Retroviral transduction of TLS-ERG initiates a leukemogenic program in normal human hematopoietic cells (original) (raw)
Abstract
Many chimeric oncogenes have been identified by virtue of the association between chromosomal translocation and specific human leukemias. However, the biological mechanism by which these oncogenes disrupt the developmental program of normal human hematopoietic cells during the initiation of the leukemogenic process is poorly understood due to the absence of an appropriate experimental system to study their function. Here, we report that retroviral transduction of TLS-ERG, a myeloid leukemia-associated fusion gene, to human cord blood cells results in altered myeloid and arrested erythroid differentiation and a dramatic increase in the proliferative and self-renewal capacity of transduced myeloid progenitors. Thus, TLS-ERG expression alone induced a leukemogenic program that exhibited similarities to the human disease associated with this translocation. These results provide an experimental examination of the early stages of the human leukemogenic process induced by a single oncogene and establish a paradigm to functionally assay putative leukemogenic genes in normal human hematopoietic cells.
The production of blood cells is tightly regulated at all stages of development from stem cells to mature cells, but genetic alterations within stem cells can perturb the developmental program resulting in a clonal outgrowth of one or more lineages (1–3). Murine studies of leukemogenesis have suggested that alterations are required in both the proliferation and differentiation of hematopoietic cells (4). In some cases, these alterations occur as a consequence of the expression of multiple genes such as co-expression of the IL-3 gene (5, 6) (which stimulates proliferation) and the HOXB8 gene (7) (which alters differentiation). In other cases, proliferation and differentiation are affected following the expression of a single gene (7–11). In these latter cases, full leukemic growth in vivo requires additional genetic alterations. In the human, analysis of invariant chromosomal translocations associated with specific leukemias has permitted the cloning of novel genes that must be involved in leukemic transformation and progression (12–14). However, knowledge of the biological mechanism whereby these leukemogenic genes disrupt the self-renewal, proliferation, and differentiation of normal human hematopoietic cells during the early stages of leukemogenesis is lacking because of the absence of experimental systems. Consequently, much of our understanding of these genes has been extrapolated from analysis of leukemic cell lines, gene overexpression studies (transgenic or retroviral gene transfer approaches), or from knockout mice (4, 15). However, cell lines typically accumulate many genetic alterations preventing the identification of the mechanism that initiates leukemogenesis. Although murine studies have contributed greatly to our understanding of oncogenes, they do not always recapitulate human disease. For example, HOX 11 and E2A-PBX are associated with T cell and pre-B cell leukemia in humans, respectively, but overexpression in mice results in myeloid leukemia (10, 16). Moreover, differences in the susceptibility of murine versus human cells to neoplastic alterations, which is likely attributed to the greater relative genomic instability of murine cells, underscores the relevance of studying human leukemogenic genes in normal human hematopoietic cells (17).
To develop an experimental leukemogenesis model in normal human hematopoietic cells, we transduced human umbilical cord blood (CB) cells with a retrovirus vector expressing the TLS-ERG oncogene. TLS-ERG is a fusion gene generated from t(16;21)(p11;q22), a recurrent chromosomal translocation associated with poor prognosis human acute myeloid leukemia (AML), secondary AML associated with myelodysplastic syndrome (MDS), and chronic myeloid leukemia (CML) in blast crisis (18, 19). TLS/FUS (hereafter designated TLS), located on chromosome 16, encodes an RNA-binding protein (20), whereas the ERG gene on chromosome 21 encodes a transcriptional activator of the ets proto-oncogene family with DNA-binding and transactivator domains (21–23). The altered transcriptional activating and DNA-binding activities of the TLS-ERG gene product are thought to be responsible for the genesis or progression of t(16;21)-associated human myeloid leukemias (24). We report that normal human hematopoietic cells expressing TLS-ERG acquire a greatly increased capacity for self-renewal and proliferation. Together with the observation of altered myeloid and arrested erythroid differentiation programs, these results provide insight into the early stages of leukemogenesis induced by a putative leukemogenic gene expressed in normal human hematopoietic cells.
MATERIALS AND METHODS
Preparation of Retroviral Producer Cells Expressing TLS-ERG.
A 1.4-kb _Bgl_II fragment encompassing the TLS-ERG ORF was liberated from the mammalian expression plasmid, pSGTLS-ERG (24), and cloned into the _Bgl_II site downstream of the 5′-long-terminal repeat and upstream of the pgk-NEO cassette in the pMSCV2.1 retroviral vector (25). Ten micrograms of pMSCVTLS-ERG was transfected into the amphotropic PA317 packaging cell line (26) using lipofectamine (as described by manufacturer, GIBCO/BRL). After 16 h, transfection supernatant was used to infect xenotropic PG13 packaging cells (27), which produce the gibbon ape leukemia virus (GALV) env protein. Subsequent selection in G418 (Sigma; 400 μg/ml) resulted in 33 clones, which were screened using a modified viral RNA dot blot assay (28) to identify potential high titre candidates. Titres for the resulting PG13MSCVTLS-ERG and PG13MSCVNEO producer lines used during this study were 3 × 105 and 1 × 106 cfu/ml, respectively. Proviral integrity of NEO and TLS-ERG expression in producer cells was confirmed by Southern and Northern blot analysis (data not shown) as well as by Western blot analysis (Fig. 4B).
