Structural Analysis of Drosophila Merlin Reveals Functional Domains Important for Growth Control and Subcellular Localization (original) (raw)
Abstract
Merlin, the product of the Neurofibromatosis type 2 (NF2) tumor-suppressor gene, is a member of the protein 4.1 superfamily that is most closely related to ezrin, radixin, and moesin (ERM). NF2 is a dominantly inherited disease characterized by the formation of bilateral acoustic schwannomas and other benign tumors associated with the central nervous system. To understand its cellular functions, we are studying a Merlin homologue in Drosophila. As is the case for NF2 tumors, Drosophila cells lacking Merlin function overproliferate relative to their neighbors. Using in vitro mutagenesis, we define functional domains within Merlin required for proper subcellular localization and for genetic rescue of lethal Merlin alleles. Remarkably, the results of these experiments demonstrate that all essential genetic functions reside in the plasma membrane– associated NH2-terminal 350 amino acids of Merlin. Removal of a seven–amino acid conserved sequence within this domain results in a dominant-negative form of Merlin that is stably associated with the plasma membrane and causes overproliferation when expressed ectopically in the wing. In addition, we provide evidence that the COOH-terminal region of Merlin has a negative regulatory role, as has been shown for ERM proteins. These results provide insights into the functions and functional organization of a novel tumor suppressor gene.
Recent studies have identified a rapidly growing number of tumor suppressor genes whose normal function in cells directly or indirectly regulates cellular proliferation. Not surprisingly, many of these genes, such as Rb and p53, have been shown to encode proteins that regulate aspects of the cell cycle (for review see Brown, 1997). However, other tumor suppressor genes appear to function in processes less clearly related to control of the cell cycle. One of the most intriguing genes of this latter class is the Neurofibromatosis type 2 (NF2)1 gene, which encodes a member of the protein 4.1 superfamily, Merlin (Rouleau et al., 1993; Trofatter et al., 1993). Members of the protein 4.1 superfamily, a large group of membrane-associated cytoplasmic proteins, include protein 4.1; talin; the ezrin, radixin, moesin (ERM) proteins; Merlin; Drosophila Expanded; several protein phosphatases; and at least two nonmuscle myosins (for review see McCartney and Fehon, 1997). The defining feature of this superfamily is a conserved region of 200–300 amino acids usually located in the NH2 terminus of the protein. This region is particularly well conserved between Merlin and the ERM proteins. The ERM proteins appear to function as molecular linkers by binding to transmembrane proteins through the NH2-terminal domain and linking them to the cortical actin cytoskeleton through a COOH-terminal actin-binding domain. Consistent with this role, ERM proteins localize to actin-rich structures such as the adherens junction and microvilli (Franck et al., 1993; Takeuchi et al., 1994).
Although the structural similarities between Merlin and the ERM proteins suggest that they may have similar functions, the exact nature of Merlin's cellular functions has not been defined. Recent studies in cultured cells indicate that expressed Merlin protein accumulates in some actin-rich membrane domains, such as membrane ruffles at the leading edge of migrating cells (Gonzalez-Agosti et al., 1996; Sainio et al., 1997), consistent with the localization of ERM proteins. However, several lines of evidence indicate that Merlin has functions that are clearly distinct from those of ERM proteins. First, although the NH2-terminal domains of Merlin and the ERM proteins are similar (the protein 4.1 superfamily domain), Merlin lacks the well- defined COOH-terminal actin-binding domain found in ERM proteins (Turunen et al., 1994; Gary and Bretscher, 1995). In addition, while the ERM proteins are functionally redundant (Takeuchi et al., 1994), there is no evidence for redundancy between the ERM proteins and Merlin. The NF2/Merlin gene can be mutated to lethality in both Drosophila and mouse (Fehon et al., 1997; McClatchey et al., 1997). Finally, in vivo studies of subcellular localization of Merlin and Moesin reveal that they can be distinct, again supporting the idea that these proteins have different functions (McCartney and Fehon, 1996).
To define the cellular functions of Merlin and the ERM proteins, we have isolated and characterized two Drosophila genes, Moesin, the sole ERM gene in Drosophila, and Merlin, a well conserved NF2 homologue (McCartney and Fehon, 1996). In the present study, we show that Merlin is essential for viability in Drosophila and is required for the proper regulation of cell proliferation. Furthermore, our analysis indicates that essential Merlin functions occur at the cytoplasmic face of the cell membrane and that all of these functions can be mediated by the conserved NH2-terminal region.
