Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer (original) (raw)
Abstract
Interferon γ (IFNγ) is the hallmark cytokine produced by T helper type 1 (Th1) cells, whereas interleukin (IL)-4 is the hallmark cytokine produced by Th2 cells. Although previous studies have revealed the roles of cytokine signaling and of transcription factors during differentiation of Th1 or Th2 cells, it is unclear how the exclusive expression pattern of each hallmark cytokine is established. The DNaseI hypersensitivity site IV within the mouse Il4 locus plays an important role in the repression of Il4 expression in Th1 cells, and it has been named the Il4 silencer. Using Cbfβ- or Runx3-deficient T cells, we show that loss of Runx complex function results in derepression of IL-4 in Th1 cells. Binding of Runx complexes to the Il4 silencer was detected in naive CD4+ T cells and Th1 cells, but not in Th2 cells. Furthermore, enforced expression of GATA-3 in Th1 cells inhibited binding of Runx complexes to the Il4 silencer. Interestingly, T cell–specific inactivation of the Cbfβ gene in mice led to elevated serum immunoglobulin E and airway infiltration. These results demonstrate critical roles of Runx complexes in regulating immune responses, at least in part, through the repression of the Il4 gene.
Upon encountering antigen, naive CD4+ Th cells differentiate into effector cell subsets that are defined by expression of distinct cytokines. Th1 cells produce IFNγ and mainly participate in cellular immune responses against intracellular pathogens, whereas Th2 cells produce IL-4, -5, and -13 and control infection with extracellular microbes (1). An inappropriate balance in Th1- and Th2-mediated responses has been proposed to be involved in various immune system disorders. For example, IL-4 and -5 are strongly implicated in atopic and allergic diseases, including asthma, through their enhancement of IgE-mediated and eosinophilic immune responses (2).
Cytokine signaling and transcription factor networks play essential roles in regulating differentiation of Th cell subsets. The transcription factors T-bet and GATA-3 are the central regulators in the induction of Th1 and Th2 differentiation, respectively (3, 4). In highly polarized Th1 and Th2 cells, each of the characteristic cytokines, IFNγ and IL-4, is reciprocally expressed. In Th1 cells, the stable repression of the Il4 gene has been ascribed to epigenetic regulation initiated by combined cis-regulatory elements (5, 6). Conserved noncoding sequences (CNSs) and DNaseI hypersensitive (HS) sites, which are often used to identify putative cis-regulatory regions, have been identified in the Il4 locus. The HS IV site is located toward the 3′ end of the Il4 locus and is well-conserved between species (7). Deletion of HS IV in the mouse genome led to increased Il4 transcription in naive CD4+ T cells and to production of IL-4 in polarized Th1 cells (7). These results identified the HS IV site as an important cis-regulatory region, the Il4 silencer, which is responsible for repressing the expression of IL-4 during differentiation of Th1 cells. To further understand the molecular mechanism of action of the Il4 silencer, it will be important to identify the key trans-acting factors.
Silencing of the Cd4 gene is another example of negative transcriptional regulation during differentiation of T lymphocytes. In thymocytes committed to differentiate toward the cytotoxic T cell lineage, the Cd4 locus is epigenetically silenced by an intronic Cd4 silencer whose function requires binding of Runx transcription factor complexes (8, 9). The Runx complexes are composed of two subunits, including one of the Runx proteins, which possess a conserved DNA-binding domain, and the unique Cbfβ protein (10). Examination of mice lacking expression of either Runx1 or Runx3 in thymocytes revealed that Runx3 plays a major role in epigenetic Cd4 silencing (9, 11). Interestingly, Runx1 was suggested to be involved in repressing Gata-3 expression during differentiation of CD4+ Th cells (12). Moreover, a transient asthma-like disease, which was characterized by infiltration of eosinophilic cells into the lung, developed in Runx3-deficient mice (13, 14). In addition, the RUNX3 locus on human chromosome 1p36 maps to a region containing susceptibility genes for asthma (15). These results suggest the involvement of Runx family members in the differentiation of CD4+ Th cells. Hence, it is important to study the function of Runx complexes in CD4+ T cell differentiation programs in mouse models.