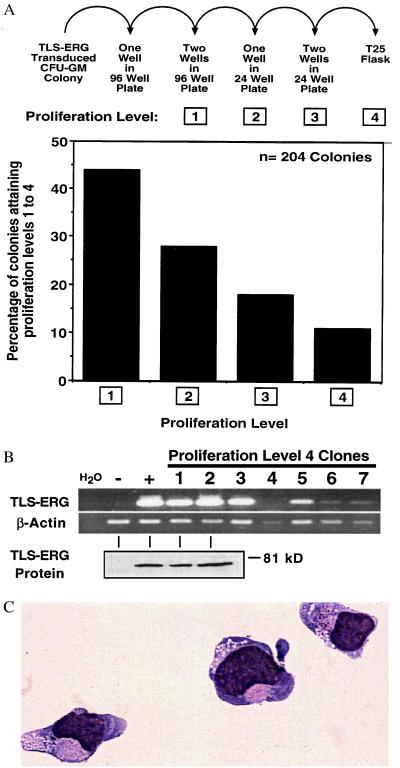
Figure 4.
TLS-ERG-transduced CFU-GM colonies proliferate in liquid culture. (A, upper) TLS-ERG-transduced G418-resistant CFU-GM colonies (from six independent experiments) were placed in liquid culture containing 300 ng/ml SCF, 300 ng/ml GM-CSF, and 50 ng/ml IL-3 and allowed to progress through 4 proliferation levels in the absence of G418. (Lower) Bar graph depicting the percentage of 204 colonies attaining a given proliferation level. (B) RT-PCR (Upper) and Western blot (Lower) analysis revealing the presence of TLS-ERG transcripts and protein in nonproliferating level 4 clones, respectively. Although H2O represents the reagent control for RT-PCR, the negative (−) and positive (+) controls for RT-PCR and Western blot analyses are from PG13MSCVNEO and PG13MSCVTLS-ERG retroviral producer cells, respectively. (C) May–Grünwald–Giemsa stained cytospin preparation of TLS-ERG-transduced CFU-GM-derived cells that had been proliferating in liquid culture for 3 weeks.
Isolation and Retroviral Infection of CD34+Lin− Human Umbilical Cord Blood Cells.
Ficoll-separated human umbilical cord blood cells were stained with a mixture of lineage-specific antibodies provided by the manufacturer (Stem Cell Technologies, Vancouver, BC), followed by addition of secondary antibody conjugated to metal colloid. Cells were then eluted through a magnetized column to enrich for CD34+ cells not expressing lineage markers (Lin−). The purity of typically obtained CD34+Lin− cells was 40–70%. In preparation for retroviral infection, 0.5–2 × 106 CD34+Lin− cells were resuspended in 2 ml IMDM supplemented with 2.5% FCS, 50 ng/ml IL-3, 20 ng/ml IL-6, 50 ng/ml SCF, 50 ng/ml FLT-3 ligand, and 20 ng/ml G-CSF (factors supplied by Amgen Biologicals and Immunex) and prestimulated for 16 h on 35-mm Petri dishes precoated with the C-terminal fibronectin fragment CH-296 (supplied by Takara Shuzo, Otsu, Japan). Following prestimulation, 2 ml of virus-containing medium from a confluent T 75-cm2 flask of either PG13MSCVNEO or PG13MSCVTLS-ERG retroviral producer cells supplemented with the aforementioned growth factors was used to infect the CD34+Lin− cells at 12-h intervals for 48 h. Infected cells were subsequently analyzed in methylcellulose progenitor cell and erythroid differentiation liquid culture assays as well as by flow cytometry.
Progenitor Assay.
To assess the effect of TLS-ERG expression on the proliferation and differentiation of human myeloid/erythroid progenitors, infected CD34+Lin− cells were plated in methylcellulose cultures as described (29) in the presence or absence of 1500 μg/ml G418. After 12–14 days, progenitor colony types were scored into granulocyte (CFU-G), macrophage (CFU-M), granulocyte/macrophage (CFU-GM), erythroid (BFU-E), and mixed (CFU-Mix) types. Morphological designation of colony type by light microscopy was confirmed by May–Grünwald–Giemsa staining of cytospin preparations.
TLS-ERG Expression Analysis.
