β-Actin Messenger RNA Localization and Protein Synthesis Augment Cell Motility (original) (raw)
Abstract
In chicken embryo fibroblasts (CEFs), β-actin mRNA localizes near an actin-rich region of cytoplasm specialized for motility, the lamellipodia. This localization is mediated by isoform-specific 3′-untranslated sequences (zipcodes) and can be inhibited by antizipcode oligodeoxynucleotides (ODNs) (Kislauskis, E.H., X.-C. Zhu, and R.H. Singer. 1994. J. Cell Biol. 127: 441–451). This inhibition of β-actin mRNA localization resulted in the disruption of fibroblast polarity and, presumably, cell motility. To investigate the role of β-actin mRNA in motility, we correlated time-lapse images of moving CEFs with the distribution of β-actin mRNA in these cells. CEFs with localized β-actin mRNA moved significantly further over the same time period than did CEFs with nonlocalized mRNA. Antizipcode ODN treatment reduced this cell translocation while control ODN treatments showed no effect. The temporal relationship of β-actin mRNA localization to cell translocation was investigated using serum addition to serum-deprived cultures. β-actin mRNA was not localized in serum-deprived cells but became localized within minutes after serum addition (Latham, V.M., E.H. Kislauskis, R.H. Singer, and A.F. Ross. 1994. J. Cell Biol. 126:1211–1219). Cell translocation increased over the next 90 min, and actin synthesis likewise increased. Puromycin reduced this cell translocation and blocked this induction in cytosolic actin content. The serum induction of cell movement was also inhibited by antizipcode ODNs. These observations support the hypothesis that β-actin mRNA localization and consequent protein synthesis augment cell motility.
Most differentiated cells are structurally and functionally polarized with regard to apical–basal, anterior–posterior, or proximal–distal axes of asymmetry. For motile cells, like fibroblasts, anterior polarity is indicated by the lamellipodium, a flattened extension of the leading edge of cytoplasm highly enriched in actin. Polymerization of actin in the lamellipodia is fundamental to the process of membrane protrusion (Wang, 1985). Conversion of protrusion into cell translocation across a surface requires coordination of the cytoskeleton, adhesion, and membrane systems (Lauffenburger and Horwitz, 1996; Mitchison and Cramer, 1996). The ability to generate and maintain this functional asymmetry involves the enrichment of actin at the lamellipodia.
mRNA sorting is one mechanism to effect the enrichment of proteins asymmetrically within a cell. We have postulated that β-actin mRNA localization results in the compartmentalized synthesis of β-actin proximal to the leading edge of the fibroblast (Lawrence and Singer, 1986) and that this localization is important for the polarity and motility of the cell (Kislauskis et al., 1995). This view is supported by our results in chicken embryo fibroblasts (CEFs)1 treated with specific antisense oligodeoxynucleotides (ODNs) that delocalized β-actin mRNA and showed a loss of cell polarity (Kislauskis et al., 1994).
Maintenance of a motile morphology requires the continuous presence of serum in media. The polymerization state of actin is sensitive to serum composition of the medium (Riddle et al., 1979); addition of serum to starved cells results in rapid actin filament formation (Ridley and Hall, 1992, 1994). Analogously, β-actin mRNA is rapidly relocalized to the developing leading lamellae of CEFs (Latham et al., 1994) or pseudopods of rat muscle cells (Hill et al., 1994) upon addition of serum or serum growth factors to quiescent cells, where the mRNA is not localized. These data suggest that the same signal transduction pathways that cause actin filaments to elongate and membranes to ruffle also regulate β-actin mRNA localization (Latham et al., 1994). That β-actin mRNA sorts rapidly to the lamellipodia suggests that protein synthesis may be important in supplying actin for cell movement.
In this work, we test the hypothesis that β-actin mRNA localization serves a physiologically significant role in cell motility. The movement of individual cells and the distribution of β-actin mRNA was assessed as a function of various treatments: serum induction, antisense inhibition, and protein synthesis inhibition. The distance and direction of cell translocation was found to correlate with the distribution of β-actin mRNA and was inhibited by ODNs that delocalized β-actin mRNA. In serum-induced cells, the increase in actin protein synthesis was significant, enough to account for a doubling of the cellular actin in 15 h. An increase in cell movement accompanied this synthesis of actin. The increase in movement was inhibited by puromycin. These data support the hypothesis that β-actin mRNA localization and the consequent localized actin synthesis contribute significantly to cell movement.
Materials and Methods
Cell Culture
Primary CEFs were prepared as previously described (Kislauskis et al., 1993), cultured for 72 h, and were replated on 0.5% gelatin-coated coverslips etched with a finder grid (model CELLocate; Eppendorf, Hamburg, Germany) for a minimum of 16 h before performing a motility analysis. Each grid square was 175 μm. Standard culture medium included 10% FBS in MEM I (GIBCO BRL, Gaithersburg, MD) unless otherwise stated. Serum-starved CEFs were cultured in OPTI–MEM I or MEM I medium (GIBCO BRL) for 24 h before any described change in medium. Cycloheximide (5 μg/ml) and puromycin (200 μg/ml) were used in the inhibitor studies. Both inhibited >95% incorporation of [35S]methionine into actin relative to untreated control as evaluated by SDS-PAGE and autoradiography (data not shown).
Motility Analysis
CEFs on coverslips with the finder grids were transferred to fresh media supplemented with various ODNs or no additives. Video images were taken at the indicated intervals before and during ODN treatment using phase contrast optics on a microscope (model Microphot-SA; Nikon, Inc., Melville, NY) connected to a black and white CCD camera (Microvideo Instruments, Inc., Avon, MN) and printed with a high resolution video graphic printer on high-density paper. Change in cell position over time was plotted on transparencies overlaid and aligned with the CELLocate grid within each video frame. The magnitude and direction of cell translocation was established by measuring a change in the position of the center of each nucleus during each interval. CEFs undergoing mitosis were excluded from the analysis. Analysis of variance between treatments in the motility analyses was performed using the Turkeys-HSD test with significance level 0.05.
