Mutational Analysis of the Structure and Localization of the Nucleolus in the Yeast Saccharomyces cerevisiae (original) (raw)
Abstract
The nucleolus in Saccharomyces cerevisiae is a crescent-shaped structure that makes extensive contact with the nuclear envelope. In different chromosomal rDNA deletion mutants that we have analyzed, the nucleolus is not organized into a crescent structure, as determined by immunofluorescence microscopy, fluorescence in situ hybridization, and electron microscopy. A strain carrying a plasmid with a single rDNA repeat transcribed by RNA polymerase I (Pol I) contained a fragmented nucleolus distributed throughout the nucleus, primarily localized at the nuclear periphery. A strain carrying a plasmid with the 35S rRNA coding region fused to the GAL7 promoter and transcribed by Pol II contained a rounded nucleolus that often lacked extensive contact with the nuclear envelope. Ultrastructurally distinct domains were observed within the round nucleolus. A similar rounded nucleolar morphology was also observed in strains carrying the Pol I plasmid in combination with mutations that affect Pol I function. In a Pol I–defective mutant strain that carried copies of the GAL7-35S rDNA fusion gene integrated into the chromosomal rDNA locus, the nucleolus exhibited a round morphology, but was more closely associated with the nuclear envelope in the form of a bulge. Thus, both the organization of the rDNA genes and the type of polymerase involved in rDNA expression strongly influence the organization and localization of the nucleolus.
Keywords: nucleus, nucleolus, nuclear envelope, ribosomal DNA (rDNA), RNA polymerases I and II
The nucleolus is the site of ribosomal DNA (rDNA)1 transcription by RNA polymerase I (Pol I), processing of rRNA transcripts and assembly of ribosomes (for reviews see Scheer and Weisenberger, 1994; Xue and Mélèse, 1994; Shaw and Jordan, 1995). Both Pol I and rDNA are present in the nucleolus together with other nucleolar proteins and small nucleolar RNAs (snoRNAs) required for these processes. The nucleolus occupies a discrete subnuclear region and has been the subject of intensive studies by cell biologists, using a variety of higher eukaryotic cell systems. In the yeast Saccharomyces cerevisiae, the nucleolus can be seen by immunofluorescence microscopy (IFM) using antibodies against suitable nucleolar proteins or by EM, as a crescent-shaped region, occupying a substantial fraction of the nucleus along the nuclear envelope. In contrast to yeast cells, the nucleolus in higher eukaryotes does not have extensive direct contact with the nuclear envelope in most systems analyzed; the nucleolus appears to contain an intranucleolar skeleton that is contiguous (and possibly identical) with the nuclear skeleton connecting to the nuclear envelope (for reviews see Bourgeois and Hubert, 1988; Hozak, 1996). It has been proposed that the fibrillar center of the nucleolus, which contains the rDNA transcriptional machinery such as Pol I, as well as rDNA, is bound to the nucleolar skeleton (Hozak, 1996; Weipoltshammer et al., 1996). However, the morphologically defined nucleolar skeleton has not been well characterized biochemically.
It has now been established, at least for salivary gland polytene nuclei in Drosophila, that a single rRNA gene copy is sufficient to organize a (mini-) nucleolus (Karpen et al., 1988). However, it is not known how the nucleolus is localized to certain locations within the nucleus, that is, to the nuclear periphery in the case of S. cerevisiae and to the interior, presumably, by virtue of the overall organization of the nuclear matrix in higher eukaryotes. It is not known whether rDNA or a nucleolar protein(s) or the transcribed rRNA plays the primary role in determining the localization of the nucleolus. However, it is clear that the nucleolus can function normally in yeast without its normal connection to the nuclear envelope (de Beus et al., 1994).
We have previously studied the nucleolar structures of a yeast mutant in which the gene (RPA135) for the second largest subunit of Pol I is deleted and rRNA is synthesized by RNA polymerase II (Pol II) from a hybrid gene consisting of the 35S rRNA coding region fused to the GAL7 promoter (“_GAL7-35S rDNA_”) on a plasmid. Using IFM and antibodies against known nucleolar proteins, we found that the intact crescent-shaped nucleolar structure is absent in this mutant; instead several granules (termed mininucleolar bodies) that stained with these antibodies were seen in the nucleus (Oakes et al., 1993). Since the rDNA template transcribed by Pol II is carried by a plasmid, the possibility has not been excluded that the absence of the intact nucleolar structure in this particular case is due to the use of the plasmid template rather than the chromosomal rDNA repeats. Nevertheless, these observations combined with other observations on different (temperature-sensitive) Pol I mutants, which do not carry rDNA plasmids, have suggested that Pol I is important in the maintenance of the intact nucleolar structure as a structural element in addition to its functional role to produce rRNA transcripts (Oakes et al., 1993).
Significant progress has been made recently in identifying molecular components of the nucleolus and characterizing their roles in relation to nucleolar functions. With respect to the initiation of rDNA transcription in the yeast S. cerevisiae, at least four transcription factors have been identified in addition to Pol I: upstream activation factor (UAF; Keys et al., 1996), core factor (CF; Keys et al., 1994; Lalo et al., 1996; Lin et al., 1996), Rrn3p (Yamamoto et al., 1996), and TATA box–binding protein (TBP; Cormach and Struhl, 1992; Schultz et al., 1992; Steffan et al., 1996). UAF and CF are multiprotein complexes. The former contains Rrn5p, Rrn9p, and Rrn10p encoded by RRN5, RRN9, and RRN10, respectively (Keys et al., 1996), and probably three additional proteins that include histones H3 and H4 (Keener et al., 1997). The CF consists of three proteins, Rrn6p, Rrn7p, and Rrn11p, encoded by RRN6, RRN7, and RRN11, respectively. It has been demonstrated that UAF interacts directly with the upstream element of the promoter and functions, together with TBP, to recruit CF, which in turn recruits Pol I with the aid of Rrn3p (Keys et al., 1996; Steffan et al., 1996). Thus, if Pol I plays a role in organizing (and localizing) the nucleolus by its interaction with rDNA, the transcription factors which mediate this interaction might also participate in this role.
Regarding the localization of the nucleolus within the nucleus, it is possible that the tandemly repeated structure of rDNA on the chromosome, including perhaps its adjacent chromosomal DNA, might be the primary factor; for example, interactions of rDNA with the nuclear envelope or nearby structures (in yeast) or with the nucleolar skeleton (in higher eukaryotes) might determine the localization of the nucleolus. Alternatively, or in addition, some nucleolar proteins, such as Pol I as suggested from previous work (Oakes et al., 1993), might play an important role in the maintenance of the nucleolar structure and localization. To study this question, we used yeast mutants in which the chromosomal rDNA repeats were deleted mostly (Chernoff et al., 1994) or completely (Wai, H., L. Vu, and M. Nomura, unpublished experiments). Such mutant strains are able to grow by transcribing rDNA carried on a plasmid. Two kinds of plasmids were used to examine the significance of Pol I in localization of the nucleolus: one carries a single native rDNA repeat that contains the 35S rRNA coding region with the intact rDNA promoter and the 5S rRNA gene (“Pol I rDNA plasmid”); the other carries the GAL7-35S rDNA fusion gene and the 5S rRNA gene (“Pol II rDNA plasmid”). It should be noted that Pol I and related factors are all maintained functionally intact in both systems. Yet rRNA is synthesized exclusively by Pol II in the second system, since there is no rDNA carrying the native Pol I promoter in the strains with the Pol II rDNA plasmids. With the strains carrying the Pol I rDNA plasmid, which is transcribed by the Pol I transcriptional machinery, the plasmid template and nucleolar proteins were detected mostly along the nuclear envelope. That is, the nucleolus (mininucleoli in this case) is predominantly localized to the nuclear periphery, as in the case of the wild-type yeast cells. In contrast, in the strains growing by virtue of the Pol II rDNA plasmid, a round nucleolus with a limited contact with the nuclear envelope was observed. These results as well as other results obtained for some other yeast mutants are presented in this paper, and we discuss factors responsible for the organization and localization of the nucleolus in this organism.