Materials and Methods
Drosophila Cultures and Stocks Used
All Drosophila cultures were maintained on standard corn meal, yeast, molasses, and agar medium. w1118 stocks were used for the transformation of UASMerlin transgenes. The Merlin alleles used in this study are described in Fehon et al. (1997).
Somatic Mosaic Analysis and Histology of Adult Eyes
Fly stocks capable of producing clones were generated by crossing w1118 sn3 Mer* P{neoFRT}19A/FM6 with y w P{w[+mC] = PiM}5A P{w[+mC] = PiM}10D P{neoFRT}19A/Y;P{hs-FLP}, Sb/TM6B. Offspring from this cross were heat shocked at either 36 or 72 h after egg laying (AEL) using one of two different heat shock regimens: 30 min at 38°C, 60 min at 25°C, and 30 min at 38°C or 60 min at 38°C, 60 min at 25°C, and 60 min at 38°C. In the adult, mutant clones were marked with w1118 in the eye and sn3 in the thorax; wild-type sister clones were marked in the eye with four copies of the mini-white transgene and in the thorax with the yellow mutation. To analyze clone size in the eye, flies were placed in vented Eppendorf tubes (Madison, WI) and flash frozen in liquid nitrogen. The treated flies were dried with dry carbon dioxide, and the heads were then dissected with a razor blade and mounted on a glass slide on double stick tape. Eyes were examined and ommatidia counted with the compound microscope using transmitted light and the 10× objective. Fixation and sectioning of adult eyes was performed as previously described (Tomlinson and Ready, 1987), with the exception that the tissue was postfixed in 2% OsO4 and embedded in Araldite resin.
Wing Measurements
Wing cuticles were prepared by first incubating the entire fly in 70% ethanol and then submerging it in a drop of water on a siliconized slide, where the wings were dissected away and mounted in ∼30 μl Aquamount (BDH Laboratory Supplies, Poole, England) on a glass slide. Only wings that had been well flattened during the mounting process were used for further analysis. Camera lucida drawings were made of the wing perimeter, wing veins, and any ectopic veination. These drawings were then scanned at 75 dots per inch using a flatbed scanner and analyzed using the “measure” tool in NIH Image. The area of the entire wing or the individual areas between the wing veins (the intervein regions) were determined and expressed in square millimeters.
Sequencing of Mutant Merlin Alleles
Genomic DNA was obtained from single first instar larvae hemizygous for each Merlin allele (marked with yellow). Merlin genomic DNA was amplified using intron-specific primers. The resulting PCR products were sequenced using the AmpliCycle sequencing kit (Perkin-Elmer Corp., Branchburg, NJ).
Construction of Truncated Merlin Proteins
A Bluescript shuttle vector was generated with an NH2-terminal myc epitope tag by annealing two primers, consmyc S (sense) 5′ AAT TCA CCA TGG AGC AAA AGC TCA TTT CTG AAG AGG ACT TGA GGC CTA A and consmyc A (antisense) 5′ GAT CTT AGG CTT CAA GTC CTC TTC AGA AAT GAG CTT TTG CTC CAT GGT G, which produce a duplex with over-hanging EcoRI and BglII ends. This fragment was subsequently cloned into an EcoRI/BamHI-cut Bluescript SK− plasmid. (All restriction enzymes were obtained from New England Biolabs, Beverly, MA.) Digestion of the modified vector with StuI allowed PCR-generated Merlin transgenes to be cloned in-frame immediately downstream of the myc epitope.
All Merlin constructs were generated by PCR amplification from a full-length Merlin cDNA clone (McCartney and Fehon, 1996). The sequence of all PCR-amplified regions was confirmed by sequencing using standard methods. To make upstream activation sequences of the yeast Gal4 transcription factor (UAS) expression constructs, a XhoI/XbaI fragment from the BSSK-myc shuttle vector was then cloned into an XhoI/XbaI-prepared pUASt transformation vector (Brand and Perrimon, 1993). Transformation of these constructs was performed as described (Rebay et al., 1993). 4–10 independent lines were established for each construct.