Phosphorothioate ODNs
Phosphorothioate-modified ODNs were synthesized (model 394 DNA synthesizer; Applied Biosystems, Inc., Foster City, CA) and purified by electrophoresis through a 20% polyacrylamide gel. Additional contaminants were removed by columns (sepack C18; Millipore, Milford, MA). The ODNs were lyophilized overnight and resuspended in diethyl pyrocarbonate-treated sterile water (Kislauskis et al., 1994). ODNs (8 μM) were included in medium containing 10% FBS, and, when appropriate, fresh media and ODNs were replaced every 4 h. Within three consecutive 4-h treatments, the phenotypic effects on lamellipodia structure, cell polarity, and actin filament distribution were evident (Kislauskis et al., 1994). The effects of specific antizipcode ODNs on steady state β-actin mRNA localization are maximal within 4–6 h of a single dose, with nearly full recovery of β-actin mRNA relocalization within 12 h (data not shown). The sequences of ODNs A–C and C+ have been previously described (Kislauskis et al., 1994). New ODNs used in this work are the control ODN Crev, which is identical to ODN C but synthesized in the reverse orientation (5′-GCATTTATGGGTTTTGTT), and B rev (5′TGTGGGTGTGGGGACACTACT), which is a control for possible nonspecific effects associated with 4Gs in ODN B (Yaswen et al., 1993; Stein and Kreig, 1995). Antisense ODN D corresponds to position 1325– 1342 in the β-actin 3′-untranslated region (stop codon position 1222) in the chicken cDNA and flanks the antizipcode sequences.
In Situ Hybridization and Microscopy
In situ hybridization was performed using nick-translated digoxigeninlabeled β-actin cDNA probes to detect endogenous β-actin mRNA as previously described (Sundell and Singer, 1990; Kislauskis et al., 1994), unless otherwise stated. Alternatively, Cy-3 fluorochrome–conjugated antisense ODN corresponding to β-actin 3′-untranslated sequences were used as probes as previously described (Kislauskis et al., 1993). Coverslips processed after alkaline phosphatase detection of in situ hybrids were mounted in GelMount aqueous/dry mounting media (Biomeda, Foster City, CA). Coverslips processed for fluorescence detection of hybridized probes were mounted in phenylenediamine (1 mg/ml) after staining with 4′,6-diamidino-2-phenylindole. Fluorescence and phase contrast microscopy were performed on a microscope (model Microphot-SA; Nikon, Inc.). mRNA was judged to be localized when most of the colored in situ signal was asymmetrically distributed in lamellae, separated from the nucleus. Signal that contacted the nucleus scored as nonlocalized.
Actin Quantitation
Approximately 0.5 × 106 CEFs were plated in 60-mm dishes and cultured in 10% FBS/MEM for 18 h. The cells were rinsed with cold Hank's buffer twice and incubated in MEM for 23.5 h, when either puromycin (200 μg/ml) or the same volume of MEM was added for 30 min. Parallel cultures were trypsinized and counted to determine cell number at the time of extraction. Each plate of CEFs was simultaneously induced with 10% FBS/ MEM, quickly rinsed with Hank's buffer, and then extracted on ice with 300 μl of cold 100 mM NaCl, 50 mM Tris, pH 7.4, 5 mM MgCl, 5 mM EDTA, and 1% Triton X-100. CEF extract was scraped from each plate, mixed by gentle inversion for 2 h in the cold, and centrifuged for 2 min at 5,000 rpm. One-quarter of the extract supernatant was electrophoresed through a standard 10% SDS-PAGE with a 3% stacking gel (National Diagnostic, Atlanta, GA) maintaining 25 mA through the stacking gel and 35 mA through the resolving gel. A range of concentrations of purified actin (No. A-0348; Sigma Chemical Co., St. Louis, MO) from 0.2–5.0 μg were loaded in the same gel as reference standards. The gel was fixed and stained overnight in 0.2% Coomassie brilliant blue, 50% EtOH, and 5% acetic acid. It was destained in 30% MeOH, 5% glycerol and soaked overnight in 25% MeOH, 5% glycerol and dried between cellophane sheets. The quantity of actin per lane was determined by scanning densitometry using ImageQuantTM Version 1.1 for Macintosh (Molecular Dynamics, Sunnyvale, CA).