Materials and Methods
Materials
The rabbit anti-A190 antibody used in this work was described previously (Wittekind et al., 1990). Antibodies against Ssb1p were provided by J. Broach (Princeton University, Princeton, NJ). The goat anti–rabbit IgG– fluorescein conjugate (FITC), goat anti–mouse IgG–rhodamine conjugate (TRITC), and horse serum were purchased from Sigma Chemical Co. (St. Louis, MO). All other chemical reagents were from Fisher Scientific (Fairlawn, NJ), or J.T. Baker Chemical Co. (Phillipsburg, NJ). BioNick labeling system was purchased from GIBCO BRL (Gaithersburg, MD). Biotinylated anti–avidin D and fluorescein avidin DCS were purchased from Vector Laboratories (Burlingame, CA).
Media, Strains, and Plasmids
YEPD medium contains 1% yeast extract, 2% bacto peptone (Difco Laboratories, Inc., Detroit, MI) and 2% d-glucose. YEP–galactose medium is the same, except that 2% d-galactose is substituted for d-glucose. Synthetic glucose (SGlu) medium (2% d-glucose, 0.67% yeast nitrogen base) (Difco Laboratories, Inc.) was supplemented with l-tryptophan and required bases as described by Sherman et al. (1986). Synthetic galactose medium (SGal) is the same as SGlu but 2% d-galactose is substituted for glucose. For making solid medium, 2% agar was added.
The yeast strains and plasmids used in this study are described in Table I (see also Table II). All genetic and cloning techniques were standard procedures (Sherman et al., 1986; Guthrie and Fink, 1991). NOY758 was constructed based on the method of Chernoff et al. (1994) in the following way. Control strain NOY505 was first transformed with pRDN-hyg1 using URA3 for selection. Plasmid pRDN-hyg1 is a 2μ plasmid carrying, in addition to URA3, an rDNA locus (RDN) with a recessive hygromycin-resistant mutation in the 18S rRNA coding region (Chernoff et al., 1994), and was a gift from Drs. Y.O. Chernoff and S.W. Liebman (University of Illinois, Chicago, IL). The transformants were directly plated on YEP–galactose medium containing 300 μg/ml hygromycin (Calbiochem-Novabiochem, La Jolla, CA). Several hygromycin-resistant mutants were isolated, grown in the absence of hygromycin repeatedly, and then tested for their hygromycin resistance. One of the stably hygromycin-resistant mutants was kept as NOY758, and deletion of most of the chromosomal rDNA repeats was confirmed by Southern analysis. The number of residual rDNA copies was estimated to be ∼5% or less relative to those of the control strain (NOY505). It is expected, as was observed, that one or a few residual copies must be present in this strain, since deletion of the chromosomal rDNA copies (hygromycin-sensitive allele) is based on unequal homologous recombination between rDNA repeats.
Table I.
Yeast Strains and Plasmids Used
| Designation | Description |
|---|---|
| Strains | |
| NOY505 | _MAT_a ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 (Nogi et al., 1993) |
| NOY758 | _MAT_a ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔ, pRDN-hyg1 |
| NOY759 | _MAT_a ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔ, pNOY353 |
| NOY777 | _MAT_a ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔ rpa12Δ::LEU2, pRDN-hyg1 |
| NOY770 | _MAT_a ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3, pRDN-hyg1 |
| NOY773 | _MAT_a ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3, pNOY353 |
| NOY780 | _MAT_a ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 rpa12Δ::LEU2, pRDN-hyg1 |
| NOY408-1a | _MAT_α ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135Δ::LEU2, pNOY102 (Nogi et al., 1991) |
| YJV100 | _MAT_α ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135Δ::LEU2 rdnΔ::pGRIM (Venema et al., 1995) |
| Plasmids | |
| pRDN-hyg1 | 2μ plasmid carrying RDN-hyg1; rdn-hyg1 rdn-ani1 leu2-d URA3 ampr (Chernoff et al., 1994) |
| pNOY102 | 2μ plasmid carrying GAL7-35S rDNA URA3 ampr (Nogi et al., 1991) |
| pNOY353 | 2μ carrying GAL7-35S rDNA 5S rDNA TRP1 ampr |
| pGRIM103 | A pUC19 derivative carrying GAL7-35S rDNA TRP1-d (Venema et al., 1995) |
Table II.
Characteristics of Yeast Strains Studied and Their Nucleolar Morphology
| rDNA | Nucleolar morphology (Fig. 7) | |||
|---|---|---|---|---|
| Strains | Chromosomal | Plasmid* | Pol I | |
| NOY505 | WT | — | WT | a |
| NOY758 | rdnΔ | p-Pol I | WT | b |
| NOY759 | rdnΔ | p-Pol II | WT | c |
| NOY777 | rdnΔ | p-Pol I | rpa12Δ(ts) | c |
| NOY770 | rdnΔΔ | p-Pol I | WT | b |
| NOY773 | rdnΔΔ | p-Pol II | WT | c |
| NOY780 | rdnΔΔ | p-Pol I | rpa12Δ(ts) | c |
| NOY408-1a | WT‡ | p-Pol II (−5S) | rpa135Δ | d |
| YJV100 | p-Pol II (−5S) integrated§ | — | rpa135Δ | e |
NOY759 was constructed by transforming NOY758 with pNOY353 and growing on SGal plates containing 5-fluoroorotic acid (5-FOA) (1 mg/ ml) to select cells that had lost pRDN-hyg1. NOY777 was constructed from NOY759 by disrupting RPA12 with LEU2 as was done previously (Nogi et al., 1993) followed by introduction of pRDN-hyg1 by transformation and subsequent screening for loss of pNOY353 after growth in the presence of l-tryptophan. NOY770 was constructed from NOY758 by a standard gene replacement method, which used the known sequences flanking the chromosomal rDNA repeats and replaced any rDNA repeats remaining in NOY758 by HIS3. (This replacement has a deletion of 297 bp of non-rDNA at the centromere-proximal side of the rDNA repeats. Non-rDNA at the telomere-proximal side of the rDNA repeat has four 3,652-bp repeats, each of which contains ASP3, an uncharacterized open reading frame and a single copy 5S rRNA gene, immediately adjacent to the rDNA repeats. The replacement procedure has deleted the first repeat and 2,908 bp of the second repeat. Details of the method and characterization of the strain will be reported elsewhere.) NOY773 was constructed by transforming NOY770 with pNOY353 and plating on SGal plates containing 5-FOA to select cells that had lost pRDN-hyg1. NOY780 was constructed by disrupting RPA12 with LEU2 in NOY773, transforming with pRDN-hyg1 and then screening for loss of pNOY353 after growth in the presence of l-tryptophan. Construction of NOY408-1a has been previously described (Nogi et al., 1991). Strain YJV100 (Venema et al., 1995) was a gift from Dr. J. Venema (Vrjie University, Amsterdam, The Netherlands). This strain is a derivative of NOY408-1a and carries the fusion gene, GAL7-35S rDNA, integrated into chromosomal rDNA repeats by a high copy integrative system. The copy number of the integrated fusion gene (and other vector genes) was estimated to be 20–25 and was expected to be tandemly arrayed from the mode of integration and amplification. Approximately equal numbers of the rDNA copies were also shown to be present (Venema et al., 1995; see also Tables I and II.
pNOY353 carries the 7,547-bp BamHI–XhoI fragment, which contains GAL7-35S rDNA (the GAL7 promoter fused to the 35S rRNA coding region) inserted between BamHI and SalI sites of pTV3, a TRP1, ARS, 2μ plasmid vector (Rose and Broach, 1991). This plasmid also contains the 1,085-bp PvuII–EcoRV fragment carrying the 5S rRNA gene inserted in the SmaI site upstream of the GAL7 promoter. Plasmid pNOY102 has been described previously (Nogi et al., 1991).