To construct BSSK-myc MerΔBB, the 5′ half of Merlin was amplified using the M13 Universal Primer of the BSSK− vector, and an antisense Merlin primer within which a PvuII site was engineered (underlined in primer sequence): 5′ GCG TCA TCT GCA GCT GAT GCG. This product was then digested with EcoRI and PvuII. The 3′ MerΔBB region was similarly generated using M13 reverse and an internal Merlin sense primer whose 5′ sequence began at codon 177: 5′ TGG GAG GAA CGG ATC AAG ACA TGG; this product was digested with PstI and ligated together with the 5′ piece into an EcoRI/PstI cut BSSK-mycMer +. Positive clones were sequenced to verify the presence of the deletion. The BSSK-mycMerBBA vector was constructed in a similar manner using sense (5′ TAC CAG ATG ACC GCG GCA GCG TGG GAG GAA CGG) and antisense (5′ CTC CCA CAT TTC CGC GGC TGC TGC GGC CTG ATC GGT CAC TCC) primers that changed amino acids 171–177 to alanines and contained a SacII site for cloning (underlined in primer sequence). To make BSSK-mycMer3, a genomic PCR fragment containing the Mer3 mutation was digested with NheI and NspI. A BSSK-mycMer + vector was prepared by a complete NheI digest and a partial NspI digest. (The site is unique in Merlin, but another site is in the BSSK− vector.) A ligation was performed using the prepared PCR product and the NheI/NspI-prepared BSSK− myc tag vector. Positive clones were sequenced to detect the presence of the point mutation.
To make Merlin green fluorescent protein (GFP) fusion proteins, UAS_mycMer_ + , UAS_mycMerΔBB_, UAS_mycMerBBA_, and UAS_mycMer3_ were digested with BglII and SacI. The SacI site is unique in Merlin and is located in the last codon of the Merlin open reading frame. A GFP fragment was excised from pGEM7GFPRS (a gift of Kevin Edwards and Dan Kiehart, Duke University) using SacI and XbaI. The SacI site lies eight codons 5′ to the start ATG of the GFP open reading frame and is in-frame to the Merlin open reading frame. A pCaSpeR-hs vector was prepared using BglII and XbaI. A three-piece ligation was performed using gel-purified fragments of Merlin, GFP, and the prepared pCaSpeR-hs vector.
Transfection of Schneider 2 Cells and In Vivo Competition Experiments
For expression of the truncated Merlin fragments in Schneider line 2 (S2) cultured cells, two sets of expression constructs were used: pCaSpeR-hs, a heat shock inducible vector, and pRmHa-3, an inducible metallothionein promoter vector. For the heat shock vectors, a BglII/XbaI fragment from the UAS Merlin truncation constructs was cloned into a BglII/XbaI-prepared pCaSpeR-hs vector. To generate pRmHa-3, a BglII/BamHI fragment from pCaSpeR-hs Merlin constructs was cloned into BamHI-prepared pRmHa-3. The maintenance, induction, and immunofluorescent analysis of S2 cells was performed as previously described (Fehon et al., 1990). In the time course experiments, transfected S2 cell cultures were induced with a 20 min/38°C heat shock; samples were then collected at time points 1, 3, and 6 h after heat shock (AHS) induction. The samples were fixed and stained with a mouse anti-myc antibody 9c10.
For the in vivo competition experiment, we cotransfected S2 cells with two different plasmids: a pRmHa-3 myc-tagged Merlin construct (either wild-type Merlin, Merlin1–600, or MerlinΔBB) and a pCaSpeR-hs non–myc-tagged Merlin GFP fusion protein construct. The transfected cells were treated with a CuSO4 solution (final concentration 0.6 mM) to induce the metallothionein promoter, incubated for 3 h, and heat shocked for 20 min/ 38°C to induce expression of the heat shock construct. Samples were collected 5 h AHS, fixed, and stained with a mouse anti-myc antibody.
Determination of In Vivo Activity
To determine the rescue activity of our UASMer transgenes, y1 w1118 Mer/ FM7; T80Gal4/T80Gal4 females were crossed to UASmycMer transgenic males. Four equal classes of flies would be expected from this cross, including males hemizygous for a Merlin mutation that lack endogenous Merlin function. To determine the percentage of rescue, the number of F1 progeny excluding the rescued hemizygous males was divided by three to yield the predicted number of mutant offspring. The number of observed mutant progeny (marked by yellow) was then divided by this number to yield the percentage of rescue. The numbers were then normalized to the rescue provided by Merlin1–600, which provided the greatest rescue.