Results
The Distribution of β-Actin mRNA Correlated with the Magnitude and Direction of Cell Translocation
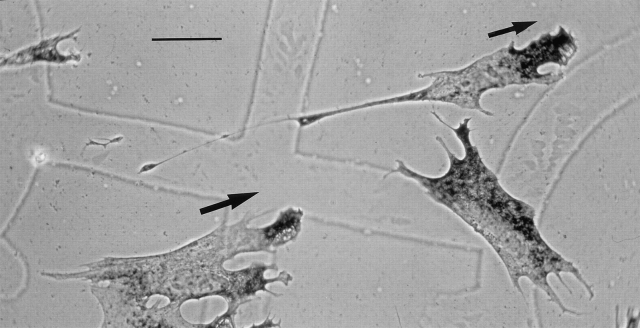
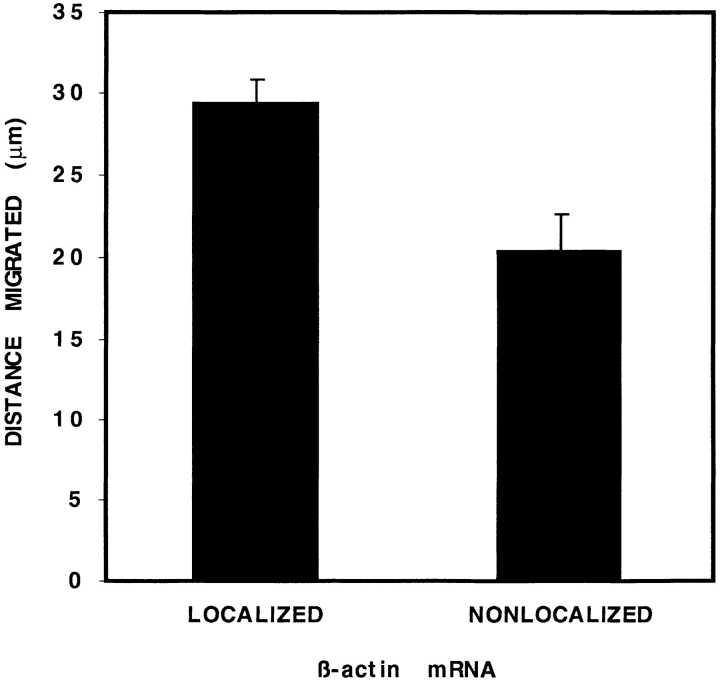
To determine whether CEF movement correlated with the distribution of β-actin mRNA, we recorded time-lapse video images of individual CEFs crawling over a finder-grid coverslip. Immediately thereafter, coverslips were fixed and subjected to in situ hybridization using probes specific to β-actin mRNA. CEFs were categorized (Fig. 1) as either “localized,” when the majority (∼80%) of mRNA signal occurred within the leading lamellae, or “nonlocalized,” when the signal was distributed throughout the cytoplasm. Each of 171 cells was evaluated and a change in position of the nucleus (μm) was determined relative to the distribution of β-actin mRNA in the same cells (Fig. 2). During the 45min interval, nearly all cells (93%) moved a measurable distance. Of these motile cells, 68% showed localized β-actin mRNA toward the leading edge in one or more cell protrusions, consistently (97%) in the direction of movement. The remainder (32%) of motile cells were categorized as having nonlocalized mRNA signal. Few nonmotile cells (24%) showed localized β-actin mRNA. Significantly, CEFs with localized mRNA translocated an average of 1.6-fold further than cells with nonlocalized mRNA (28.1 compared to 17.6 μm). This differential in motility between CEFs with localized and nonlocalized β-actin mRNA was highly significant (P ⩽ 0.0001). Thus, the distribution of β-actin mRNA correlated with the magnitude and direction of cell translocation.
Figure 1.
Direction of cell motility correlates with β-actin distribution. CEFs cultured on gelatin-coated, gridded coverslips were video recorded twice, 45 min apart, and then fixed and processed to detect β-actin mRNA by in situ hybridization. Alkaline phosphatase activity (dark intracellular staining) corresponded to the distribution of endogenous β-actin mRNA at the time of fixation. Signal is well-localized in the lamellae of two CEFs (in the direction of motility, arrows) and not in a third. Grid square is 175 μm. Bar, 30 μm.
Figure 2.
Speed correlates with localization of β-actin mRNA. CEF speed was calculated for 171 CEFs from the magnitude of the distance each CEF migrated (translocated) during the 45–60min period. Nearly all CEFs (93%) moved during that period. β-actin mRNA was localized (predominantly peripheral) in 68% of CEFs. CEFs with localized β-actin mRNA migrated 1.7-fold faster than nonlocalized. Error bars indicate the standard error of the mean for each data point.
Inhibition of β-Actin mRNA Localization Reduced Cell Motility
In our previous study, ODNs complementary to zipcode sequences, but not flanking 3′ UTR sequences, delocalized endogenous β-actin mRNA and affected cell morphology (Kislauskis et al., 1994). ODN treatments did not reduce steady-state levels of actin mRNA nor the ability of β-actin mRNA to be translated (Kislauskis et al., 1994). Hence, the effect of ODN treatment on CEF shape was proposed to have resulted from the mislocalized synthesis of β-actin throughout the cytoplasm. To extend these observations and probe the role of β-actin mRNA localization in cell motility, the same protocol was used and cell translocation evaluated over the subsequent 60-min interval (Fig. 3).
Figure 3.
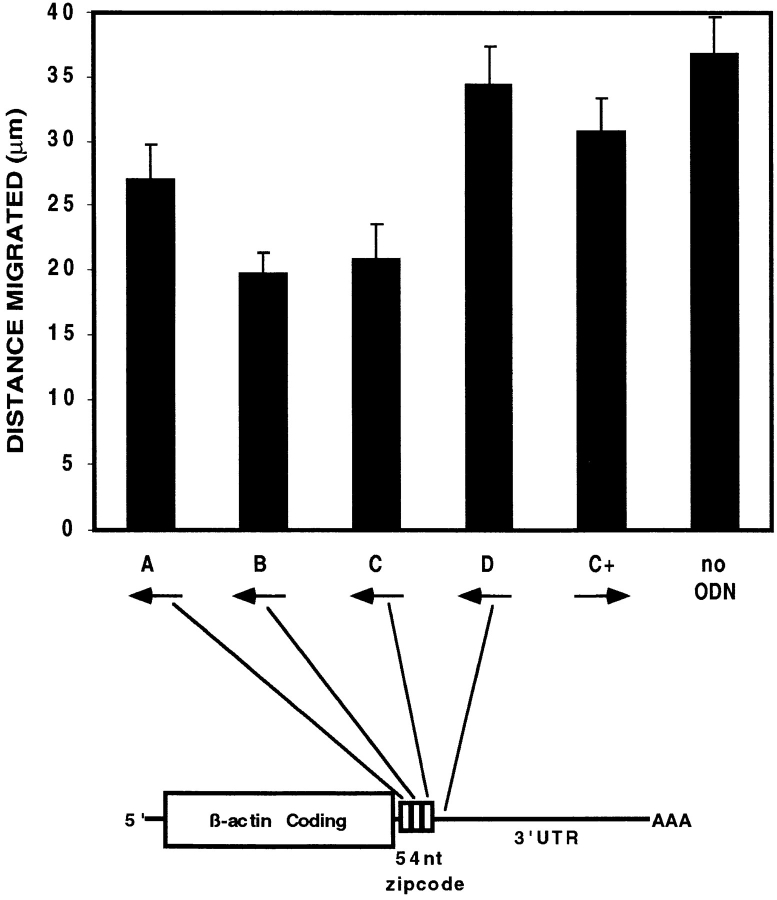
Antizipcode oligodeoxynucleotides inhibit cell motility. CEFs were treated for 12 h with various phosphorothioate ODNs corresponding to antisense (A–D), sense (C+), or no ODNs, as depicted schematically. A significant reduction in translocation over a 60-min period was observed with ODNs complementary to the middle and distal third of the 54-nucleotide zipcode, only (B or C). Error bars indicate the standard error of the mean for each data point.