Reverse transcription (RT)-PCR was performed to detect TLS-ERG and β-actin mRNA transcripts in TLS-ERG-transduced CFU-GM-derived cells. Poly(A)+ RNA was isolated using the Pharmacia micro mRNA purification kit as described by the manufacturer, and cDNA was synthesized from 1 μg of mRNA template in a 20-μl reaction (50 mM Tris-HCl, pH 8.3/75 mM KCl/3 mM MgCl2/10 mM DTT/0.5 mM dNTP) using 200 units of Superscript RNase H-M-MLV Reverse Transcriptase (GIBCO) and 20 ng of random hexamer primers. Subsequent PCR reactions were performed with 0.4–2 μl of cDNA first strand reaction in a 50-μl volume [10 mM Tris·HCl, pH 8.3/50 mM KCl/1.5 mM MgCl2/0.2 mM each dNTP/2.5 units of Taq DNA polymerase (BMC)] using 0.5 μM of the T (5′-GGTGGCTATGAACCCAGAGGT-3′) and E (5′-CCTCGTCGGGATCCGTCATCT-3′) primers to detect TLS-ERG transcripts and the Beta1 (5′-GATCCACATCTGCTGGAAGG-3′) and Beta2 (5′-AAGTGTGACGTTGACATCCG-3′) primers to detect β-actin transcripts. TLS-ERG (35 amplification cycles: denaturation 95°C, 30 s; annealing 58°C, 30 s; extension 72°C, 90 s) and β-actin (35 amplification cycles: denaturation 94°C, 45 s; annealing 52°C, 45 s; extension 72°C, 120 s) PCRs were performed using a RoboCycler Gradient Temperature Cycler (Stratagene). The 211-bp TLS-ERG and 222-bp β-actin amplification products were analyzed on a 1% agarose gel.
TLS-ERG protein expression was assessed by resolving whole cell extracts on 10% SDS/PAGE followed by standard Western blot/ECL analysis using ERG antibody (Red-1).
Liquid Culture for Erythroid Differentiation.
Human umbilical cord blood CD34+Lin− cells were isolated and infected as described above and cultured in StemPro-34 serum-free medium (GIBCO/BRL) with 1 mg/ml G418, 20 ng/ml SCF (Amgen), 2 ng/ml IL-3 (Amgen), 1 ng/ml G-CSF (Amgen), and 1 unit/ml erythropoietin (Eprex) as described (30). Infected cells were seeded at an initial concentration of 105 cells per ml in 25-cm2 suspension culture flasks, and samples were taken to detect CD36, glycophorin-A (gly-A), and CD33-expressing cells by flow cytometric analysis, cytospin preparation, and methylcellulose culture at 2–5-day intervals.
Flow Cytometry.
Cell surface marker expression on expanded cells derived from TLS-ERG-transduced progenitors was assessed by flow cytometry. Cells were washed in phosphate-buffered saline (PBS) containing 5% FCS. Approximately 106 cells were resuspended in 1 ml of PBS + 5% FCS containing 5% human serum (to block Fc receptors) for 30 min at 4°C, washed, and then incubated with monoclonal antibodies at a concentration of 5 μg/ml for 30 min at 4°C. The antibodies used, purchased from either Becton Dickinson or Coulter, were as follows: anti-CD3-fluorescein isothiocyanate (FITC), anti-CD4-FITC, anti-CD7-phycoerythrin (PE), anti-CD8-PE, anti-CD13-PE, anti-CD14-FITC, anti-CD15-FITC, anti-CD19-PE, anti-CD20-FITC, anti-CD33-PE, anti-CD34-FITC, anti-CD36-PE, anti-CD38-PE, and anti-glycophorin A. Cells were then washed three times in PBS + 5% FCS and analyzed on a FACScalibur (BD). An aliquot of cells was also stained with mouse IgG conjugated to FITC and PE as an isotype control. Fluorescence levels excluding greater than 99% of the cells in these negative controls were considered to be positive and specific for human cells.
RESULTS
Frequency and Lineage Distribution of Progenitors Transduced by TLS-ERG.