Results
Characterization of Mutations within the Drosophila Merlin Gene
The Drosophila Merlin gene is located on the X chromosome at cytological position 18D-E. Four mutations within Merlin have been previously identified (Fehon et al., 1997). Although none of these alleles causes embryonic lethality, three of these mutations cause larval and pupal lethality without strong distinguishing phenotypes. The three lethal Merlin alleles encode putative truncated proteins as a result of nonsense mutations: Merlin1 (Mer1), Gln324 to stop; Mer2, Gln318 to stop; Mer4, Gln170 to stop. A single viable allele, Mer3, was recovered from these screens and is due to a missense mutation of Met177 to Ile. Flies homozygous for Mer3 survive as viable, sterile adults and display a broadened wing phenotype along with low and variably penetrant expression of weakly roughened eyes and the development of abnormal head cuticle structures (data not shown). Although no genetic deficiency exists to test for residual function of these alleles, we conclude that Mer4 is likely a null mutation based on the severity of the truncation predicted for this allele.
To characterize the cellular phenotypes of Merlin, we performed a somatic mosaic analysis using the yeast 2 micron Flipase enzyme/Flipase Recognition Target site (FLP/FRT) system (Golic and Lindquist, 1989; Xu and Rubin, 1993). The parental chromosome of all four Mer alleles carries an FRT site at cytological position 19A. Although previous experiments mapped Merlin distal to this FRT at 18D-E, we confirmed that Mer− clones were generated after heat shock FLP induction by staining mosaic tissue with the Merlin antibody (data not shown). In the adult eye, mutant clones were identified by the lack of pigment due to the presence of the white mutation; wild-type sister clones were marked by a dark orange eye color, and heterozygous ommatidia were pale orange. The overall morphology of the eye within the mutant clone appeared normal (Fig. 1 B), although mutant clones occasionally displayed a very weak roughened phenotype (data not shown). Histological sections of these clones revealed essentially normal differentiation of ommatidia with only a few disruptions in the organization of ommatidia when compared with the neighboring wild-type cells (Fig. 1 C).
Figure 1.
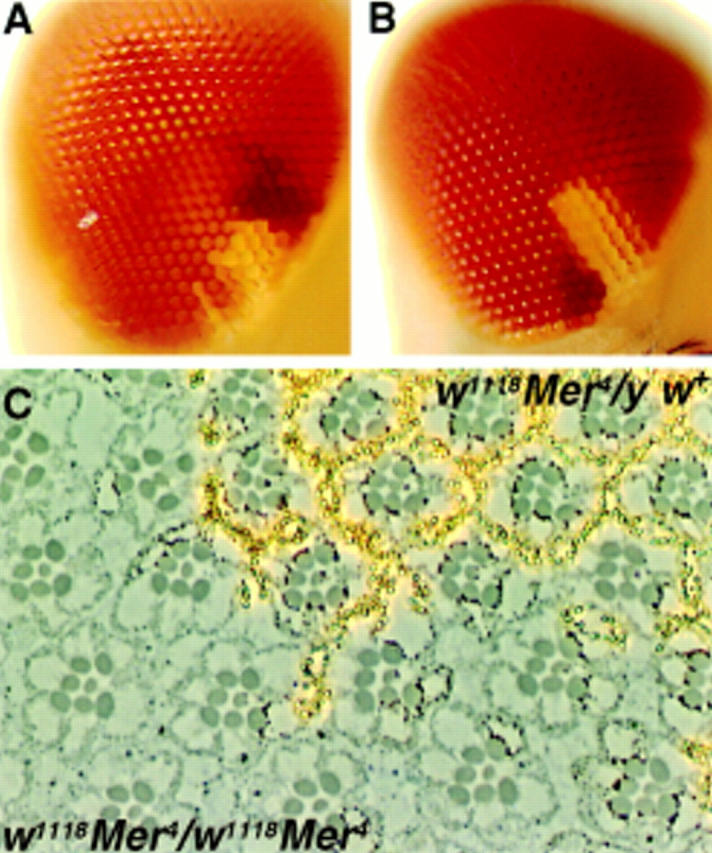
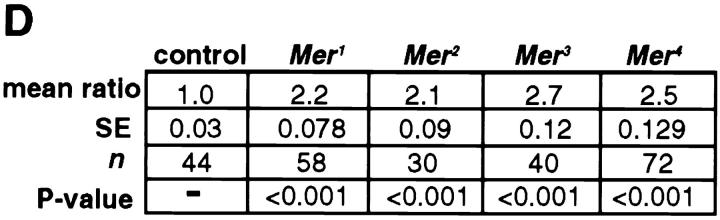
Mer mutant clones in the eye overproliferate relative to control clones. (A) Control clone w1118 sn3 19AFRT and (B) mutant clone w1118 sn3 Mer319AFRT induced at 72 h AEL. In all cases, the mutant or control clone is marked with w1118 (white), and the wild-type sister clone is marked with four copies of mini-w + (red). The heterozygous background is marked with two copies of mini-w + (orange). Comparison of the mutant clone to either its sister clone or to the control clone reveals that the Merlin mutant clone is consistently larger, containing more ommatidia. Histological sections of a w1118 sn3 Mer4 clone induced at 36 h AEL (C) reveal essentially no abnormal phenotypes in the ommatidial morphology of the homozygous mutant tissue. This tissue can be distinguished from the neighboring heterozygous or homozygous wild-type tissue by the lack of pigment granules (yellowish spots) due to the w1118 marker. (D) Summary of the proliferation analysis of Merlin mutant clones. When the t test was applied to these data, the difference in ratio between the control and all four Merlin mutants was found to be significant (P < 0.001).