In this study, we show that T cell–specific inactivation of the Cbfβ gene led to spontaneous development of asthma-related symptoms, including elevated serum IgE and airway infiltration. In cells cultured under Th1 differentiation conditions, derepressed IL-4 production was detected in IFNγ-producing Th1 cells in the absence of Cbfβ or Runx3 protein. Furthermore, we show that binding of Runx complexes to the Il4 silencer correlated with IL-4 repression and was antagonized by GATA-3. These results demonstrate that Runx complexes play an important role in repressing IL-4 expression during Th cell differentiation and in the regulation of immune responses.
RESULTS AND DISCUSSION
Expression of Runx1 and Runx3 proteins during Th cell differentiation
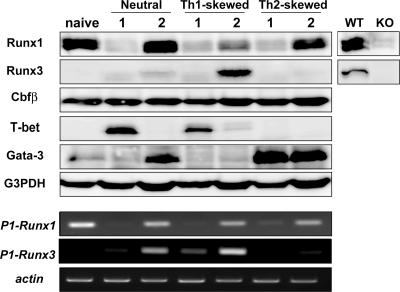
We first examined expression of Runx proteins during differentiation of CD4+ Th cells. Purified CD4+CD25−CD62L+ naive T cells were stimulated with immobilized anti-CD3 and soluble anti-CD28 antibodies. 2 d after stimulation, Runx1 protein was substantially decreased (Fig. 1). After another 4 d of culture, although expression of Runx1 protein was restored and detected in both Th1 and Th2 cells, Runx3 protein was detected almost specifically in Th1 cells (Fig. 1). Thus, both Runx1 and Runx3 proteins are expressed in polarized Th1 cells. Expression of distal (P1) promoter–derived Runx1 or Runx3 transcript was well correlated with that of Runx1 or Runx3 protein, suggesting that activation of a distal promoter is important for regulated expression of Runx proteins. Considering the redundant function of Runx1 and Runx3 in Cd4 silencing in CD8+ T cells (11), it is also possible that these two transcription factors function redundantly in CD4+ T cells. Because association with the nonredundant Cbfβ protein is essential for the function of both Runx1 and Runx3, we analyzed the effect of loss of Cbfβ on Th cell differentiation (10, 16).
Figure 1.
Expression of Runx1 and Runx3 protein during Th cell differentiation. Naive CD4+ T cells stimulated with immobilized anti-CD3 antibody and soluble anti-CD28 antibody were cultured with no additional supplement (neutral) and with specific combinations of cytokine and antibody for inducing Th1 (Th1-skewed) or Th2 (Th2-skewed) differentiation. At 2 d (lane 1) and 6 d (lane 2) after stimulation, expression of Runx1, Runx 3, Cbfβ, T-bet, and Gata-3 proteins were examined (top). (bottom) Expression of distal promoter–derived Runx1 and Runx3 transcripts are shown. Data are representative of three independent experiments.
Generation of T cell–specific, Cbfβ-deficient mice
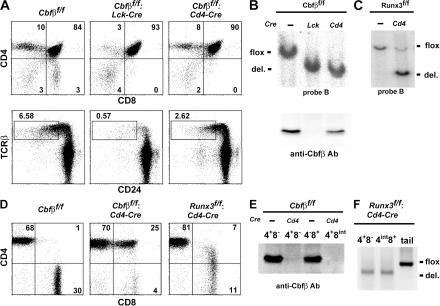
Because germline-null mutations of Cbfβ and Runx3 result in embryonic and neonatal lethality, respectively (16–18), we generated _Loxp_-flanked Cbfβfllox(Cbfβf) and Runx3flox (Runx3f) mutant alleles by gene targeting (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20062373/DC1). Mice harboring either Cbfβf or Runx3f alleles were crossed with Lck-Cre or Cd4-Cre transgenic mice, to inactivate the targeted genes at CD4−CD8− DN or CD4+CD8+ DP stages, respectively. Whereas inactivation of Runx1 at the DN stage resulted in a more than fivefold reduction in the number of total thymocytes (9), the reduction was only approximately twofold in Cbfβf/f: Lck mice, although development of mature thymocytes was severely impaired (Fig. 2 A and Fig. S2). In contrast, the number of mature thymocytes was only moderately reduced in Cbfβf/f: Cd4 mice (Fig. 2 A and Fig. S2). Although Cre-mediated recombination of the Cbfβf allele appeared to be very efficient in DP thymocytes by both Lck- and Cd4-Cre transgene, a significant amount of Cbfβ protein could be detected in those cells from the Cbfβf/f: Cd4 mice (Fig. 2 B). However, in the peripheral TCRαβ cells from Cbfβf/f: Cd4 mice, no Cbfβ protein was detected (Fig. 2 E), indicating that Cbfβ protein was gradually lost after inactivation of the gene. Similarly efficient inactivation of Runx3flox allele by Cd4-Cre transgene in thymus resulted in a loss of Runx3flox allele in peripheral T cells (Fig. 2, C and F), which is consistent with loss of Runx3 protein in CD8+ T cells from Runx3f/f: Cd4 mice (Fig. 1).