IFM and Fluorescence In Situ Hybridization (FISH)
Yeast strains were grown in YEP–galactose liquid medium at 25°C to an A 600 of between 0.1 and 0.3. Cells were fixed in 3.7% formaldehyde and processed for indirect immunofluorescence microscopy as described previously (Oakes et al., 1993). Cells were then stained with a 1:500 dilution of rabbit IgG solution containing anti–yeast Pol I A190 subunit and a 1:1,000 dilution of mouse YN2Cl serum containing anti-Ssb1p. The anti-A190 staining was revealed by a 1:2,000 dilution of goat anti–rabbit IgG– FITC conjugate. The anti-Ssb1p staining was revealed by a 1:2,000 dilution of goat anti–mouse-TRITC conjugate. DNA was stained with DAPI (4′6-diamidino-2-phenylindole). The protocol used for FISH was as described (Guacci et al., 1994; Castano et al., 1996). Plasmids pRDN-hyg1 and pNOY353 were used as probes to detect rDNA. The DNA preparations were digested with restriction enzymes followed by biotinylation using the BioNick labeling system. Hybridized probes were detected by successive incubations in FITC–avidin (5 μg/ml), biotinylated anti-avidin (5 μg/ml), and finally FITC–avidin (5 μg/ml). IFM and FISH were performed with a Zeiss Axioskop or Axioplan (Carl Zeiss Inc., Oberkochen, Germany) equipped with a SenSys camera (Photometrics, Tucson, AZ), using filters for fluorescein, rhodamine, and UV detection. Pictures were taken digitally or with Kodak T-Max ASA 400 black and white film. Black and white negatives were scanned into Photoshop (Adobe System Corp., Mountain View, CA) using a slide scanner (Polaroid Sprintscan 35; Polaroid, Pennfield, NY). Digital images were pseudocolored and superimposed.
Electron Microscopy
Starting with a fresh patch of cells, yeast strains were grown in YEPD or YEP-galactose medium to an A 600 value of ∼0.5 and embedded in Spurr's epoxy resin as described (Byers and Goetsch, 1991) with the following modifications. After fixation, cells were incubated in pretreatment solution (Byers and Goetsch, 1991) for 15 min at ∼25°C. Removal of cell walls was done with 0.5 mg of Zymolyase 100T (ICN Biomedicals, Costa Mesa, CA) per A 600 unit of cells for 1–2 h at ∼25°C. Sections were post-stained with 1% uranyl acetate and lead citrate using standard methods. Photomicrographs were taken on a JEOL 100CX electron microscope.
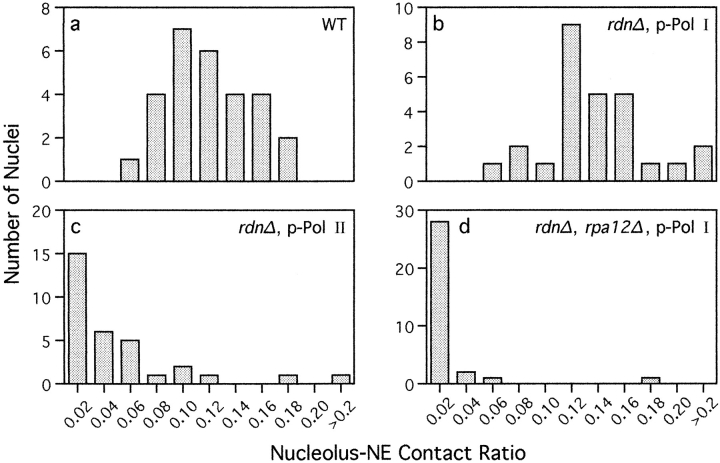
Morphometric analyses was performed using a digital planimeter. EM negatives were selected solely on the basis of exhibiting sufficient contrast to visualize the nucleolus and nuclear envelope. After 5–10-fold enlargement, prints were marked as follows: the perimeter of the nucleolus in contact with the nuclear envelope with red, and the perimeter not in contact with blue. The linear distance of contact with the nuclear envelope (length of red trace) was divided by the area of the nucleolus (determined from red plus blue traces) to arrive at the “nucleolus–nuclear envelope contact ratio.” Data were collected on a per nucleus basis. In cells that contained nucleolar granules not associated with the nucleolus, the data for areas not connected and connected were summed, so that for each nucleus the total linear distance of contact was divided by the total nucleolar area. A total of 119 nuclei from four strains were analyzed (see Fig. 4).
Figure 4.
Morphometric analysis of the extent of contact between the nucleolus and the nuclear envelope. The nucleolus– nuclear envelope (NE) contact ratio is the linear distance of contact between the nucleolus and nuclear envelope divided by the cross-sectional area of the nucleolus (see Materials and Methods). A low value indicates less contact of the nucleolus with the nuclear envelope. The distribution of ratios is shown for NOY505 (control; panel a), NOY758 (rdnΔ, pRDN-hyg1; panel b), NOY759 (rdnΔ, pNOY353; panel c), and NOY777 (rdnΔ, rpa12Δ, pRDN-hyg1; panel d). The number of individual nuclei analyzed from each strain was (n =) 28, 27, 32, and 32, respectively. Panels a–d correspond to a–d in Fig. 3.
Results
IFM and FISH Analyses of Nucleolar Structures in Yeast Mutants in Which the Chromosomal rDNA Repeats Are Deleted
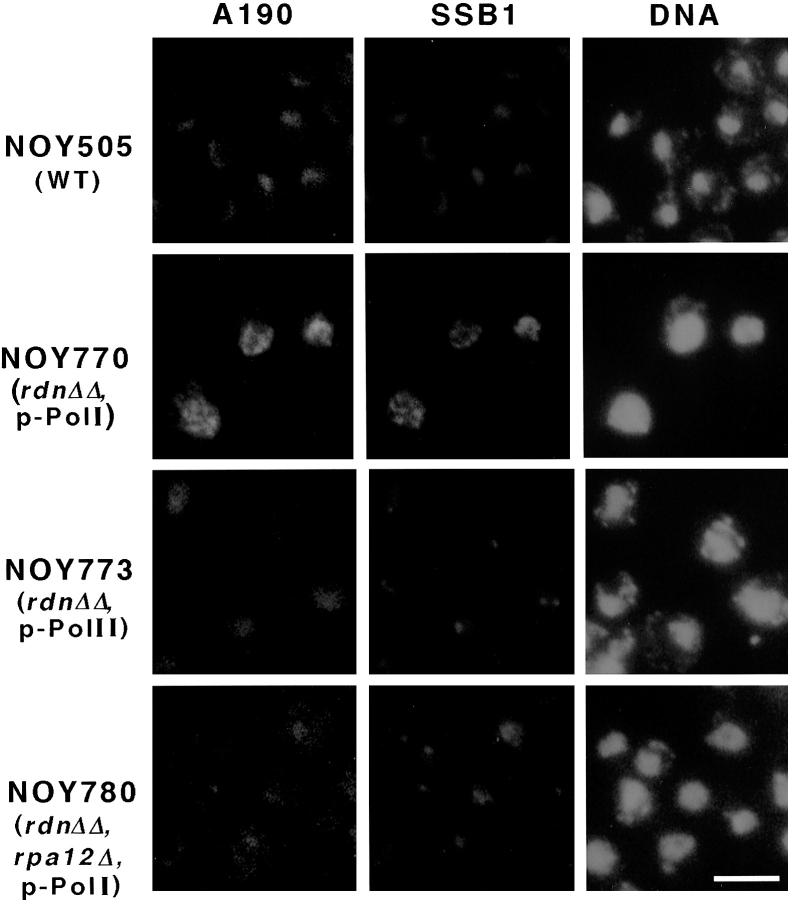
We initially constructed yeast mutants (“_rdnΔ_”) in which most of the chromosomal rDNA repeats were deleted according to the method described by Chernoff et al. (1994; see Materials and Methods). Two such rdnΔ mutant strains were constructed (see Table II): one strain (NOY758) carries a single native rDNA copy on a plasmid (pRDN-hyg1); another strain (NOY759) carries a 35S rRNA coding region fused to the GAL7 promoter (“_GAL7- 35S rDNA_”) together with the native 5S rRNA gene on a plasmid (pNOY353). Nucleolar structures in these two strains were studied by IFM using antibodies against the largest A190 subunit of Pol I and those against the nucleolar protein Ssb1p (Clark et al., 1990), and compared with the nucleolar structure of the parent strain (NOY505) without an rdn deletion. We observed that in NOY758, which carries the Pol I rDNA plasmid, both Pol I and Ssb1p were detected as several fluorescent foci mostly along the nuclear envelope. In contrast, in NOY759, which carries the Pol II rDNA plasmid, Ssb1p was seen as a single and occasionally two (but rarely more) fluorescent foci that were present with minimal contact with the nuclear envelope (data not shown; see Fig. 1 and below). (It should be noted that the Pol II plasmid used here [pNOY353] carries the 5S rRNA gene in addition to the GAL7-35S rDNA fusion gene to complement the chromosomal rdn deletion, and is different from plasmid pNOY102. The latter plasmid, which carries the GAL7-35S rDNA gene but not the 5S rRNA gene, was used to allow the growth of strains that are defective for Pol I [Nogi et al., 1991; Oakes et al., 1993; see later sections].)