Comparison of genetically marked, wild-type (control), and Mer− mutant clones in the eye suggested that the mutant clones were consistently larger than their wild-type sisters. To address this possibility, we compared the number of ommatidia within each mutant clone to the number of ommatidia in its sister clone and generated a ratio of mutant clone size to wild-type sister clone size. When a control clone was generated, the area of the white −-marked clone was equal to that of its white + sister (Fig. 1 A and D). In contrast, Merlin mutant clones ranged from 2.1 to 2.7 times the size of their wild-type sisters, depending on the allele of Merlin examined (Fig. 1, B and D). This observation suggests that the Merlin mutant cells either proliferate more rapidly than their wild-type neighbors or that they continue proliferating later in development. Another possible explanation is that the mutant cells have a defect in cell death leading to apparent overproliferation. Acridine orange staining of Merlin mutant imaginal discs did not indicate any changes in the level of cell death, however (data not shown). Because loss of Merlin function in clones results in overproliferation without any gross morphological defects, we conclude that Merlin functions in a process that specifically affects the regulation of proliferation.
In Vitro Mutagenesis
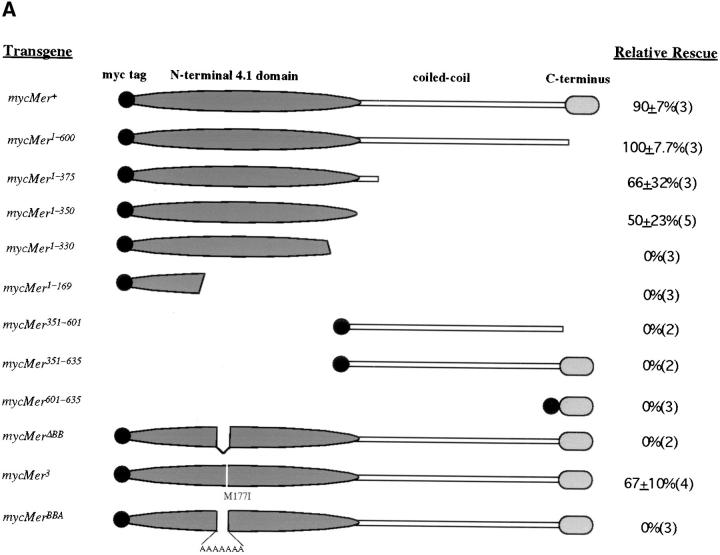
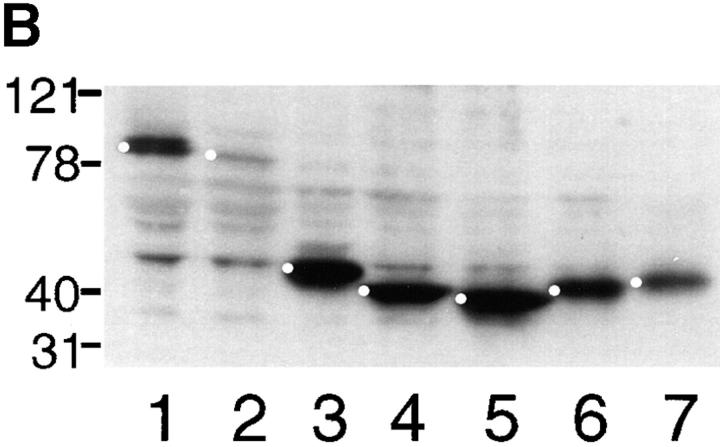
To further investigate its cellular roles, we generated NH2- and COOH-terminal truncations of the Merlin protein (Fig. 2 A). These mutations were used to perform two sets of experiments: First, we examined the subcellular localization of these mutant Merlin proteins when expressed in cultured cells and in tissue, and second, we examined their in vivo genetic functions. The fragments of Merlin generated for the structure/function analysis were selected based on comparisons with human Merlin and with the ERM proteins (McCartney and Fehon, 1996), with the assumption that regions that are highly conserved are likely to have functional significance. The molecular organization of Merlin is similar to that of ERM family members and consists of an NH2-terminal protein 4.1 domain, a putative coiled-coil