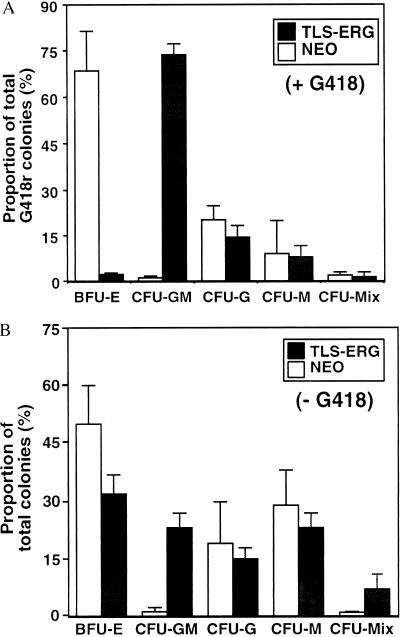
CD34+Lin− cells were isolated from CB and transduced with a retrovirus vector expressing the TLS-ERG cDNA (24). To evaluate the effect of TLS-ERG expression on the proliferative and differentiative capacity of myeloid, erythroid, and multilineage progenitors, the transduced cells were plated in methylcellulose assays as described (29). Although the efficiency of gene transfer into colony-forming cells with the TLS-ERG vector was comparable (32%, n = 7) with the control NEO vector (43%, n = 6), there was a large difference in the lineage distribution of the transduced progenitors. We observed a 62-fold increase in the frequency of bipotent granulocyte/macrophage progenitors (CFU-GM) and a 31-fold decrease in the frequency of immature erythroid progenitors (BFU-E) in TLS-ERG- versus NEO-transduced cells (Fig. 1A). The frequency of TLS-ERG- and NEO-transduced granulocyte (CFU-G), macrophage (CFU-M), and CFU-Mix colonies was comparable. When TLS-ERG-transduced cells were plated in the absence of G418, a 20-fold increase in the frequency of CFU-GM and a 1.5-fold decrease in the frequency of BFU-E was observed, indicating that TLS-ERG affected progenitor growth in the absence of G418 selection (Fig. 1B).
Figure 1.
Comparison of the frequency and distribution of TLS-ERG- and NEO-transduced progenitors. Following transduction of TLS-ERG and NEO, CD34+Lin− human CB cells were plated in methylcellulose cultures as previously described (29). The transduced progenitors were selected in 1500 μg/ml G418 (A) or not (B). The frequency and distribution of myeloid (CFU-GM, CFU-G, CFU-M), erythroid (BFU-E), and myeloid/erythroid (CFU-Mix) progenitor colonies was scored after 14 days. Colony designations were confirmed by May–Grünwald–Giemsa staining of cytospin preparations. Mean frequencies and standard deviations are graphically represented from seven independent experiments performed with seven different CB samples.
Increased Proliferation and Self-Renewal of TLS-ERG-Transduced Progenitors.
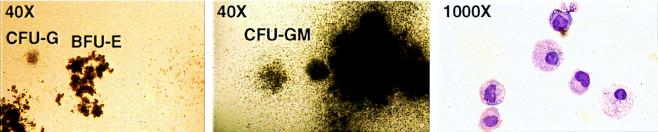
In addition to increasing the frequency of CFU-GM, overexpression of TLS-ERG dramatically increased their proliferation, resulting in macroscopic colonies in methylcellulose cultures. In comparison to typical NEO-transduced CFU-G and BFU-E colonies shown at 40× magnification in Fig. 2 Left, the majority of TLS-ERG-transduced CFU-GM colonies were dense compact colonies surrounded by a diffuse halo of cells and grew to a large diameter (0.5–3.0 mm, containing 2 × 104-2 × 105 cells/colony) (Fig. 2 Middle). Cytological analysis of cells dissociated from a TLS-ERG-transduced CFU-GM colony confirmed the presence of granulocytes and macrophages (Fig. 2 Right). PCR analysis confirmed that all the G418R CFU-GM-derived colonies contained integrated TLS-ERG cDNA sequences (data not shown).
Figure 2.
TLS-ERG enhances the proliferative capacity of CFU-GM. (Left) Photomicrographs of NEO-transduced colonies revealing typically sized day 14 CFU-G and BFU-E colonies. (×40.) (Middle) TLS-ERG-transduced colonies showing a large CFU-GM colony after 14 days in culture. (×40.) (Right) May–Grünwald–Giemsa stained cytospin preparation of cells derived from a large TLS-ERG-transduced CFU-GM. (×1,000.)
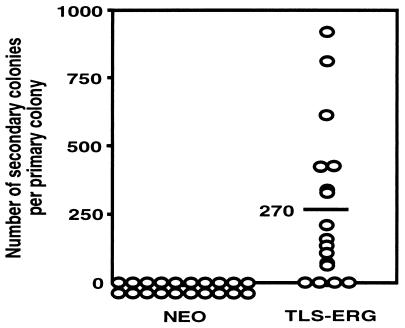
Although enhancement of self-renewal capacity is a hallmark of leukemic stem and progenitor cells, normal progenitors have very limited self-renewal capacity as measured by secondary replating of primary colonies (31). To evaluate whether TLS-ERG had affected the self-renewal potential of transduced progenitors, individual well isolated TLS-ERG- and NEO-transduced G418R colonies were randomly plucked from primary methylcellulose cultures on day 12 and replated in secondary cultures under the same conditions. As expected, NEO-transduced colonies did not generate secondary colonies. In contrast, 80% of primary TLS-ERG-transduced colonies gave rise to secondary colonies, producing a mean of 270 secondary colonies per primary colony (Fig. 3). Thus, TLS-ERG expression had a dramatic effect both on the proliferation and on the self-renewal capacity of transduced progenitors.
Figure 3.