Figure 2.
Effect of stage-specific inactivation of the Cbfβ gene on differentiation of TCR+ T cells. (A) Representative FACS profile of CD4/CD8 (top) and TCRβ/CD24 (HSA; bottom) expression on total thymocytes from indicated mice. (B and C) Southern blot analyses to assess the efficiency of Cre-mediated recombination in DP thymocytes (B) and total thymocytes (C) from indicated mice are shown. Immunoblot analysis of Cbfβ protein in DP thymocytes is shown in B (bottom). (D) Representative FACS profiles of CD4/CD8 expression in TCRαβ+ lymph node cells from indicated mice. (E) Immunoblot analysis of Cbfβ expression in peripheral T cells from Cbfβf/f: Cd4 and control mice. (F) DNA-PCR analyses confirmed Cre-mediated inactivation of the Runx3 gene in peripheral mature T cell.
In peripheral lymphoid tissues from Cbfβf/f: Cd4 mice, mature TCR_αβ_ T cells consisted of two major subsets, CD4+CD8− and CD4+CD8int cells (Fig. 2 D). Perforin expression in CD4+CD8int cells was comparable to that in wild-type CD8+ T cells (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20062373/DC1), which is consistent with the CD4+CD8int phenotype resulting from the loss of Cd4 silencing in CD8+ cytotoxic-lineage cells in the absence of Cbfβ protein and Runx complexes (9).
Development of asthma-related symptoms after T cell–specific inactivation of the Cbfβ gene
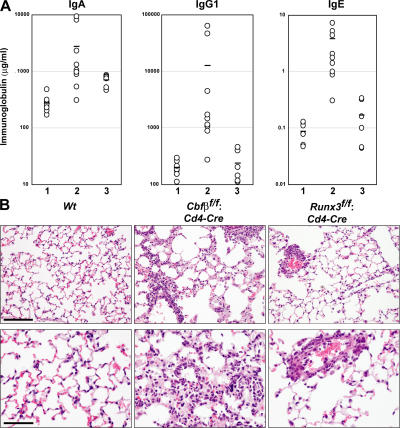
It has been shown that outbred Runx3-deficient mice develop a transient inflammatory infiltrate in their lungs and elevated serum IgE (13, 14). In Cbfβf/f: Cd4 mice, serum IgA, IgG1, and IgE titers were significantly elevated (Fig. 3 A). Numerous lymphocytes and eosinophils were found to infiltrate bronchioles, perivascular space, and alveolar septae in the lung from all Cbfβf/f: Cd4 mice examined (Fig. 3 B). Mild infiltration of lymphoid cells in the bronchioles and perivascular space, but not in alveolar septae, was also observed in about one-third of Runx3f/f: Cd4 mice (Fig. 4). Thus, T cell–specific loss of Cbfβ protein led to spontaneous development of asthma-related features, and a similar, but milder, disease developed in mice lacking Runx3 in T cells. Because such asthma-related findings are often correlated with enhanced Th2 responses, we next examined cytokine production and differentiation of CD4+ T cells in the absence of Cbfβ.
Figure 3.