ODN treatments corresponding to the middle or distal third of the zipcode (ODN B or C, respectively) had marked effects on translocation relative to untreated cells (no ODN) or control ODN treatments (C+ or D). Migration distance was reduced by 55% with ODN B (13.6 μm) and by 38% with ODN C (18.9 μm) relative to untreated CEFs (30.4 μm), while the effect of antizipcode A, corresponding to the first third of the 54-nucleotide zipcode, was not significantly different from the control ODN treatments (P > 0.05). The conclusion from this analysis was that inhibition of β-actin mRNA localization resulted in significantly reduced cell translocation.
Serum-induced Relocalization of β-Actin mRNA and Translocation Are Mediated Through the Zipcode
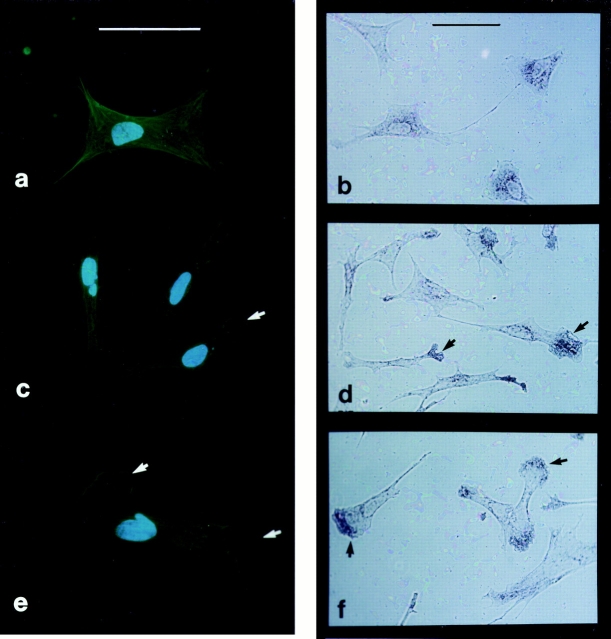
Rapid changes in the distribution of actin mRNA and protein occur within minutes of serum addition to serumstarved (24 h) CEFs (Fig. 4 a). Lamellipodia are induced by serum replacement and become enriched in phalloidinstained actin filaments (Fig. 4, c and e), and β-actin mRNA changes from nonlocalized (Fig. 4 b) to localized (Fig. 4, d and f).
Figure 4.
Serum-induced relocalization of β-actin protein and its mRNA. CEFs cultured on gelatin coverslips were serum deprived for 24 h and induced with 10% FBS and fixed after 2 and 5 min. Representative examples of the distribution of β-actin–specific FITCimmunocytochemistry (a, c, and e) and β-actin mRNA by in situ hybridization (b, d, and f) are shown. a and b, 24 h starved control; c and d, 2 min induced; and e and f, 5 min induced. Arrowheads show lamellipodia staining and membrane protrusion. Bar, 20 μm.
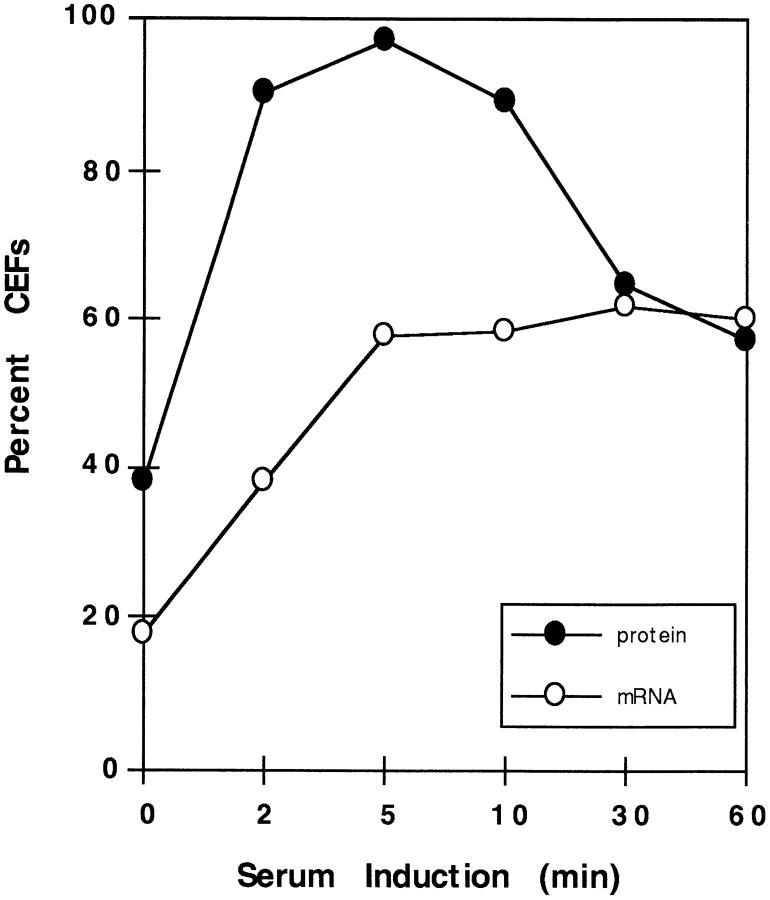
Both actin and protein localization occurred rapidly upon addition of serum (Fig. 5). Within 2 min, the percentage of CEFs with phalloidin-labeled lamellipodia increased from 38 to 92%, while those with localized β-actin mRNA approached half-maximum, from 18 to 39%. By 5 min, the percentage of CEFs with actin-rich lamellipodia peaked at 97%, while CEFs with localized mRNA reached a steadystate percentage (60%). After 1 h, the percentage of CEFs with lamellipodia declined to nearly 60%, the same percentage showing localized β-actin mRNA. Therefore, rapid relocalization of β-actin mRNA in response to serum correlated temporally with protrusion of lamellipodia, an early event in motility.