Individual TLS-ERG-transduced CFU-GM colonies self-renew and form secondary colonies upon replating. Well isolated TLS-ERG- and NEO-transduced colonies derived from day 12 primary G418-selected methylcellulose cultures were randomly plucked and replated in secondary cultures under the same conditions used to establish the primary cultures. Each open dot represents the number of secondary colonies generated per primary colony. The horizontal line indicates the mean.
TLS-ERG-Transduced Progenitors Proliferate in Liquid Culture.
To determine whether TLS-ERG expression was able to increase the proliferation and self-renewal capacity of the transduced progenitors to enable their establishment in liquid culture, a total of 204 G418R TLS-ERG-transduced CFU-GM-derived colonies (2 × 104-2 × 105 cells per colony) from six separate experiments were individually isolated and allowed to progress through four proliferation levels in the absence of G418 (Fig. 4A). Colonies were initially seeded into a single well of a 96-well plate. In 44% of these wells, proliferation necessitated splitting into two wells of a 96-well plate (proliferation level 1). Colonies that continued to proliferate (28%) were seeded into one well of a 24-well plate (level 2) and then into two wells of a 24-well plate (18%; level 3). Finally, 11% of the colonies progressed to level 4, which involved the proliferation of 1–40 × 106 cells in T25 suspension flasks. The clonagenicity of four level 4 cell groups tested was confirmed by analysis of retroviral integration sites using Southern blot (data not shown). As expected, NEO-transduced G418R myeloid colonies were unable to achieve proliferation level 1. Thus, by three independent assays (colony size, secondary plating, and liquid culture), we have conclusively shown that TLS-ERG expression, alone, dramatically affects the proliferation and self-renewal of normal human CFU-GM progenitors.
Although 11% of the clones could proliferate in liquid culture for extended periods of time, none were able to grow indefinitely. Regardless of proliferation level, most TLS-ERG-transduced clones ceased their proliferation because of terminal differentiation, whereas the rest ceased to proliferate but remained blocked at the promyelocytic stage (see below). DNA laddering analysis of the latter clones did not reveal evidence of apoptotic death (data not shown), a finding consistent with recent data demonstrating that TLS-ERG inhibits apoptosis (32). To determine whether the cessation of proliferation was associated with a loss of TLS-ERG expression, RT-PCR analysis was performed. TLS-ERG transcripts were detected in six of seven clones (Fig. 4B Upper), suggesting that a decline in proliferative capacity was not associated with a loss of TLS-ERG expression. Western blot analysis confirmed the expression of full-length TLS-ERG protein in two of the clones (1 and 2) tested (Fig. 4B Lower). We conclude that TLS-ERG expression is insufficient for indefinite proliferation, suggesting that it alone cannot completely immortalize. In addition, it should be noted that our observations are very similar to those obtained from growing primary human leukemic cells in culture. It is extremely rare to isolate permanently growing AML cell lines from leukemic blood or bone marrow samples, which in most cases stop proliferating after several weeks in culture (1, 2, 33). In fact, the proliferation we have observed from a single transduced colony is far greater than that of clonogenic AML progenitors. Only in rare circumstances can permanently growing human cell lines be isolated after long periods of time and after a crisis event in culture.
We also assessed several of the highly proliferative clones for their responsiveness to growth factors. In all cases, the clones were factor-dependent, showing growth responses to SCF, IL-3, and GM-CSF, with the strongest response to all three factors together (data not shown). No response was seen with FLT-3 ligand or G-CSF, alone or in combination. Thus, TLS-ERG expression did not relieve the growth factor dependence of the transduced progenitors. Interestingly, IRTA17 and IRTA21, two cell lines established from the bone marrow of an AML (M2) patient carrying t(16;21) as the sole gross chromosomal abnormality, were dependent on SCF, GM-CSF, IL-3, and G-CSF for their growth (34).
Characterization of TLS-ERG-Transduced Cells Proliferating in Liquid Culture.
The cell surface phenotype and morphology of proliferating TLS-ERG-transduced cells that reached proliferation level 4 were determined by flow cytometry and cytology. May–Grünwald–Giemsa staining of cytospin preparations revealed morphologic features that were characteristic of promyelocytes: coarse cytoplasmic granulation, slight nuclear indentation, and an appropriate nuclear:cytoplasmic ratio (Fig. 4C). Expression of the CD13, CD15, and CD33 myeloid markers but not the monocytic marker, CD14, confirmed the myelocytic nature of these cells (data not shown) and suggested that their differentiation program was arrested at the promyelocytic stage, rarely proceeding to mature neutrophils despite culture with GM-CSF and G-CSF. Lymphoid markers (CD3, CD4, CD7, CD8, CD19, and CD20), erythroid markers (CD36, glycophorin A), and CD34, a hematopoietic stem and progenitor cell marker, were not detected (data not shown). The cell surface phenotype was similar to that observed in primary leukemic cells obtained directly from patients with t(16;21)(p11;q22) and from cell lines (34). Chromosomal analysis of a TLS-ERG-transduced CFU-GM-derived clone revealed a normal karyotype (data not shown), suggesting that the proliferative advantage conferred by TLS-ERG occurred in the absence of gross chromosomal aberrations.