Figure 1.
IFM analysis of nucleolar proteins in yeast mutants in which the chromosomal rDNA repeats are deleted. Yeast strains NOY505 (control), NOY770 (rdnΔΔ, pRDN-hyg1), NOY773 (rdnΔΔ, pNOY353) and NOY780 (rdnΔΔ, rpa12Δ, pRDN-hyg1) were analyzed for the Pol I A190 subunit, nucleolar protein Ssb1p, and DNA as described in Materials and Methods. Bar, 5 μm.
We carried out similar analyses using a corresponding pair in which the chromosomal rDNA repeats are completely deleted by the use of a standard gene replacement technique starting from a rdnΔ strain used in the above experiments. These complete deletion strains (“_rdnΔΔ_” strains) use the same plasmid systems (Table II): one strain (NOY770) carries the Pol I rDNA plasmid (pRDN-hyg1) and the other strain (NOY773) carries the Pol II rDNA plasmid (pNOY353). Fig. 1 shows the results of IFM analysis to localize Pol I and the nucleolar protein Ssb1p in the rdnΔΔ strains and the control strain (NOY505) without the rDNA deletion. It is to be noted that all the strains were grown in the galactose medium (to allow for the growth of the strains carrying the Pol II rDNA plasmid) and at 25°C (to allow the growth of temperature-sensitive mutants, described below). The control strain (NOY505) showed localization of both A190 and Ssb1p at the nuclear periphery in the form of a typical crescent-shaped nucleolar structure. NOY770, which uses the Pol I rDNA plasmid, showed a punctate pattern often at the nuclear periphery for both A190 and Ssb1p, and the two proteins appeared to be colocalized. In contrast, NOY773, which uses the Pol II rDNA plasmid, showed a single and occasionally two (but rarely more) foci for Ssb1p. However, anti-A190 antibodies showed a weak staining of most of the area of the nucleus and did not colocalize with Ssb1p. (The Pol I localization in this strain will be discussed further below.)
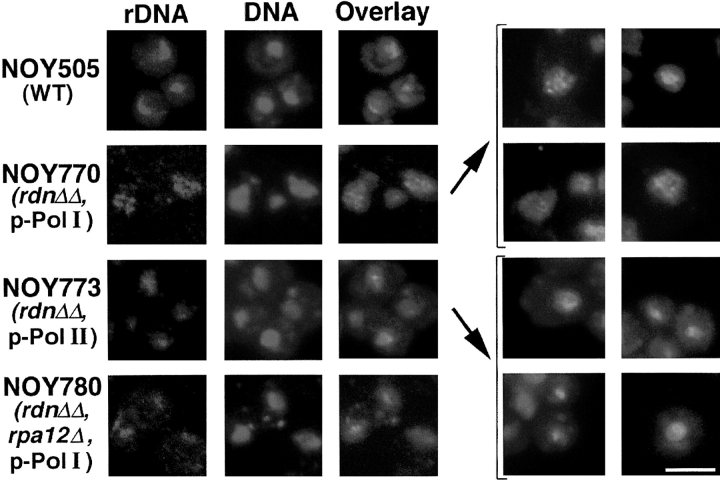
To examine localization of the plasmids carrying rDNA genes in these strains, FISH analysis was carried out using the corresponding plasmid DNAs as hybridization probes (see Materials and Methods). The results are shown in Fig. 2. Samples of the control strain (NOY505) showed staining of a crescent- or bar-shaped (or sometimes a dot-shaped) region that appeared to be at or near the nuclear periphery. NOY770, which uses the Pol I rDNA plasmid, showed punctate staining mostly at the nuclear periphery, as in the case of IFM analysis of A190 and Ssb1p. NOY773, which uses the Pol II rDNA plasmid, showed one or a few foci without extensive contact with the nuclear envelope, a pattern similar to that observed for Ssb1p by IFM.
Figure 2.
FISH analysis of rDNA in yeast mutants in which chromosomal rDNA repeats are deleted. Yeast strains NOY505 (control), NOY770 (rdnΔΔ, pRDN-hyg1), NOY773 (rdnΔΔ, pNOY353) and NOY780 (rdnΔΔ, rpa12Δ, pRDN-hyg1) were analyzed for rDNA and DNA as described in Materials and Methods. Additional images of NOY770 and NOY773 are indicated in brackets. Images of rDNA and DNA were pseudocolored red and blue, respectively. Individual images were overlayed and regions of colocalization appear as pink staining. Bar, 5 μm.
We have not analyzed localization of plasmid DNA (by FISH) and that of nucleolar proteins (by IFM) simultaneously for the same cells. Nevertheless, combining the results shown in Figs. 1 and 2, we conclude that in chromosomal rDNA deletion strain NOY770 (and NOY758), the Pol I rDNA plasmid, Pol I (and presumably other proteins required for transcription), and Ssb1p (and presumably other nucleolar components required for rRNA processing and modifications) are all colocalized mostly at the nuclear periphery, forming many mini-nucleoli. In contrast, in chromosomal rDNA deletion strain NOY773 (and NOY759), the Pol II rDNA plasmid is localized without extensive contact with the nuclear envelope. Ssb1p (and presumably other nucleolar components required for rRNA processing and modifications) is colocalized with this plasmid template, forming nucleoli that must also contain Pol II and other proteins required for transcription. The Pol I rDNA plasmid (in NOY770) and the Pol II rDNA plasmid (in NOY773) are both present at ∼90 copies per cell (Wai, H., unpublished experiments). Thus, the results of both IFM and FISH analyses indicate that many mininucleoli coalesce, forming one or a few nucleoli per cell in NOY773.
It should be noted that in strain NOY773 Pol I is present in the nucleus, but it does not localize to the “Pol II nucleolus,” nor does it localize to the nuclear periphery. We measured the cellular amount of A135, the second largest subunit of Pol I, in this strain by SDS-PAGE followed by immunoblot analysis. The amount found was comparable to that in the control wild-type strain (data not shown). In addition, extracts prepared from this strain had specific Pol I transcription activity, indicating the presence of an assembled Pol I in cell extracts (data not shown). Thus, the predominant localization of Pol I to the nuclear periphery appears to require the presence of the intact rDNA gene on the Pol I plasmid.
EM Analysis of Nucleolar Structures in Yeast Mutants in Which the Chromosomal rDNA Repeats Are Deleted
Nucleolar structures of the rdnΔ and rdnΔΔ strains described above were also studied by EM analysis of thin sections of these yeast cells (Fig. 3). Compared with the electron-dense crescent structure adjacent to the nuclear envelope in the control strain (NOY505; Fig. 3 a), the electron-dense structure corresponding to the nucleolus in the rdnΔ strain carrying the Pol II rDNA plasmid (NOY759) clearly has less contact with the nuclear envelope (Fig. 3 c). In addition, the nucleolus in this rdnΔ strain appears to be differentiated into two regions, one with greater electron density and the other with relatively less electron density. The rdnΔ strain (NOY758) carrying the Pol I rDNA plasmid showed a clearly different pattern. Here many small electron-dense foci that do not coalesce into a single nucleolar structure are seen, and some of them appear to be at or near the nuclear periphery (Fig. 3 b). The electron density of the nucleolar materials in this strain appears to be relatively uniform, and unlike the rdnΔ strain with the Pol II rDNA plasmid, no clear indication of two subregions with different electron densities was noted. We do not know whether there is any correspondence between these two regions in the strain with the Pol II rDNA plasmid (Fig. 3 c) and subnucleolar regions defined in the nucleolus of higher eukaryotes, e.g., the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC).
Figure 3.