Figure 5.
The course of serum-induced relocalization of β-actin protein and its mRNA. CEFs were induced with 10% FBS after a 24 h serum starvation (Fig. 4). Coverslips with CEFs were removed 0, 2, 5, 10, 30, and 60 min after induction. The percent of CEFs is represented with localized mRNA (open circles) or with lamellipodia staining by β-actin immunocytochemistry (closed circles). The experimental mean was calculated from three separate experiments for each point except 60 min (single experiment).
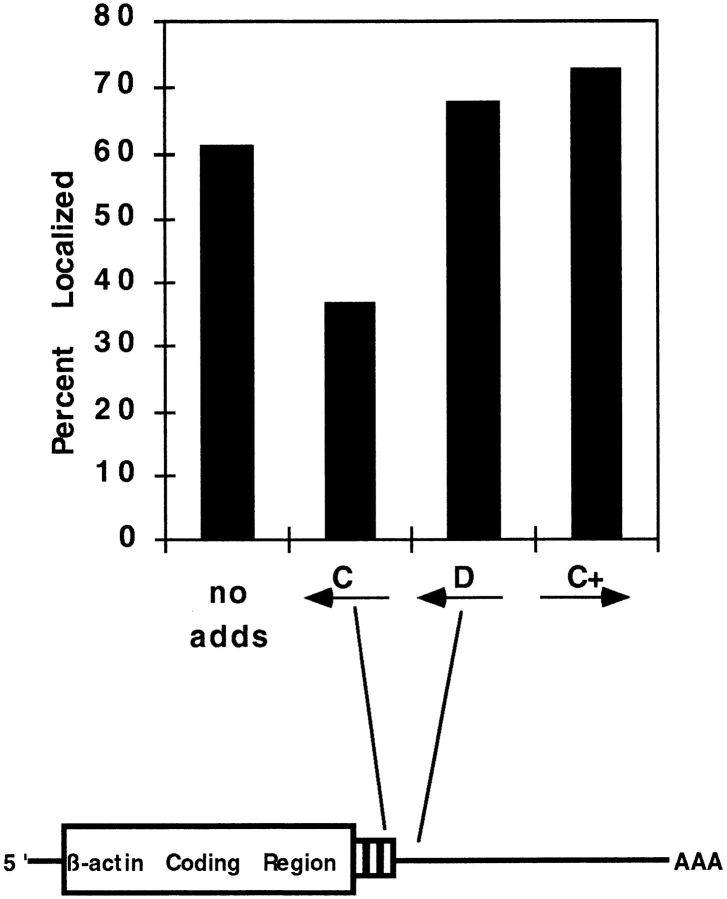
Next, we added serum and one of various ODNs simultaneously, or no additive, to serum-starved cells to evaluate the consequences of ODN treatment on the rapid induction of β-actin mRNA localization and cell translocation (Fig. 6). Because ODNs enter the cell rapidly and hybridize to their targets within minutes (Politz et al., 1995), it was possible that antisense treatment could block the transport and anchoring of β-actin mRNA in the leading lamellae. After a 30-min treatment, the CEFs were fixed and processed to detect β-actin mRNA by in situ hybridization. In this experiment, β-actin mRNA was localized in 23% of the serum-starved CEFs (Fig. 6). Within 30 min of serum treatment, 61% showed localized β-actin mRNA (Latham et al., 1994). Treatment with either antizipcode ODN B or C impeded this localization. β-Actin mRNA was localized in 37% of CEFs treated with serum containing antizipcode C, whereas localization was comparable to untreated cells after treatment with control ODNs (D = 68% and C+ = 73%, respectively). Therefore, the zipcode sequence in the 3′ UTR, which had previously been identified by other methods (Kislauskis et al., 1994), also mediated the serum-induced relocalization.
Figure 6.
Serum-induced relocalization of β-actin mRNA is mediated through the β-actin zipcode. CEFs were serum deprived for 24 h and induced for 30 min with 10% FBS containing the indicated ODNs or no ODNs. β-Actin mRNA localization in the population was evaluated by in situ hybridization as described in Materials and Methods. The effects of ODNs on the percentage of CEF with localized β-actin mRNA signal is shown. The serumstarved level of localization was 23%.
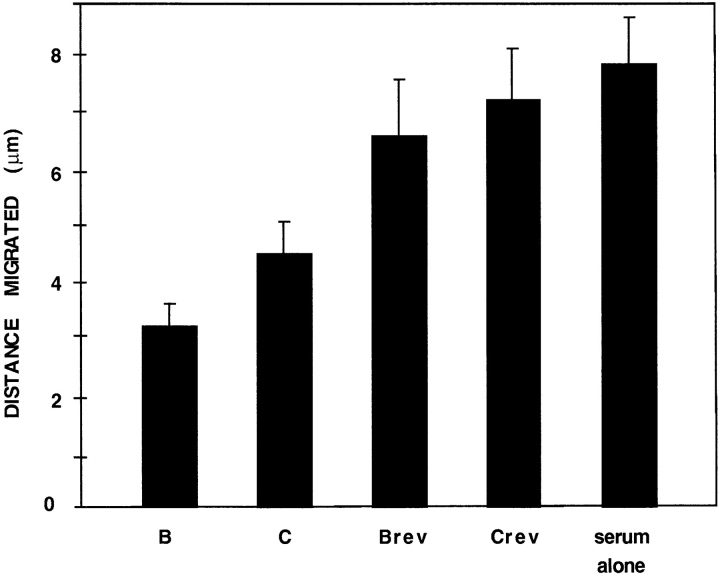
To examine whether β-actin mRNA relocalization was important for translocation, serum-starved CEFs were treated as above, in the presence or absence of antizipcode or control ODN treatments, and photographed over 90 min before being fixed and stained with FITC-phalloidin. Actin polymerization or protrusion of lamellipodia was not inhibited by ODN treatment nor was translocation after 30 min of treatment (data not shown). However, after a 90min treatment, translocation in response to serum was inhibited with ODN B and C (52 and 38%, respectively) but not the corresponding control ODNs, which had the same sequences in the reverse orientation (Fig. 7). These results indicate that the same _cis_-acting sequences (zipcodes) that regulate β-actin mRNA localization in the steady state mediate serum-induced relocalization of β-actin mRNA. Furthermore, these results suggest that β-actin mRNA localization contributed to processes in motility that followed the initial actin polymerization and protrusion of the lamellipodia.