TLS-ERG Expression Blocks Erythroid Differentiation.
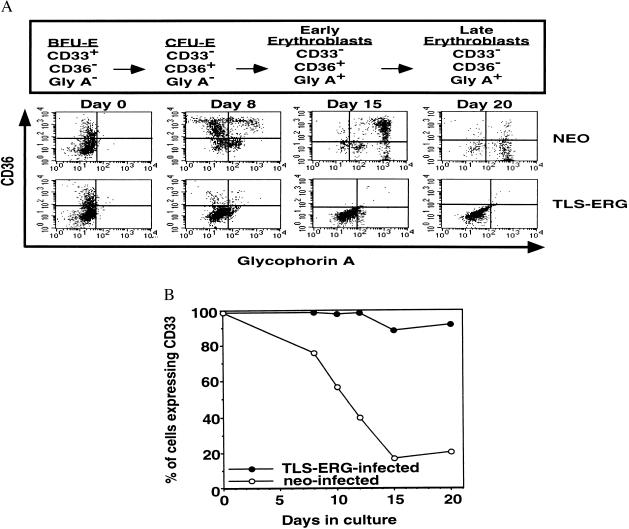
The marked reduction in the frequency of erythroid colonies we observed following methylcellulose culture of TLS-ERG-infected CD34+Lin− cells suggested that TLS-ERG plays a role in impairing the normal erythroid differentiation program at some stage prior to the BFU-E progenitor. To determine the stage at which this impairment occurred, we employed a liquid culture system that promoted erythroid differentiation and utilized sensitive flow cytometric analysis to compare the ability of TLS-ERG- and NEO-transduced CD34+Lin− cells to undergo erythroid differentiation (30). The flow cytometric analysis involved measuring changes in the expression of CD36, gly-A, and CD33 cell surface markers, which change expression during erythroid differentiation as shown in Fig. 5A Upper (34). The NEO-transduced cells exhibited a CD36, gly-A, and CD33 expression profile characteristic of erythroid development after 20 days in culture (Fig. 5A Lower and B). In contrast, TLS-ERG-transduced cells failed to express CD36 or gly-A (Fig. 5A Lower) but continued to express CD33 (Fig. 5B), indicating that their ability to undergo erythroid differentiation beyond the BFU-E stage was completely arrested. Interestingly, using a similar erythropoiesis culture, a mutant N-Ras gene, which is prevalent in MDS and AML, was also recently shown to induce erythroid dysplasia in human CD34+ cells (35). Detailed analysis of the CD33-expressing TLS-ERG-transduced cells indicated that they were not immature erythroid cells but relatively immature myeloid cells (CD13+, CD15+, and CD38+ but CD14− or CD34−) (data not shown). Unlike the NEO-transduced cells, which did not proliferate beyond day 22 due to terminal differentiation, these cells proliferated for approximately 70 days and gave rise to clonogenic progenitors that were exclusively large CFU-GM (data not shown), providing further independent evidence that TLS-ERG enhances the proliferative capacity of myeloid progenitors.
Figure 5.
Establishment of TLS-ERG- and NEO-transduced CD34+Lin− human CB cells in erythroid liquid culture assays. (A, upper) Scheme to assess erythroid differentiation. Maturation proceeds from left to right and can be followed by monitoring the cell surface expression pattern of CD33, CD36, and glycophorin-A (40). (Lower) Flow cytometric analysis of TLS-ERG- and NEO-transduced cells in liquid culture in the presence of G418 (1 mg/ml) after 20 days under conditions that promote erythroid differentiation. (B) Percentage of CD33-expressing cells during erythropoiesis culture.