Development of an asthma-like phenotype after T cell–specific inactivation of the Cbfβ gene. (A) Concentrations of serum IgA, IgG1, and IgE from 8–10-wk-old Cbfβf/f (lane 1), Cbfβf/f: Cd4 (lane 2), and Runx3f/f: Cd4 (lane 3) mice. Horizontal lines represent averages from each group. (B) Representative results of H&E-stained sections of lung from the indicated 8–10-wk-old mice are shown using low (top) and high (bottom) magnification. Lymphocytes and eosinophils infiltrate the bronchioles, perivascular space, and alveolar septa in Cbfβf/f: Cd4 mice, whereas lymphoid cells mainly infiltrate the perivascular space in Runx3f/f: Cd4-Cre mice. Bars: (B, top) 50 μm; (B, bottom) 100 μm.
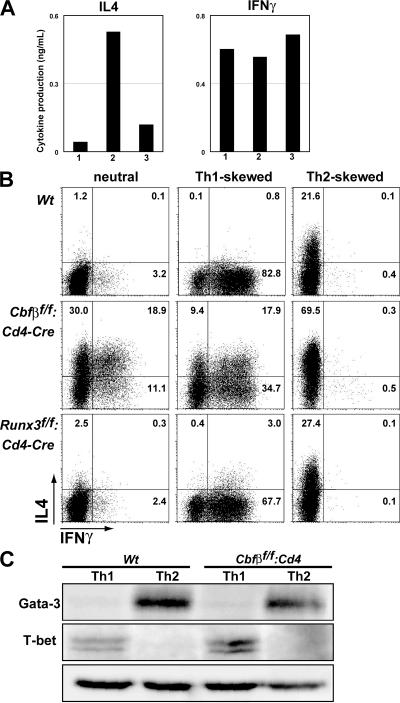
Figure 4.
Derepression of IL-4 in Th1 cells after the loss of Runx complexes. (A) Levels of secreted IL-4 and IFNγ from naive CD4+ T cells within 2 d after TCR and CD28 cross-linking. Lanes 1, 2, or 3 represents results from Cbfβf/f, Cbfβf/f: Cd4, and Runx3f/f: Cd4, respectively. Data are representative of three independent experiments. (B) Intracellular staining of IL-4 and IFNγ in restimulated cells from the indicated strains after 6 d of culture in neutral, Th1-skewed, or Th2-skewed conditions. Numbers in plots indicate the percentage of single cells in each quadrant. Data are representative of three independent experiments. (C) Immunoblot analysis of Gata-3 and T-bet expression in cells from control or Cbfβf/f: Cd4 mice cultured under Th1- or Th2-skewed condition.
Derepression of the Il4 gene in nonpolarized and Th1 cells in the absence of Runx complexes
Purified naive T cells from Cbfβf/f, Cbfβf/f: Cd4, and Runx3f/f: Cd4 mice were stimulated with anti-CD3 and -CD28 antibodies. After 48 h, the level of IL-4 secreted from Cbfβ-deficient cells was 10-fold higher than that from control cells (Fig. 4 A). After an additional 4 d of culture with IL-2, cells were analyzed by intracellular IL-4 and IFNγ staining. Consistent with the higher IL-4 production observed after 2 d, IL-4–producing cells, including IL4/IFNγ double producers, were differentiated efficiently from Cbfβ-deficient naive CD4+ T cells (Fig. 4 B). The differentiation of IFNγ-producing Th1 cells was also enhanced by Cbfβ-deficiency by yet uncharacterized mechanisms.
When Cbfβ-deficient naive CD4+ T cells were cultured under Th1 polarizing conditions, in the presence of IL-4 neutralizing antibody, cells producing both IL-4 and IFNγ were detected (Fig. 4 B). These IL-4/IFNγ double producers were, thus, differentiated independently of IL-4 signaling. In contrast, only IL-4–producing Th2 cells were differentiated under Th2-skewed conditions. These results indicate that expression of both IL-4 and IFNγ in the same cell is an outcome of IL-4 derepression in Th1 cells, rather than IFNγ derepression in Th2 cells. Previous studies showed that constitutive expression of Gata-3 induced derepression of the Il4 gene in Th1 cells (19, 20). However, we observed no difference in the level of induced T-bet or Gata-3 (Fig. 4 C), indicating that IL-4 derepression was not a consequence of dysregulation of these transcription factors.