Figure 7.
Serum induction of motility is inhibited by antisense ODNs. CEFs were serum deprived for 24 h and induced with 10% FBS containing the indicated ODN for 90 min as indicated. Motility (translocation) was measured over a 30-min interval after treatment. Similar to the results in Fig. 3, antizipcode ODN B significantly inhibited translocation, while control ODN Brev did not.
Role of Protein Synthesis in Translocation
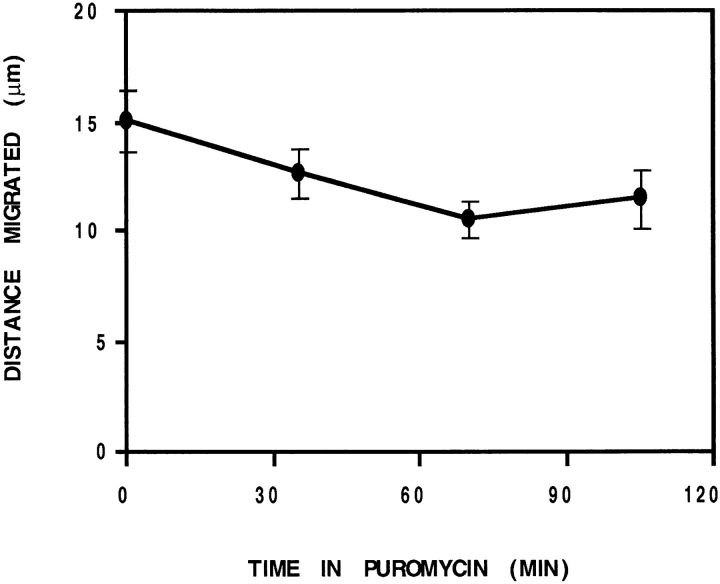
To investigate the role of protein synthesis in cell motility, CEFs were treated with an inhibitor of polysome integrity (puromycin). Analysis of translocation distance was determined within 105 min of treatment to minimize collateral effects of general inhibition of protein synthesis. In a steady-state culture (Fig. 8), translocation was suppressed by 15% after 35 min of treatment and an additional 15% over the next 35-min interval. Thereafter, translocation levels off at ∼70% of controls.
Figure 8.
Inhibition of protein synthesis suppresses motility. Analysis of translocation distance for a steady-state culture of CEFs over consecutive 30-min periods after treatment with puromycin (200 μg/ml). Control cells were evaluated before treatment (time 0). Translocation was suppressed by 30% after 70 min in puromycin.
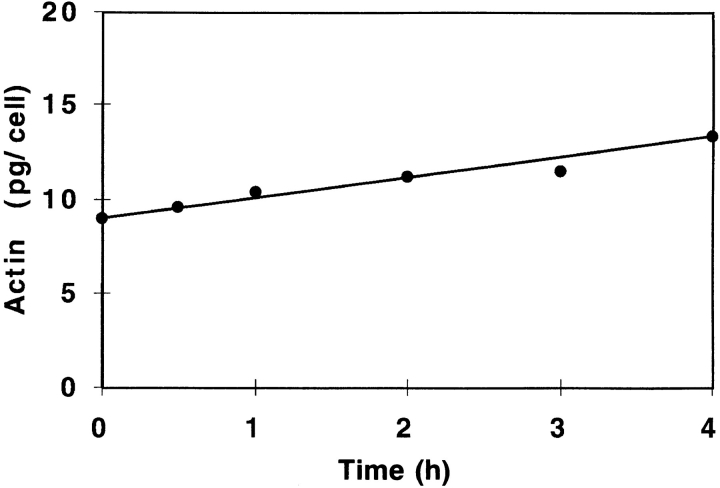
To determine whether actin synthesis could account for the increased motility in response to serum, we measured total cytosolic actin content after the addition of serum to serum-starved cells (Fig. 9). Serum-starved cells contained 9 pg actin per cell, while CEFs cultured in serum (steadystate) contained ∼18 pg. Within 30 min of serum addition, an increase in actin synthesis was detected. By 4 h, actin content increased to 13.2 pg/cell, at a rate of 1.67 pg/h. Puromycin treatment effectively blocked this induction (data not shown). These results indicate that actin protein synthesis can play a significant role in increasing the cellular actin content over time.
Figure 9.
Cytoplasmic actin content after serum induction. Line plot of the increase in actin content in serum-starved CEFs before and after the addition of serum. Coomassie-stained gels were used for quantitation.
Discussion
The results presented herein are consistent with the hypothesis that β-actin mRNA localization has a physiologically significant role in fibroblast motility. Presumably, the mRNA augments cell motility by providing synthesis of new actin monomers for polymerization at the leading edge. Several results support this interpretation. First, cells with β-actin mRNA localized in the leading lamellae moved farther than cells that did not, previous to fixation and in situ hybridization. Second, inhibition of this localization with specific antizipcode ODNs retarded translocation distances relative to controls, independent of the growth conditions of the cells. Third, serum addition to starved cells rapidly induced both mRNA localization and motility. Fourth, the amount of actin per cell increased significantly after serum stimulation. Fifth, this increase was inhibited by puromycin, as was the increase in cell motility.
Because F-actin and actin mRNA appear in the lamellipodia and leading lamellae, respectively, within 2 min of serum addition, both the actin protein and mRNA appear to sort simultaneously. That β-actin mRNA localization does not require protein synthesis (Sundell and Singer, 1990) and puromycin did not inhibit formation of the lamellipod shows that these events are independent, initially. Later (>30 min), disruption of actin mRNA localization and inhibition of protein synthesis did have an effect on cell translocation. Therefore, it is reasonable to propose that translation of localized β-actin mRNA is important to achieve maximal translocation.