DISCUSSION
To provide insight into human leukemogenesis, we have established an experimental system where the biological consequences of putative leukemogenic genes can be assessed following their expression in normal human hematopoietic cells. By transducing human umbilical cord blood cells with TLS-ERG, a myeloid leukemia-associated fusion gene, we have demonstrated that a leukemogenic program can be initiated in normal human hematopoietic cells. This program, which altered both the proliferation and differentiation of lineage-committed progenitors, was characterized by a marked increase in the frequency, self-renewal, and proliferation of granulocyte/macrophage progenitors and an impairment of erythroid differentiation. Remarkably, TLS-ERG alone was responsible for initiating these diverse biological effects because the abnormal phenotype was evident within the first 7 to 14 days of methylcellulose culture, making it unlikely that additional genetic alterations had accumulated. This result is consistent with the clinical observation that half of all TLS-ERG patients carry t(16;21)(p11;q22) as the sole gross chromosomal abnormality (18, 19). Although there was heterogeneity in the extent of proliferation and self-renewal among different progenitors (80% gave rise to a range of 50 to 900 secondary colonies and growth of transduced CFU-GM in liquid culture was possible for up to 10 weeks), TLS-ERG expression caused a large proportion of transduced progenitors to exhibit an abnormal phenotype, indicating that TLS-ERG was very potent. Differences in TLS-ERG expression levels or in the nature of the target cells that were transduced may account for the heterogeneity. Further support for the leukemogenic potency of TLS-ERG was evident from its ability to markedly decrease the frequency of transduced erythroid colonies to 2% and to prevent the appearance of differentiated CD36+ or gly-A+ erythroid cells following the incubation of TLS-ERG-infected CD34+Lin− cells in a liquid culture assay for erythroid differentiation. These experiments indicate that TLS-ERG can either induce a block in erythroid development at or prior to the BFU-E stage or impair the survival of these cells. Although TLS-ERG affected the growth and differentiation of primary human progenitor cells, we were unable to determine its effect on primitive repopulating cells because of their resistance to retroviral infection (36). In addition, it is clear that expression of TLS-ERG alone did not induce complete immortalization because transduced cells could not proliferate permanently in culture or in NOD/SCID mice (data not shown).
Recently, we have demonstrated that human AML stem cells, as assayed by initiation of leukemia in NOD/SCID mice (37), originate from primitive human hematopoietic cells rather than committed progenitors for both blastic and myelomonocytic subtypes (38). This finding, together with the clinical observation that the erythroid lineage is often not part of the leukemic clone in AML and many subtypes of MDS, supports the hypothesis that one effect of the human leukemogenic process in these myeloid disorders is to prevent normal differentiation along other lineages including the erythroid lineage (39). Thus, the arrested erythroid development we observed in TLS-ERG-transduced cells provides the basis for experimental validation of this hypothesis.
In closing, the experimental system we have described provides an in vitro approach to assay the biological function of putative leukemogenic genes, either singly or in combination, in normal human hematopoietic cells. This should aid in gaining a greater understanding of human leukemogenesis and in the identification of molecules that mediate or inhibit leukemogenic gene function.
Acknowledgments
We thank A. Bernstein, N. Iscove, M. Minden, and the members of our laboratory for critically reviewing this manuscript as well as B. Patterson and X.-Y. Wu for assistance with cytologic and karyotypic analyses. This work was supported by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society, the Canadian Genetic Diseases Network of the National Centers of Excellence (to J.E.D.), the Medical Research Council (to J.E.D.), an MRC Scientist award (to J.E.D.), and postdoctoral fellowships from the National Institutes of Health (1F32CA72193) (to C.Y.I.); the National Cancer Institute of Canada supported this work with funds provided by the Terry Fox Run (to D.S.P.). V.N.R. and E.S.P.R. were supported by National Institutes of Health grants.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CB, cord blood; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome; GALV, gibbon ape leukemia virus; SCF, stem cell factor; FITC, fluorescein isothiocyanate; PE, pycoerythrin; CFU-GM, granulocyte/macrophage progenitor; CFU-G, granulocyte progenitor; CFU-M, macrophage progenitor; BFU-E, blast-forming unit erythroid progenitor; RT, reverse transcription; FCS, fetal calf serum; gly-A, glycophorin-A.