Association of Runx complexes with the Il4 silencer
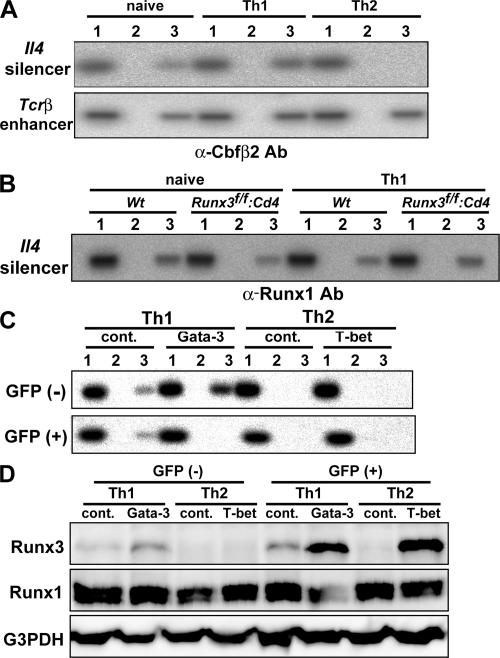
Because derepression of the Il4 gene was induced in Th1 cells upon deletion of the HS IV Il4 silencer, which contains a putative Runx recognition motif (5′-ACCRCA-3′) (7), we next examined whether Runx complexes directly associate with the Il4 silencer by chromatin immunoprecipitation (ChIP) assays. The Il4 silencer region was efficiently amplified from DNA precipitated with anti-Cbfβ2 antibody, but not with control antibody, from both naive CD4+ T cells and Th1 cells (Fig. 5 A). In sharp contrast, anti-Cbfβ2 antibody failed to precipitate the Il4 silencer from Th2 cells, although the antibody precipitated control Tcrβ enhancer, which is known to be regulated by Runx complexes, from both Th1 and Th2 cells. Thus, binding of Runx complexes is well correlated with the specificity of Il4 silencer activity.
Figure 5.
Binding of Runx complexes to the Il4 silencer in naive and Th1 cells. (A) ChIP analysis of Runx complex association with the Il4 silencer. PCR amplification was conducted with 1% of nonprecipitated input DNA (lane 1) and with DNA prepared from chromatin of naive CD4+ T cells, Th1 cells, or Th2 cells and precipitated with control (lane 2) and anti-Cbfβ2 (lane 3) antibody. Data are representative of three independent experiments. (B) ChIP analysis with anti-Runx1 antibody was performed as shown in B. Cells deficient for Runx3 protein were used to eliminate possible cross-reactivity of anti-Runx1 antibody with Runx3 protein. (C and D) Cells cultured under Th1 or Th2 skewed condition for 3 d were infected with either control vector or vector encoding Gata-3 or T-bet, respectively. Retrovirally transduced (GFP+) and nontransduced (GFP−) cells were separated and were analyzed for Runx complex binding to the Il4 silencer by ChIP analysis (C), as shown in A, and analyzed for Runx1 or Runx3 protein expression (D). Data are representative of two independent experiments.
The level of IL-4 production and the severity of asthma-related symptoms were higher in Cbfβf/f: Cd4 mice than in the Runx3f/f: Cd4 mice (Figs. 3 and Figs.4). This discrepancy suggests a compensatory function within Runx family members in the regulation of Il4 silencer activity. Indeed, Runx1 protein is expressed in naive CD4+ T cells and Th1 cells (Fig. 1). Therefore, we analyzed whether Runx1 protein binds to the Il4 silencer by using an anti-Runx1 antibody in ChIP assays. To eliminate possible cross-reactivity of the Runx1 antibody with Runx3 protein, we used Runx3-deficient cells as a control. The Il4 silencer region was precipitated from both control cells and Runx3-deficient cells by the anti-Runx1 antibody (Fig. 5 B). Thus, it is likely that Runx1 is involved in regulating Il4 silencer function, at least when Runx3 is not present.