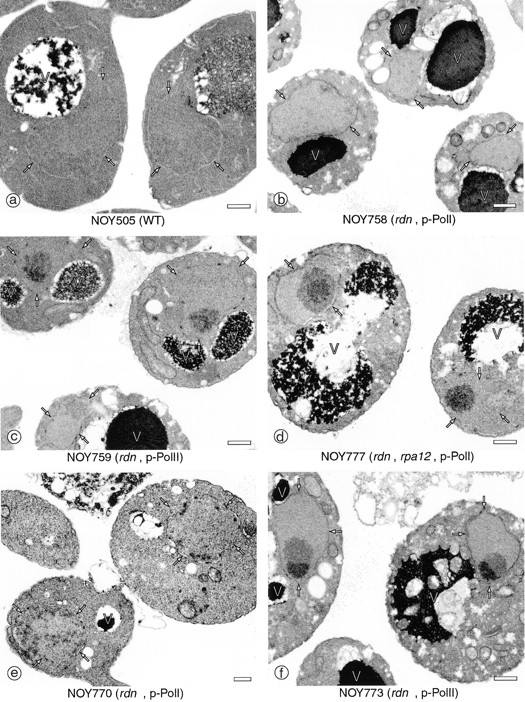
EM analysis of yeast mutants in which chromosomal rDNA repeats are deleted. Yeast strains NOY505 (control), NOY758 (rdnΔ, pRDN-hyg1), NOY759 (rdnΔ, pNOY353), NOY777 (rdnΔ, rpa12Δ, pRDN-hyg1), NOY770 (rdnΔΔ, pRDN-hyg1), and NOY773 (rdnΔΔ, pNOY353) were grown in YEP–galactose at 25°C to an A 600 of ∼0.5 and prepared for EM as described in Materials and Methods. Representative fields of cells in which nuclei and nucleoli are visible are shown. The electron-dense material representing the normal nucleolus is located at the lower portion of the two nuclei shown in a, NOY505, WT. The nuclear envelope is marked with arrows to serve as a point of reference. V, vacuole. Bars, 0.5 μm.
EM analyses of nucleolar structures were also carried out using the rdnΔΔ strains described above. The nucleolar structures and localizations are very similar to those seen for the rdnΔ pair. NOY770, which carries the Pol I rDNA plasmid, showed many small foci localized mostly at or near the nuclear periphery (Fig. 3 e), as in the case of the corresponding rdnΔ strain NOY758 (Fig. 3 b). NOY773, which carries the Pol II rDNA plasmid, showed a rounded single nucleolus that consisted of two subnucleolar regions (Fig. 3 f) as in the case of the corresponding rdnΔ strain NOY759 (Fig. 3 c).
To establish the differences between the rdnΔ strain with the Pol I rDNA plasmid and the rdnΔ strain with the Pol II rDNA plasmid with regard to the degree of contact of the nucleolus with the nuclear envelope, we measured nucleolus–nuclear envelope contact ratios as described in Materials and Methods. The ratio of the linear distance of contact of the nucleolus with the nuclear envelope to the area of the nucleolus seen on electron micrographs is defined as nucleolus–nuclear envelope contact ratio. This ratio, presented in arbitrary units, is an accurate measure of the localization of the nucleolus within the nucleus. The results are shown in Fig. 4, a–c. A comparison of the two rdnΔ strains reveals that NOY758 carrying the Pol I rDNA plasmid (Fig. 4 b) has much higher contacts with the nuclear envelope than NOY759 carrying the Pol II rDNA plasmid (Fig. 4 c). The control strain (NOY505; Fig. 4 a), which transcribes the chromosomal rDNA by Pol I, also shows much higher contact ratio than the Pol II plasmid strain (NOY759) and resembles the Pol I plasmid strain (NOY758), though it is somewhat lower than this latter strain.
After we completed the present work, Nierras et al. (1997) published a paper in which they described IFM analysis of a nucleolar protein, Nop1p, in a strain corresponding to our rdnΔ strain carrying the Pol I rDNA plasmid and stated that Nop1p is spread throughout the nucleus. However, inspection of the published picture suggests that Nop1p is perhaps localized predominantly at the nuclear periphery forming punctate nucleolar structures in at least some cells. In fact, electron microscopy of this rdnΔ/Pol I rDNA plasmid strain (L-1521; Nierras et al., 1997) revealed a nucleolus that was not localized to a single electron-dense region and appeared as numerous smaller areas, many of which were associated with the nuclear periphery (Aris, J.P., unpublished results). Thus, the nucleolar ultrastructure in this rdnΔ/Pol I rDNA plasmid strain was indistinguishable from the rdnΔ/Pol I rDNA plasmid strain NOY758 (see Fig. 3 b).
Effects of rpa12Δ Mutation on Nucleolar Localization
In both rdnΔ and rdnΔΔ strains, the nucleolar localization as well as nucleolar structure is different between the strains carrying the Pol I rDNA plasmid (pRDN-hyg1), and those carrying the Pol II rDNA plasmid (pNOY353), as described above. There are two differences between these systems. First, the machinery to transcribe rDNA is different; the former using Pol I and Pol I–specific transcription factors, whereas the latter uses Pol II and Pol II– related transcription factors. Second, the plasmid templates are different in the promoter region; the former uses the native rDNA promoter and the latter uses the GAL7 promoter, although both use the same rRNA-coding region, producing the same rRNA transcript. To determine whether the difference in polymerase is responsible for the observed difference in nucleolar localization (and structure), we introduced a rpa12Δ::LEU2 mutation in the RPA12 locus in rdnΔ (NOY758) and rdnΔΔ (NOY770) strains carrying the Pol I rDNA plasmid, yielding NOY777 and NOY780, respectively (see Table II). It has been demonstrated previously that RPA12 encoding the A12 subunit of Pol I is not an essential gene, but the rpa12Δ::LEU2 mutation causes a temperature-sensitive phenotype (Nogi et al., 1993). We grew these two rpa12Δ strains at 25°C in YEP–galactose medium for EM and IFM analyses, the same growth condition used for all other strains.
Using IFM, we found that in strain NOY780, which has the genotype rpa12Δ::LEU2 rdnΔΔ and carries the Pol I rDNA plasmid, Ssb1p is localized without extensive contact with the nuclear envelope, forming mostly a single (and occasionally two) nucleolar structure(s) that resembles that seen for the rdnΔΔ strain carrying the Pol II rDNA plasmid (NOY773). No punctate pattern at the nuclear periphery was observed for Ssb1p or Pol I (Fig. 1). As for Pol I in this strain (NOY780), IFM using anti A190 antibodies showed an apparent colocalization with Ssb1p (Fig. 1), which would be expected from transcription of rDNA by the mutant Pol I. However, the staining was weak and quite a few cells did not show a clear signal above the background. It was previously observed that in rpa12Δ strains growing at permissive temperatures, the cellular concentration of A190 was lower than in the control RPA12 strain, and that Pol I activity in extracts was also much reduced (Nogi et al., 1993). The Pol I rDNA plasmid in this strain (NOY780) was also detected by FISH as one or a few foci as was observed for the Pol II plasmid strain; no punctate pattern at the nuclear periphery was observed (Fig. 2). The same results were also obtained for the strain NOY777, which is rdnΔ rpa12Δ and carries the Pol I rDNA plasmid. NOY777 showed a localization pattern different from the isogenic RPA12 strain, NOY758, and similar to that for the rdnΔ RPA12 strain carrying the Pol II plasmid (NOY759) (data not shown). Thus, the Pol I rDNA plasmid and Ssb1p are localized together, forming one or more nucleoli without extensive contact with the nuclear envelope.
The same conclusion on the effects of the rpa12Δ mutation on nucleolar localization was also obtained by analyzing the rdnΔ rpa12Δ strain carrying the Pol I plasmid (NOY777) by EM, as shown in Figs. 3 and 4. Even though this strain (NOY777) was grown at 25°C and transcribed the Pol I rDNA plasmid using Pol I, a single rounded nucleolus was observed that had two subnucleolar regions with different electron density (Fig. 3 d). Often the nucleolus of this strain (NOY777) had minimal contact with the nuclear envelope (Fig. 4 d). Thus, the nucleolus in this strain resembles the nucleolus observed for the rdnΔ strain carrying the Pol II rDNA plasmid (NOY759; Fig. 3 c). Many mininucleoli that must have been formed as in the case of the isogenic RPA12 strain (NOY758; Fig. 3 b), appear to have coalesced mostly into a single nucleolus away from the nuclear periphery.