References
- 1.Fearon E, Burke P, Schiffer C, Zehnbauer B, Vogelstein B. N Engl J Med. 1986;315:15–24. doi: 10.1056/NEJM198607033150103. [DOI] [PubMed] [Google Scholar]
- 2.Keinanen M, Bloomfield C, Machnicki J, del la Chapelle A. N Engl J Med. 1988;318:1153–1158. doi: 10.1056/NEJM198805053181803. [DOI] [PubMed] [Google Scholar]
- 3.Sawyers C L, Denny C T, Witte O N. Cell. 1991;64:337–350. doi: 10.1016/0092-8674(91)90643-d. [DOI] [PubMed] [Google Scholar]
- 4.Adams J M, Cory S. Cancer Surv. 1992;15:119–141. [PubMed] [Google Scholar]
- 5.Chang J M, Metcalf D, Lang R A, Gonda T J, Johnson G R. Blood. 1989;73:1487–1497. [PubMed] [Google Scholar]
- 6.Wong P M C, Chung S-W, Dunbar C E, Bodine D N, Ruscetti S, Nienhuis A W. Mol Cell Biol. 1989;9:798–808. doi: 10.1128/mcb.9.2.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perkins A C, Cory S. EMBO J. 1993;12:3835–3846. doi: 10.1002/j.1460-2075.1993.tb06062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawley R G, Fong A Z C, Reis M D, Zhang N M L, Hawley T S. Cancer Res. 1997;57:337–345. [PubMed] [Google Scholar]
- 9.Lavau C, Szilvassy S J, Slany R, Cleary M L. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawley R G, Fong A Z, Lu M, Hawley T S. Oncogene. 1994;9:1–12. [PubMed] [Google Scholar]
- 11.Thorsteinsdottir U, Sauvageau G, Hough M R, Dragowska W, Lansdorp P M, Lawrence H J, Largman C, Humphries R K. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowley J D. Semin Hematol. 1990;27:122–136. [PubMed] [Google Scholar]
- 13.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 14.Drexler H G, MacLeod R A F, Borkhardt A, Janssen J W G. Leukemia. 1995;9:480–500. [PubMed] [Google Scholar]
- 15.Adams J M, Cory S. Science. 1991;254:1161–1167. doi: 10.1126/science.1957168. [DOI] [PubMed] [Google Scholar]
- 16.Kamps M P, Baltimore D. Mol Cell Biol. 1993;13:351–357. doi: 10.1128/mcb.13.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zakian V A. Cell. 1997;91:1–3. doi: 10.1016/s0092-8674(01)80001-5. [DOI] [PubMed] [Google Scholar]
- 18.Hiyoshi M, Koh K-R, Yamane T, Tatsumi N. Clin Lab Haematol. 1995;17:243–246. [PubMed] [Google Scholar]
- 19.Kong X-T, Ida K, Ichikawa H, Shimizu K, Ohki M, Maseki N, Kaneko Y, Sako M, Kobayashi Y, Tojou A, Miura I, Kakuda H, Funabiki T, Horibe K, Hamaguchi H, Akiyama Y, Bessho F, Yanagisawa M, Hayashi Y. Blood. 1997;90:1192–1199. [PubMed] [Google Scholar]
- 20.Ichikawa H, Shimizu K, Hayashi Y, Ohki M. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- 21.Reddy E S P, Rao V N, Papas T S. Proc Natl Acad Sci USA. 1987;84:6131–6135. doi: 10.1073/pnas.84.17.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu K, Ichikawa H, Tojo A, Kaneko Y, Maseki N, Hayashi Y, Ohira M, Asano S, Ohki M. Proc Natl Acad Sci USA. 1993;90:10280–10284. doi: 10.1073/pnas.90.21.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hart A H, Corrick C M, Tymms M J, Hertzog P J, Kola I. Oncogene. 1995;10:1423–1430. [PubMed] [Google Scholar]
- 24.Prasad D D K, Ouchida M, Lee L, Rao V N, Reddy E S P. Oncogene. 1994;9:3717–3729. [PubMed] [Google Scholar]
- 25.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 26.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murdoch B, Pereira D S, Wu X, Dick J E, Ellis J. Gene Ther. 1997;4:744–749. doi: 10.1038/sj.gt.3300448. [DOI] [PubMed] [Google Scholar]
- 29.Lapidot T, Pflumio F, Doedens M, Murdoch B, Williams D E, Dick J E. Science. 1992;255:1137–1141. doi: 10.1126/science.1372131. [DOI] [PubMed] [Google Scholar]
- 30.Warren M, Rose W, Beall L, Cone J. Stem Cells. 1995;13:167–174. doi: 10.1002/stem.5530130208. [DOI] [PubMed] [Google Scholar]
- 31.Nakahata T, Ogawa M. J Clin Invest. 1982;70:1324–1328. doi: 10.1172/JCI110734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi H-K, Fujimura Y, Ouchida M, Prasad D D K, Rao V N, Reddy E S P. Oncogene. 1997;14:1259–1268. doi: 10.1038/sj.onc.1201099. [DOI] [PubMed] [Google Scholar]
- 33.Griffin J, Lowenberg B. Blood. 1986;68:1185–1195. [PubMed] [Google Scholar]
- 34.Hiyoshi M, Yamane T, Hirai M, Tagawa S, Hattori H, Nakao Y, Yasui Y, Koh K-R, Hino M, Tatsumi N. Br J Haematol. 1995;90:417–424. doi: 10.1111/j.1365-2141.1995.tb05168.x. [DOI] [PubMed] [Google Scholar]
- 35.Darley R L, Hoy T G, Baines P, Padua R A, Burnett A K. J Exp Med. 1997;185:1337–1347. doi: 10.1084/jem.185.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larochelle A, Vormoor J, Hanenberg H, Wang J C Y, Bhatia M, Lapidot T, Moritz T, Murdoch B, Xiao X L, Kato I, Williams D A, Dick J E. Nat Med. 1996;2:1329–1337. doi: 10.1038/nm1296-1329. [DOI] [PubMed] [Google Scholar]
- 37.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiurl M A, Dick J E. Nature (London) 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 38.Bonnet D, Dick J E. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 39.McCulloch E. Blood. 1983;62:1–13. [PubMed] [Google Scholar]
- 40.Okumura N, Tsuji K, Nakahata T. Blood. 1992;80:642–650. [PubMed] [Google Scholar]