Contemporary models of cell motility have not seriously considered a role for translation (Lauffenburger and Horwitz, 1996; Mitchison and Cramer, 1996). This is in part because protein synthesis inhibitors were not observed to completely prevent motility or protrusion of the lamellipodia (Spooner et al., 1971; Albrecht-Buhler, 1980). Over time, quantitation of translocation distance demonstrated that inhibition of protein synthesis resulted in a partial effect (∼50%) on motility in primary fibroblasts.
How could protein synthesis influence the dynamics of the cellular actin pool? In response to serum stimulation, protein synthesis rates have been estimated to increase fourfold. At the peak rate of synthesis, 4–6 h after serum addition, actin synthesis accounts for 15% of the total cell constituents (Riddle et al., 1979). If ribosomes are spaced 15 nucleotides apart, synthesis of one actin/s/mRNA is well within the established translation rate of a polysome/ mRNA complex, estimated to be about five amino acid residues per second (Darnell et al., 1995). This would result in the synthesis of about 150,000 actin molecules/min/cell, assuming ∼2,500 mRNAs/cell (Latham et al., 1994). We have measured the average actin content per cell to be 10.5 pg, which represents ∼1.5 × 108 actin molecules/cell. This is also the number of actin molecules calculated per cell by another independent method, based on ∼1.8 pl volume/cell and an actin concentration of 135 μM. At a rate of synthesis of 150,000 actin/min, a cell could increase its actin content by 6% per hour (0.9 × 107 actin molecules/ h). Growing CEFs divide approximately every 20 h, allowing sufficient time for the actin content to double. These calculations are consistent with the amount of actin synthesis we actually observed per cell. We observe that actin content after serum stimulation increased at the rate of 1 pg/h/cell, or ∼1.4 × 107 molecules/h, nearly a 10% increase per hour. Based on an estimated 2,500 mRNAs, this is the equivalent of 3,900 actin molecules synthesized per second, or ∼1.5 actins/s/mRNA. Most important, however, is the obvious conclusion that all the synthesis is restricted to where the mRNA is localized (i.e., the lamella). The generation of over 105 actin molecules/min in a cytoplasmic compartment that represents only a few percent of the total cell volume may have significant consequences for this region of the cell, the region most involved in cell motility (see below).
The serum induction of mRNA localization, and subsequent actin synthesis, provides the basis for a model of how translation may promote cell motility. Serum induces a burst of rapid actin polymerization in the leading lamella that precedes protrusion of the lamellipod. In mammary adenocarcinoma cells stimulated with an upshift of EGF concentration, polymerization is nucleated from severed actin filaments, and actin monomers are estimated to add to the leading edge at the rate of between 60,000 and 600,000 per second (Chan, A.Y., S. Raft, M. Bailly, J.B. Wyckoff, J.E. Segall, and J.S. Condeelis, manuscript submitted for publication), in an area within 1.5 μm of the leading edge membrane (Condeelis, 1993; Segall et al., 1996). This polymerization persists for 1 min, possibly using as much as 36 million monomers. Because the leading lamella is a minor portion of the total cytoplasm, this rapid reduction of actin monomers may deplete the local concentration, requiring either sorting of recycled actin or new synthesis. In cells moving in a gradient of EGF or spontaneously in culture, repetitive cycles of actin polymerization at the leading edge are expected. Localized actin synthesis could influence this cyclical reaction over time by augmenting the supply of free actin monomers necessary for preferential polymerization at the leading edge. This continuing, highly localized synthesis may become significant in maintaining the persistence of movement. In this model, it would follow that inhibition of mRNA localization, or of actin translation, would reduce the constant asymmetric supply of monomers, eventually affecting the translocation of the cell. The data presented here are consistent with this model since the initial protrusion events (polymerization of actin in lamellipodia) upon serum induction were unaffected by either the antizipcode ODNs or puromycin treatment (data not shown). However, after 60 min, inhibition of protein synthesis began to negatively affect motility, suggesting that the continued supply of new actin monomer replenishes the leading edge.
β-Actin mRNA localization could affect other structures that are important in motility (Farmer et al., 1983; Bershadsky et al., 1995). Focal adhesions influence locomotion by providing traction for the contractile force. Synthesis of actin (and other proteins) in association with this structure likewise may provide an anchoring point for filament elongation during protrusion. Fibroblasts, although slow in their migration, can generate very high traction forces because of their strong substrate adherence (Lauffenburger and Horwitz, 1996). The localized supply of actin monomers at the leading edge could also facilitate the synthesis and assembly of proteins involved in forming the focal adhesions.
This work has focused on the β-isoform of actin, implicated in cell motility (Hoock et al., 1991; Herman, 1993). Other actin isoforms such as α- and γ- are also components of the cytoskeleton in particular cell types (Otey et al., 1988). The ratio of the various isoforms within the cell has a profound effect on cell structure and function (Schevzov et al., 1992; Lloyd and Gunning, 1993). While each actin isoform is highly conserved in amino acid sequence, the 3′ UTRs of their respective mRNAs differ. Possibly, each isoform is synthesized in its respective cytoplasmic compartment, and protein sorting results (von Arx et al., 1995), in part, from the distribution of their respective mRNAs. mRNA localization therefore represents a spatial component of gene expression in the cytoplasm dictating where a protein will be synthesized. This level of control of gene expression could be as significant to cell structure and function as which protein is expressed.
Abbreviations used in this paper
CEF
chicken embryo fibroblasts
ODN
oligodeoxynucleotides
Footnotes
Essential conceptual contributions were made by John Condeelis (Albert Einstein College of Medicine), and critical comments on early drafts were made by Yu-Li Wang (Worcester Foundation for Biomedical Research). The authors thank Dr. Peter Quesenberry for access to his videomicroscope. Helpful comments related to this manuscript also were made by members of the laboratory: Tony Ross, Joan Politz, Krishan Taneja, and Chris Powers. We appreciate Birgit Koppetsch's skill at preparing primary CEF cultures and the secretarial help provided by Terri O'Toole. The essence of much of this work was published in abstract form (1995. Mol. Biol. Cell. 6[Suppl.]:309a). Funding for this work was provided to E.H. Kislauskis by a Muscular Dystrophy Research Grant and to R.H. Singer from National Institutes of Health (grant AR41480).
