycomb-group proteins repress floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 2006;20:1667–1678. doi: 10.1101/gad.377206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Kohler C. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7:947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006 doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4146–4155. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu C-D, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 33.Coen ES, Meyerowitz EM. The war of whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 34.Alvarez-Venegas R, Avramova Z. SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 35.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Z-B, Anderson M, Diaz MO, Zeleznik-Le N. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the co-repressor C-terminal-binding protein. Proc. Natl Acad. Sci. USA. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papp B, Mueller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]