Our results indicated that Runx complexes dissociate from the Il4 silencer in Th2 cells, despite its expression. This result suggests that there is either a mechanism that inhibits Runx binding to the Il4 silencer in Th2 cells or one that permits binding only in Th1 cells. Therefore, we examined the effect of enforced expression of Th2- and Th1-specific factors, Gata-3 and T-bet, on the binding of Runx complex to the Il4 silencer. Although Gata-3 expression in Th1 cells induced dissociation of Runx complexes from the Il4 silencer, T-bet expression in Th2 cells only induced Runx3 expression, but not Runx complex association with the Il4 silencer (Fig. 5, C and D). Furthermore, Gata-3 induced IL4 expression in polarized Th1 cells (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20062373/DC1), as previously reported (20). These results demonstrate that Gata-3 functions to inhibit binding of Runx complexes to the Il4 silencer.
Our results are consistent with the recent description of T-bet–dependent Runx3 expression in Th1 cells and Runx3 binding to the Il4 silencer (21). However, based on the expression patterns of Runx1 and Runx3 proteins, we propose that Runx1 is mainly involved in repressing the Il4 gene in naive CD4+ T cells. When Runx1 expression is reduced after encounter with antigen, newly expressed Runx3 plays a role in initiating Il4 repression in the early phase of Th1 differentiation. This would be followed by maintenance of Il4 repression by both Runx1 and Runx3 proteins. In contrast, during Th2 cell differentiation, Gata-3 induces dissociation of Runx complexes from the Il4 silencer by a yet uncharacterized mechanism and induces IL4 expression.
Collectively, our results demonstrate that loss of Runx complex function in CD4+ Th cells leads to spontaneous development of asthma-related symptoms caused by enhanced Th2 responses that are caused, at least in part, by failure of Il4 silencing. It is not documented whether Il4 silencer-deficient mice spontaneously develop similar symptoms, although impaired Th1-mediated immunity upon Leishmania major infection of these mice was reported (7). It is possible that, in addition to loss of Il4 silencing, additional mechanisms caused by loss of Runx complex function in T cells facilitate disease development. Alternatively, because an asthma-like phenotype was also attributed to loss of Runx3 function in dendritic cells (13, 14), we must consider the potential involvement of cells other than T cells in disease development. Because the human RUNX3 locus is closely linked to one of the asthma-susceptibility loci (15), dysfunction of Runx complexes may be involved in the human disease. A better understanding of Runx complex function during Th cell differentiation should provide important additional insights into the pathogenesis of allergic diseases.
MATERIALS AND METHODS
Mice.
The targeting vectors for generating Cbfβflox and Runx3flox alleles were constructed in the pL2-Neo plasmid (8), with genomic fragments containing the loxP-flanked coding exon and neoR gene inserted within the short 5′ side homology region (Fig. S1). The vector for the Cbfβ genomic fragment was obtained from S.-C. Bae (Chungbuk University, Cheongju, South Korea). Transfection into E14 ES cells was performed as previously reported (9). The Lck-Cre and Cd4-Cre transgenic mice were provided by C. Wilson (University of Washington, Seattle, WA). Mouse colonies were maintained in an animal facility in the Research Center for Allergy and Immunology RIKEN Institute, and experiments were performed according to the institutional guidelines for animal care.
Antibodies.
Anti-Runx3 antibody was provided by Y. Ito (Institute of Molecular and Cell Biology, Singapore) (22). Anti-Runx1 and -Cbfβ2 antibodies were generated by immunizing rabbits with peptides corresponding to the N terminus of the distal promoter–derived Runx1 protein and to the C-terminal end of Cbfβ2, respectively. Anti-Gata3 (HG3-31) and −T-bet (4B10) antibodies were purchased from Santa Cruz Biotechnologies, and all monoclonal antibodies used for staining cells were obtained from BD Biosciences.
Isolation and culture of naive T cells.