Address all correspondence to Edward H. Kislauskis, Department of Cell Biology, University of Massachusetts Medical School, 55 Lake Avenue, Worcester, MA 01655. Tel.: (508) 856-4230. Fax: (508) 856-5612. E-mail: ehk@insitu.ummed.edu
References
- Albrecht-Buhler G. Autonomous movements of cytoplasmic fragments. Proc Natl Acad Sci USA. 1980;77:6639–6643. doi: 10.1073/pnas.77.11.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershadsky AD, Gluck U, Denisenko ON, Sklyarova TV, Spector I, Ben-Ze'ev A. The state of actin assembly regulates actin and vinculin expression by a feedback loop. J Cell Sci. 1995;108:1183–1193. doi: 10.1242/jcs.108.3.1183. [DOI] [PubMed] [Google Scholar]
- Condeelis J. Life at the leading edge: the formation of cell protrusions. Annu Rev Cell Biol. 1993;9:411–444. doi: 10.1146/annurev.cb.09.110193.002211. [DOI] [PubMed] [Google Scholar]
- Darnell, J.E., H. Lodish, D. Baltimore, A. Berk, S.L. Zipursky, and P. Matsudaira. 1995. Molecular Cell Biology. W. H. Freeman, editor. Scientific American Books, Inc., New York. 129.
- Farmer SR, Wan KM, Ben-Ze'ev A, Penman S. Regulation of actin mRNA levels and translation responds to changes in cell configuration. Mol Cell Biol. 1983;3:182–189. doi: 10.1128/mcb.3.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman IM. Actin isoforms. Curr Opin Cell Biol. 1993;5:48–55. doi: 10.1016/s0955-0674(05)80007-9. [DOI] [PubMed] [Google Scholar]
- Hill MA, Schedlich L, Gunning P. Serum induced signal transduction determines the peripheral location of β-actin within the cell. J Cell Biol. 1994;126:1221–1230. doi: 10.1083/jcb.126.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoock TC, Newcomb PM, Herman IM. β-actin and its mRNA are localized at the plasma membrane and the regions of moving cytoplasm during the cellular response to injury. J Cell Biol. 1991;112:653–664. doi: 10.1083/jcb.112.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3′untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X-C, Singer RH. Sequences required for intracellular localization of β-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis, E.H., A. Ross, V.L. Latham, X.-C. Zhu, G. Bassell, K.L. Taneja, and R.H. Singer. 1995. The mechanism of mRNA localization: its effect on cell polarity. In Localized RNA. H. Lipshitz, editor. Springer, New York.
- Latham VM, Kislauskis EH, Singer RH, Ross AF. β-Actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. Intracellular localization of messenger RNA for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- Lloyd C, Gunning P. Noncoding regions of the γ-actin gene influence the impact of the gene on myoblast morphology. J Cell Biol. 1993;121:73–82. doi: 10.1083/jcb.121.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Otey CA, Kalnoski MH, Bulinski JC. Immunolocalization of muscle and nonmuscle of actins in myogenic cells and adult skeletal muscle. Cell Motil Cytoskel. 1988;9:337–348. doi: 10.1002/cm.970090406. [DOI] [PubMed] [Google Scholar]
- Politz JC, Taneja KL, Singer RH. Characterization of hybridization between synthetic oligonucleotides and RNA in living cells. Nucleic Acids Res. 1995;23:4946–4953. doi: 10.1093/nar/23.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle VGH, Dubrow R, Pardee AB. Changes in the synthesis of actin and other cell proteins after stimulation of serum-arrested cells. Proc Natl Acad Sci USA. 1979;76:1298–1302. doi: 10.1073/pnas.76.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. Signal transduction pathways regulating rhomediated stress fiber formation: Requirement for a tyrosine kinase. EMBO (Eur Mol Biol Organ) J. 1994;13:2600–2610. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Lloyd C, Gunning P. High level expression of transfected β- and γ-actin genes differentially impacts on myoblast architecture. J Cell Biol. 1992;117:775–785. doi: 10.1083/jcb.117.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall JE, Tyerech S, Boselli L, Masseling S, Helft J, Chan A, Jones J, Condeelis J. EGF stimulates lamellipod extension in metastastic mammary adenocarcinoma by an actin-dependent mechanism. Clin Exp Metastasis. 1996;14:61–72. doi: 10.1007/BF00157687. [DOI] [PubMed] [Google Scholar]
- Spooner BS, Yamada KM, Wessells NK. Microfilaments and cell locomotion. J Cell Biol. 1971;49:595–613. doi: 10.1083/jcb.49.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA, Kreig AM. Phosphorothiate oligonucleotides: antisense or anti-protein. Antisense Res Dev. 1995;5:241. doi: 10.1089/ard.1995.5.241. [DOI] [PubMed] [Google Scholar]
- Sundell CL, Singer RH. Actin mRNA localizes in the absence of protein synthesis. J Cell Biol. 1990;111:2397–2403. doi: 10.1083/jcb.111.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arx P, Bantle S, Soldati T, Perriard J-C. Dominant negative effect of cytoplasmic actin isoprotein on cardiomyocyte cytoarchitecture and function. J Cell Biol. 1995;131:1759–1773. doi: 10.1083/jcb.131.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-L. Exchange of actin subunits at the leading edge of living fibroblasts: possible role on treadmilling. J Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaswen P, Stampfer MR, Ghosh K, Cohen JS. Effects of sequence of thioated oligonucleotides on cultured human mammary epithelial cells. Antisense Res Dev. 1993;3:67–77. doi: 10.1089/ard.1993.3.67. [DOI] [PubMed] [Google Scholar]