It should be noted that the two rpa12Δ::LEU2 strains grew more slowly than the corresponding control strains under the growth condition used in these experiments. For the rdnΔΔ strains, the decrease in growth rate caused by the rpa12Δ mutation was ∼20% (the doubling times on YEP–galactose medium at 25°C for NOY770 and NOY780 were 7.3 and 8.8 hours, respectively). For the rdnΔ strains, the decrease was ∼40% (the doubling times on YEP–galactose medium at 25°C for NOY758 and NOY777 were 5 and 7 h, respectively). However, the effect of the rpa12Δ mutation on growth rate does not account for the striking alteration in the structure and localization of the nucleolus. NOY777, which is rdnΔ and carries rpa12Δ, grew at about the same growth rate as NOY770, which is rdnΔΔ and RPA12, and yet the former showed a nucleolar localization/structure very different from the latter (and from its control RPA12 strain, NOY758) as described above.
These observations demonstrate that the predominant localization of the (mini-) nucleolus (and Pol I) to the nuclear periphery requires the presence of an intact Pol I. Deletion of the gene for the A12 subunit prevents the occurrence of many separate mininucleolar foci and their predominant localization to the nuclear periphery even under conditions in which Pol I is sufficiently functional to allow cells to grow at a rate close to that of the wild type.
Nucleolar Structures in a Strain Using Pol II to Transcribe rDNA Template in Chromosome XII
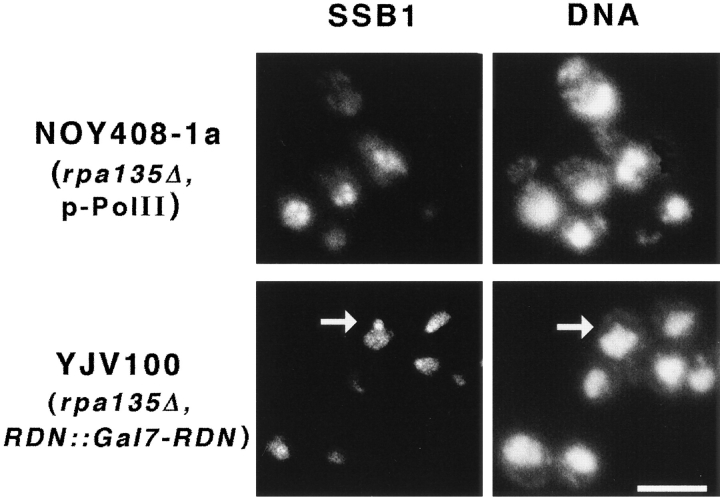
The results described in the previous sections demonstrated the essential role of the transcriptional machinery in the localization and organization of the nucleolus. The crescent-shaped nucleolus in normal yeast cells is obviously different from the mininucleoli seen in rdnΔ or rdnΔΔ strains carrying the Pol I rDNA plasmid. Mininucleoli are distributed through the nucleoplasm, although they are predominantly localized at the nuclear periphery. Thus, the chromosomal rDNA repeats must exert an influence on nucleolar morphology that is absent in the Pol I or Pol II rDNA plasmid systems. For example, clustering of rRNA genes in a single locus is expected to prevent the formation of many independent mininucleoli, and the presence of DNA flanking the rDNA repeats might have a role in the nucleolar localization. To study this question, we used strain YJV100, which is a derivative of NOY408-1a and carries the GAL7-35S rDNA fusion gene integrated into the chromosomal rDNA repeats (Venema et al., 1995). Like NOY408-1a (Nogi et al., 1991), the strain carries a deletion (rpa135Δ::LEU2) in the essential gene encoding the A135 subunit of Pol I and can grow only in galactose medium by transcribing the GAL7-35S rDNA gene using Pol II. The copy number of this hybrid gene integrated into the chromosomal rDNA in this strain was previously estimated to be 20–25 and about an equal number of the native rDNA repeats was also present (Venema et al., 1995). As shown in Fig. 5 (bottom panel), the nucleolus revealed by IFM using anti-Ssb1p antibodies was a single dot localized at the nuclear periphery (see for example, Fig. 5, arrows; see also the results of EM analysis to be described below). It is clearly different from the crescent structure seen for normal yeast cells (Fig. 1, NOY505). The structure is also different from that seen for NOY408-1a; in this strain several separate granules, previously called mininucleolar bodies (Oakes et al., 1993), were observed (Fig. 5, top panel). Both the original plasmid (pGRIM) integrated into the chromosomal rDNA repeats in YJV100 and the plasmid (pNOY102) carried by NOY408-1a contain the GAL7-35S rDNA for Pol II transcription, and their copy numbers are similar. Therefore, the difference in the nucleolar structure between YJV100 and NOY408-1a must be due to the intranuclear state of the plasmids. The integrated plasmid copies are probably physically close together and mininucleoli formed from individual hybrid genes may have coalesced into a single nucleolar structure at the nuclear periphery. In contrast, the non-integrated plasmid copies in NOY408-1a may not have such topological restrictions and may be able to form several separate mininucleolar bodies away from the nuclear periphery.
Figure 5.
IFM analysis of nucleolar structures in strains transcribing the GAL7-35S rDNA fusion gene on a plasmid (NOY408-1a) or the same fusion gene integrated into chromosome XII at the RDN locus (YJV100) by use of Pol II. Nucleolar protein Ssb1p was analyzed by IFM and DNA was stained by DAPI as described in Materials and Methods. Arrows indicate an example of the nucleolus as revealed by anti-Ssb1p that shows a special structure protruding from the main body of the nucleus. Bar, 5 μm.
The difference between pNOY408-1a and YJV100 was also clearly demonstrated by EM analysis of these strains. As was seen in previous work (Oakes et al., 1993), one or a few mininucleolar bodies were seen mostly away from the nuclear periphery in thin sections of NOY408-1a cells (Fig. 6 a). The rounded nucleolar bodies contained two ultrastructurally distinct subnucleolar regions, similar to the Pol II rDNA plasmid systems, and lacked extensive contact with the nuclear envelope (Fig. 6 a). In YJV100 cells, the nucleolus was a single rounded body similar to that seen for the Pol II rDNA plasmid systems, showing in many instances a segregation of a very electron-dense area and a less electron-dense area (Fig. 6 b), and was clearly different from the normal crescent-shaped nucleolus (Fig. 3 a). However, in YJV100 cells, there is a bulge or outpocketing of the nuclear envelope in the area of the nucleolus (Fig. 6 b). This feature could also be seen often in IFM analysis of YJV100 cells (see Fig. 5, arrows). It appears that although the polymerase system clearly plays an important role, the tandemly repeated chromosomal rDNA structure has, presumably through its connection to flanking chromosomal DNA regions, an influence on association of the nucleolus with the nuclear envelope.
Figure 6.
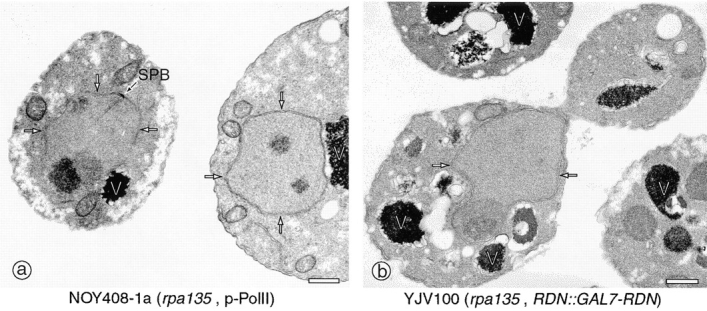
EM analysis of yeast mutants. Yeast strains NOY408-1a (rpa135Δ, p-Pol II) (a) and YJV100 (RDN::GAL7-RDN) (b) were analyzed as described in Fig. 3. Representative fields of cells in which nuclei and nucleoli are visible are shown. The nuclear envelope is marked with arrows as a point of reference. SPB, spindle pole body. Bars, 0.5 μm.