Naive CD4+CD25−CD62L+ T cells were sorted by flow cytometry. Differentiation of CD4+ T cells was induced as previously described (12). In brief, for inducing Th1 or Th2 differentiation, the naive cells stimulated with 2 μg/ml of immobilized anti-CD3 and 2 μg/ml of soluble anti-CD28 antibody were cultured in the presence of 5 ng/ml IL-12 and 1 μg/ml anti–IL-4 antibody or 10 ng/ml IL-4 and 1 μg/ml anti-IFNγ antibody and 1 μg/ml anti–IL-12 antibody, respectively, during the first 2 d. Cells were then maintained in the medium supplemented with 20 U/ml rIL-2 for an additional 4 d before staining for intracellular cytokines (12).
Western blot and ELISA.
Whole-cell lysates were resolved by SDS-PAGE and transferred to Hybond-P membranes (GE Healthcare). The membranes were probed with an appropriate primary antibody, and immunocomplexes were detected using ECL reagents (GE Healthcare). Cytokine and serum immunoglobulin levels were assessed by ELISA using Quantikine (R&D Systems) and Mouse Ig ELISA Quantitation kits (Bethyl), respectively.
ChIP assay.
ChIP assays were performed according to protocols provided for the ChIP Assay kit (Millipore). Chromatin DNA was fragmented by sonication to a mean length of 500 bp, and was immunoprecipitated with control, anti-Cbfβ2, or -Runx1 antibody. The precipitated DNA was subjected to PCR amplification. The primer sequences used in ChIP assay and RT-PCR are described in Fig. S5.
Retrovirus infection.
The pMigRI–T-bet and the pMX–Gata-3 vectors were provided by S.L. Reiner (University of Pennsylvania, Philadelphia, PA) and M. Kubo (RIKEN Research Center for Allergy and Immunology, Yokohama, Japan), respectively. Naive T cells were stimulated in Th1 or Th2 conditions for 3 d, infected with retroviruses, and sorted 2 d after infection for analyses.
Online supplemental material.
Fig. S1 shows the targeting strategy used to generate Cbfβflox and Runx3flox alleles. Fig. S2 shows the decreased number of total and mature thymocytes in Cbfβf/f: Lck and Cbfβf/f: Cd4 mice. Fig. S3 shows the expression of perforin in peripheral T cells from Cbfβf/f: Cd4 mice. Fig. S4 shows the induction of IL4 expression in Th1 cells by Gata-3 transduction. Fig. S5 provides primer sequences used in this study. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20062373/DC1.
Supplemental Material
[Supplemental Material Index]
Acknowledgments
We thank Chieko Tezuka for the genotyping of mice. We are grateful to Dr. Suk-Chul Bae for providing the vector containing the Cbfβ genomic fragment, and Dr. Yoshiaki Ito and Dr. Kosei Ito for providing anti-Runx3 antibody. We also thank Dr. Takeshi Egawa for his critical reading of the manuscript.
This work was supported by grants from Precursory Research for Embryonic Sciences and Technology, Japan Science and Technology Agency (I. Taniuchi), and the Sandler Program for Asthma Research (D.R. Littman).
The authors have no conflicting financial interests.
Abbreviations used: ChIP, chromatin immunoprecipitation; CNS, conserved noncoding sequence; HS, hypersensitive.
References
- 1.Reiner, S.L., and R.M. Locksley. 1995. The regulation of immunity to Leishmania major. Annu. Rev. Immunol. 13:151–177. [DOI] [PubMed] [Google Scholar]
- 2.Rudikoff, D., and M. Lebwohl. 1998. Atopic dermatitis. Lancet. 351:1715–1721. [DOI] [PubMed] [Google Scholar]
- 3.Szabo, S.J., S.T. Kim, G.L. Costa, X. Zhang, C.G. Fathman, and L.H. Glimcher. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. [DOI] [PubMed] [Google Scholar]
- 4.Zheng, W., and R.A. Flavell. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 89:587–596. [DOI] [PubMed] [Google Scholar]
- 5.Kubo, M., J. Ransom, D. Webb, Y. Hashimoto, T. Tada, and T. Nakayama. 1997. T-cell subset-specific expression of the IL-4 gene is regulated by a silencer element and STAT6. EMBO J. 16:4007–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, G.R., S.T. Kim, C.G. Spilianakis, P.E. Fields, and R.A. Flavell. 2006. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 24:369–379. [DOI] [PubMed] [Google Scholar]
- 7.Ansel, K.M., R.J. Greenwald, S. Agarwal, C.H. Bassing, S. Monticelli, J. Interlandi, I.M. Djuretic, D.U. Lee, A.H. Sharpe, F.W. Alt, and A. Rao. 2004. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 5:1251–1259. [DOI] [PubMed] [Google Scholar]
- 8.Zou, Y.R., M.J. Sunshine, I. Taniuchi, F. Hatam, N. Killeen, and D.R. Littman. 2001. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat. Genet. 29:332–336. [DOI] [PubMed] [Google Scholar]
- 9.Taniuchi, I., M. Osato, T. Egawa, M.J. Sunshine, S.C. Bae, T. Komori, Y. Ito, and D.R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. [DOI] [PubMed] [Google Scholar]
- 10.Speck, N.A. 2001. Core binding factor and its role in normal hematopoietic development. Curr. Opin. Hematol. 8:192–196. [DOI] [PubMed] [Google Scholar]
- 11.Woolf, E., C. Xiao, O. Fainaru, J. Lotem, D. Rosen, V. Negreanu, Y. Bernstein, D. Goldenberg, O. Brenner, G. Berke, et al. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA. 100:7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komine, O., K. Hayashi, W. Natsume, T. Watanabe, Y. Seki, N. Seki, R. Yagi, W. Sukzuki, H. Tamauchi, K. Hozumi, et al. 2003. The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J. Exp. Med. 198:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fainaru, O., E. Woolf, J. Lotem, M. Yarmus, O. Brenner, D. Goldenberg, V. Negreanu, Y. Bernstein, D. Levanon, S. Jung, and Y. Groner. 2004. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 23:969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fainaru, O., D. Shseyov, S. Hantisteanu, and Y. Groner. 2005. Accelerated chemokine receptor 7-mediated dendritic cell migration in Runx3 knockout mice and the spontaneous development of asthma-like disease. Proc. Natl. Acad. Sci. USA. 102:10598–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haagerup, A., T. Bjerke, P.O. Schiotz, H.G. Binderup, R. Dahl, and T.A. Kruse. 2002. Asthma and atopy - a total genome scan for susceptibility genes. Allergy. 57:680–686. [DOI] [PubMed] [Google Scholar]
- 16.Wang, Q., T. Stacy, J.D. Miller, A.F. Lewis, T.L. Gu, X. Huang, J.H. Bushweller, J.C. Bories, F.W. Alt, G. Ryan, et al. 1996. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 87:697–708. [DOI] [PubMed] [Google Scholar]
- 17.Li, Q.L., K. Ito, C. Sakakura, H. Fukamachi, K. Inoue, X.Z. Chi, K.Y. Lee, S. Nomura, C.W. Lee, S.B. Han, et al. 2002. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 109:113–124. [DOI] [PubMed] [Google Scholar]
- 18.Levanon, D., D. Bettoun, C. Harris-Cerruti, E. Woolf, V. Negreanu, R. Eilam, Y. Bernstein, D. Goldenberg, C. Xiao, M. Fliegauf, et al. 2002. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 21:3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouyang, W., S.H. Ranganath, K. Weindel, D. Bhattacharya, T.L. Murphy, W.C. Sha, and K.M. Murphy. 1998. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 9:745–755. [DOI] [PubMed] [Google Scholar]
- 20.Lee, H.J., N. Takemoto, H. Kurata, Y. Kamogawa, S. Miyatake, A. O'Garra, and N. Arai. 2000. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 192:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djuretic, I.M., D. Levanon, V. Negreanu, Y. Groner, A. Rao, and K.M. Ansel. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8:145–153. [DOI] [PubMed] [Google Scholar]
- 22.Yano, T., K. Ito, H. Fukamachi, X.Z. Chi, H.J. Wee, K. Inoue, H. Ida, P. Bouillet, A. Strasser, S.C. Bae, and Y. Ito. 2006. The RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells undergoing transforming growth factor beta-induced apoptosis. Mol. Cell. Biol. 26:4474–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]