Discussion
Pol I Transcription Machinery Is Important for Localization of the Nucleolus to the Nuclear Periphery
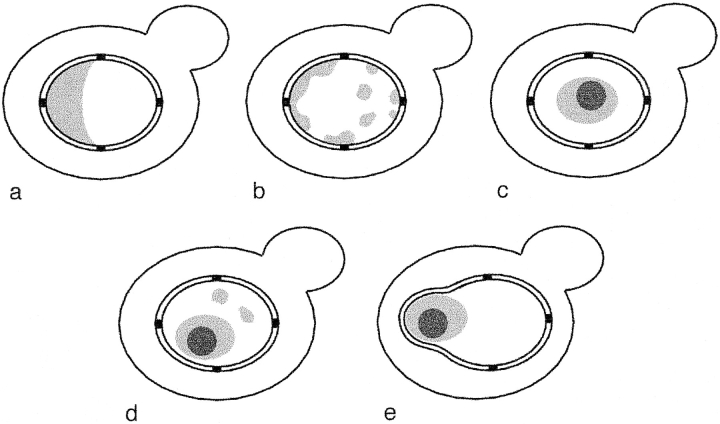
We have used yeast strains with a chromosomal rDNA deletion and carrying rDNA plasmids to study how the nucleolus is spatially organized within the nucleus. The various kinds of nucleolar morphology/localization observed in the present investigation are schematically shown in Fig. 7 and are summarized in Table II. Comparison of the two types of strains, one carrying the Pol I rDNA plasmid and the other carrying the Pol II rDNA plasmid, has demonstrated a clear difference in the nucleolar structures and localization. The former strains contained many mininucleoli distributed throughout the nucleus, predominantly localized at the nuclear periphery, while the latter strains contained a single (and possibly two but rarely more) rounded nucleolus without an extensive contact with the nuclear envelope. This difference in the degree of nucleolar coalescence and localization is not due to differences in plasmid copy numbers, since both types of strains carry a comparable number of plasmids (∼90).
Figure 7.
Schematic representation of nucleolar morphologies in yeast mutants. (a) Crescent-shaped nucleolus observed in wild-type cells. (b) Dispersed nucleolus observed in cells deleted for rDNA and bearing rDNA on a multicopy plasmid. (c) Condensed nucleolus observed in cells deleted for rDNA and bearing a multicopy plasmid expressing rDNA from a Pol II promoter. (d) Nucleolus observed in an rpa135Δ mutant that lacks functional Pol I and carries a multicopy plasmid that expresses rDNA from a Pol II promoter. (e) Nuclear envelope associated nucleolus observed in an rpa135Δ mutant containing 20–25 repeats at the rDNA locus that express rDNA from a Pol II promoter. The panels a–e correspond to EM images obtained with strains: NOY505 (a); NOY758 and NOY770 (b); NOY759, NOY773, and NOY777 (c); NOY408-1a (d); YJV100 (e). Characteristics of these strains are summarized in Table II.
There are two main differences between the Pol I rDNA plasmid and the Pol II rDNA plasmid systems. First, the transcription machinery is different. Second, the cis elements, specifically the promoters, on rDNA plasmids are different. However, the rRNA coding region and the rRNA transcript are identical between the two systems. Thus, models invoking some specific affinity between the DNA encoding rRNA or the rRNA transcript and some structures at the nuclear periphery cannot explain localization of the nucleus to the nuclear periphery. Furthermore, the results obtained for the strains with the rpa12Δ mutation and carrying the Pol I plasmid (Fig. 7 c) indicate that the presence of the intact rDNA with the intact cis elements alone is not sufficient for predominant localization to the nuclear periphery. The difference in the nucleolar morphology/localization between NOY758 (rdnΔ, RPA12, Pol I rDNA plasmid) and NOY777 (the same, but rpa12Δ) is striking (Fig. 7, b and c). This demonstrates the importance of the intact structure of Pol I for nucleolar structure and localization. One possibility to explain differences between these two strains is that mininucleoli formed on individual plasmid molecules have an inherent tendency to coalesce, and that interactions between the intact Pol I and the nuclear periphery prevent coalescence of mininucleoli into a single large nucleolar structure.
Pol I plays an essential role in organizing nucleolar structure and localization. It is known that interaction of Pol I with rDNA requires specific transcription factors such as UAF and CF (see Introduction). Thus, it is reasonable to assume that these transcription factors also play an important role in nucleolar organization and localization. It should be noted that purified UAF contains histones H3 and H4 (Keener et al., 1997). UAF might be part of a special chromatin structure unique to the rDNA locus. Certain evidence suggests that rDNA chromatin can assume two (or more) different structures that can be distinguished by their ability to silence Pol II activity in the rDNA locus (Bryk et al., 1997; Smith and Boeke, 1997; Fritze et al., 1997). Our recent experiments showed that these structures are interchangeable by epigenetic events; the form able to silence Pol II activity is stabilized by UAF, Pol I, and perhaps other Pol I–specific transcription factors (Vu, L., M. Oakes, J.P. Aris, and M. Nomura, unpublished experiments). Such chromatin structures may be responsible for localization of the nucleolus to the nuclear periphery.
Role of the Chromosomal Context of rDNA in the Localization of the Nucleolus
The localization of mininucleoli to the nuclear periphery observed in rdn deletion strains carrying the Pol I rDNA plasmid (Fig. 7 b) may reflect the localization of the crescent-shaped nucleolus adjacent to the nuclear envelope in normal yeast strains (Fig. 7 a). The chromosomal rDNA is clustered as a tandem repeat of 100–150 genes on chromosome XII (Petes, 1979). This clustering may limit contact of the nucleolus to a part of the nuclear envelope, thus forming the crescent structure, which is different from the nucleolar morphology (Fig. 7 b) seen for the Pol I plasmid system. In addition, the results (Fig. 7 e) obtained with strain YJV100 in which multi-copies of the GAL7-35S rDNA fusion gene are integrated into the chromosomal rDNA repeats, suggest that, in addition to the proposed interaction between the Pol I–specific rDNA chromatin structure and some structures at the nuclear periphery, there may be additional interactions perhaps between DNA elements flanking rDNA repeats and structures at the nuclear periphery.
Nucleolar Structures in Strains in Which Pol II Transcribes rDNA
In the rdn deletion strains carrying the Pol II rDNA plasmid, in which the GAL7-35S rDNA hybrid is transcribed by Pol II, both the template plasmid (and Pol II engaged in rRNA synthesis) and nucleolar protein Ssb1p are colocalized to regions that do not have extensive contact with the nuclear periphery. Regarding the Pol II rDNA plasmid, it is now generally accepted that mRNA transcription does not take place uniformly in the nucleus, but at many specific places (“transcription foci” or “transcription factories”) (Lawrence et al., 1989, 1993; Jackson et al., 1993; Spector et al., 1993; Wansink et al., 1993; Iborra et al., 1996; for review see Cook, 1994). Perhaps, Pol II molecules present in these transcription factories may be responsible for binding and transcribing the Pol II rDNA plasmid. For nucleolar proteins, many—including Ssb1p— are complexed with snoRNAs, forming small nucleolar RNPs (snoRNPs), which interact with precursor rRNA presumably through rRNA–snoRNA base pairing, as demonstrated by recent studies (Kiss-László et al., 1996; Ganot et al., 1997; Ni et al., 1997; for review see Smith and Steitz, 1997) (Ssb1p is associated with Box H/ACA snoRNAs, snR10 and snR11, as described by Clark et al., 1990). Thus, in the yeast cells that grow by synthesizing rRNA by Pol II, nucleolar proteins (and snoRNA) engaged in rRNA modification, processing, and perhaps ribosome assembly, will be localized to Pol II transcription factories synthesizing rRNA. However, Pol I would not be expected to be colocalized with these nucleolar components, as observed in the present work. It is evident that some nucleolar proteins, such as those involved in specific transcription, Pol I, UAF, and CF, play a primary role in determining the nucleolar localization, whereas others, such as those involved in rRNA processing, do not.
Possible Significance of the Nucleolar Localization to the Nuclear Periphery
What is the significance of the nucleolar localization to the nuclear periphery? The synthesis of ribosomes, the major nucleolar function, requires extensive nuclear–cytoplasmic transport of macromolecules, such as the nuclear import of many ribosomal proteins and the export of ribosomes. Localization of the nucleolus adjacent to the nuclear envelope, thus, might be advantageous for efficient nuclear– cytoplasmic transport. However, the rpa12 deletion that prevented the nucleolar localization to the nuclear periphery in rdn deletion strains growing at permissive temperature, caused only a small decrease (20–40%) in growth rate, as described in this paper. Similarly, it was observed previously that overproduction of Nop2p, a nucleolar protein, causes the nucleolus to become detached from the nuclear envelope without causing any decrease in growth rate (de Beus et al., 1994). Thus, the significance of the nucleolar localization to the nuclear periphery is not clear at the moment. Perhaps there are other important nucleolar functions that cannot be assessed by the simple measurement of growth rate. For example, recent studies have shown a correlation between redistribution of certain silencing proteins from telomeres to the nucleolus and lengthening of life span (Kennedy et al., 1997). A correlation between structural alterations of the nucleolus and aging of yeast cells has also been observed (Sinclair et al., 1997). These observations suggest a role of the nucleolus in the maintenance of normal aging (for review see Guarente, 1997). In addition, silencing of some Pol II genes inserted into the chromosomal rDNA repeats and its dependence on proteins such as Sir2p have been reported recently (Bryk et al., 1997; Fritze et al., 1997; Smith and Boeke, 1997; our unpublished work described above). Sir2 protein is also known to repress mitotic and meiotic recombination between the tandem rDNA repeats within the nucleolus (Gottlieb and Esposito, 1989). It is possible that the nucleolar localization at the nuclear periphery might be important in such less well-explored (or other unexplored) nucleolar functions. In this connection, it may be noted that yeast telomeres have been observed to localize to certain regions of the nuclear periphery in clusters. Silencing of Pol II genes near telomeres (for review see Loo and Rine, 1995) might be related to the nuclear location of telomeres (Klein et al., 1992; Palladino et al., 1993; Pillus and Grunstein, 1995). With several yeast mutant strains with different nucleolar localizations as characterized in this work, it should now be possible to study the question of nucleolar localization in connection with nucleolar events such as those related to silencing and cell aging.
Acknowledgments
We thank Drs. S. Liebman, J.R. Warner, and J. Venema for providing plasmid pRDN-hyg1, strains L1521 and YJV100, respectively; and Drs. S.M. Arfin and T. Pederson for critical reading of the manuscript; and D. Semanko for help in preparation of the manuscript.
This work was supported by U.S. Public Health Grants GM35949 (M. Nomura) and GM48586 (J.P. Aris) from the National Institutes of Health.
Abbreviations used in this paper
CF
core factor
FISH
fluorescence in situ hybridization
IFM
immunofluorescence microscopy
Pol I and Pol II
polymerase I and II
rDNA
ribosomal DNA
snoRNA
small nucleolar RNA
UAF
upstream activation factor
Footnotes
Address all correspondence to Masayasu Nomura, University of California, Irvine, Department of Biological Chemistry, Irvine, CA 92697-1700. Tel.: (949) 824-4564. Fax: (949) 824-3201. E-mail: mnomura@uci.edu
References
- Bourgeois CA, Hubert J. Spatial relationship between the nucleolus and the nuclear envelope: structural aspects and functional significance. Int Rev Cytol. 1988;111:1–52. doi: 10.1016/s0074-7696(08)61730-1. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banjeree M, Murphy M, Knidsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topisomerase I in Saccharomyces cerevisiae. . Genes Dev. 1996;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Vincent A, Liebman SW. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO (Eur Mol Biol Organ) J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MW, Yip MLR, Campbell J, Abelson J. SSB-1 of the yeast Saccharomyces cerevisiaeis a nucleolar-specific, silver binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J Cell Biol. 1990;111:1741–1751. doi: 10.1083/jcb.111.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR. RNA polymerase: structural determinant of the chromatin loop and the chromosome. Bioessays. 1994;16:425–430. doi: 10.1002/bies.950160611. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- de Beus E, Brockenbrough JS, Hong B, Aris JP. Yeast NOP2encodes an essential nucleolar protein with homology to a human proliferation marker. J Cell Biol. 1994;127:1799–1813. doi: 10.1083/jcb.127.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Stritch R, Esposito RE. Direct evidence for SIR2modulation of chromatin structure in yeast rDNA. EMBO (Eur Mol Biol Organ) J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;15:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Link between aging and the nucleolus. Genes Dev. 1997;11:2449–2455. doi: 10.1101/gad.11.19.2449. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Methods Enzymol. 1991;194:1–183. [PubMed] [Google Scholar]
- Hozak P. The nucleoskeleton and attached activities. Exp Cell Res. 1996;229:267–271. doi: 10.1006/excr.1996.0370. [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Biol. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO (Eur Mol Biol Organ) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. A DrosophilarRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- Keener J, Dodd DA, Lalo D, Nomura M. Histones H3 and H4 are components of upstream activation factor (UAF) required for the high level transcription of yeast rDNA by RNA polymerase I. Proc Natl Acad Sci USA. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. . Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- Keys DA, Vu L, Steffan JS, Dodd JA, Yamamoto R, Nogi Y, Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. . Genes Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- Keys DA, Lee B-S, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF-X, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo D, Steffan JS, Dodd JA, Nomura M. RRN11encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J Biol Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localised tracks of specific transcripts within interphase nuclei visualized by in situhybridisation. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Carter KC, Xing X. Probing functional organization within the nucleus: Is genome structure integrated with RNA metabolism? . Cold Spring Harbor Symp Quant Biol. 1993;58:807–818. doi: 10.1101/sqb.1993.058.01.088. [DOI] [PubMed] [Google Scholar]
- Lin CW, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder RH. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. . Mol Cell Biol. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Nierras CR, Liebman SW, Warner JR. Does Saccharomyces need an organized nucleolus? . Chromosoma (Basel) 1997;105:444–451. [PubMed] [Google Scholar]
- Nogi Y, Yano R, Nomura M. Synthesis of large ribosomal RNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Dodd J, Carles C, Nomura M. Gene RRN4 in Saccharomyces cerevisiaeencodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Nogi Y, Clark MW, Nomura M. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiaedefective in RNA Polymerase I. Mol Cell Biol. 1993;13:2441–2455. doi: 10.1128/mcb.13.4.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Petes T. Yeast ribosomal DNA genes are located in chromosome XII. Proc Natl Acad Sci USA. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus, L., and M. Grunstein. 1995. Chromatin structure and epigenetic regulation in yeast. In Chromatin Structure and Gene Expression. S.C.R. Elgin, editor. IRL Press, Oxford Univeristy Press, New York. 123–146.
- Rose MD, Broach JR. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Schultz MC, Reeder RH, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Spector DL, O'Keefe RT, Jimenez-Garcia LF. Dynamics of transcription and pre-mRNA splicing within mammalian cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;58:799–805. doi: 10.1101/sqb.1993.058.01.087. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Keys DA, Dodd JA, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream-activation-factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- Venema J, Dirks-Mulder A, Faber AW, Raué HA. Development and application of an in vivosystem to study yeast ribosomal RNA biogenesis and function. Yeast. 1995;11:145–156. doi: 10.1002/yea.320110206. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weipoltshammer K, Schofer C, Wachtler F, Hozak P. The transcription unit of ribosomal genes is attached to the nuclear skeleton. Exp Cell Res. 1996;227:374–379. doi: 10.1006/excr.1996.0287. [DOI] [PubMed] [Google Scholar]
- Wittekind M, Kolb JM, Dodd JA, Yamagishi M, Mémet S, Buhler J-M, Nomura M. Conditional expression of RPA190, the gene encoding the largest subunit of yeast polymerase I: effects of decreased rRNA synthesis on ribosomal protein synthesis. Mol Cell Biol. 1990;10:2049–2059. doi: 10.1128/mcb.10.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Mélèse T. Nucleolar proteins that bind NLSs: a role in nuclear import or ribosome biogenesis? . Trends Cell Biol. 1994;4:414–417. doi: 10.1016/0962-8924(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto RT, Nogi Y, Dodd JA, Nomura M. RRN3 gene of Saccharomyces cerevisiaeencodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO (Eur Mol Biol Organ) J. 1996;15:3964–3973. [PMC free article] [PubMed] [Google Scholar]