domain, and a COOH-terminal region (Fig. 2 A). Because of the apparent modular nature of Merlin, we generated several Merlin truncations that contained portions of the NH2- or COOH-terminal halves of the protein and a truncation containing only the central coiled-coiled region. The size and stability of each Merlin truncation was verified by immunoblot (Fig. 2 B). In addition, we examined the conserved NH2-terminal region (CNTR) in greater detail. The CNTR is nearly 60% identical between Merlin and the ERM proteins; however, a closer examination of this region revealed seven amino acids (170YQMTPEM177) that are identical in human and Drosophila Merlin but are divergent from the ERM proteins (McCartney and Fehon, 1996). The possible functional significance of this region is further supported by the presence of the Mer3 missense mutation at amino acid 177. We have named this region “the Blue Box” (BB). To investigate the functional significance of the BB, we engineered two Merlin proteins, one lacking the BB region (mycMerΔBB) and one in which the BB is replaced by a polyalanine stretch (mycMerBBA). Furthermore, to define the nature and the activity of the original Mer3 allele, we generated a myc-tagged version of this allele (mycMer3).
Figure 2.
(A) Schematic representation of myc-tagged forms of Merlin and their ability to rescue Mer4 lethality. The name of each Mer allele corresponds to the amino acid residues present in the construct. Relative rescue was defined as the percentage of Mer4 hemizygous males that survive compared with expected (calculated from sibling classes, see Materials and Methods). The data is represented as an average of at least three separate experiments using two to five independent insertion lines (number shown in parenthesis). (B) Immunoblot of whole S2 cell extract transfected with myc-tagged Merlin heat shock transgenes, probed with anti-myc antibody 9c10: lane 1, mycMer+; lane 2, mycMer1–600; lane 3, mycMer1–375; lane 4, mycMer 1–350; lane 5, mycMer1–330; lane 6, mycMer351–600; lane 7, mycMer 351–635. Bullets denote each Merlin fragment.
The Subcellular Localization of Merlin In Vitro Mutants
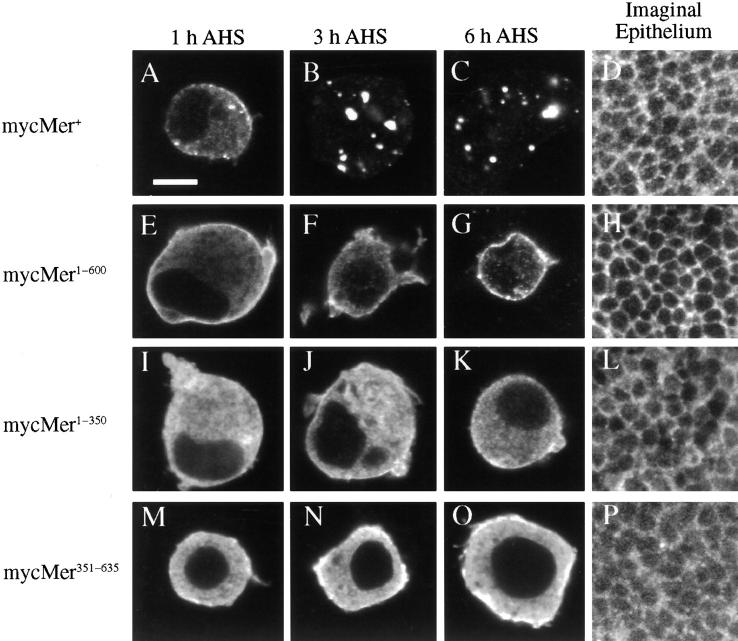
We examined the subcellular localization of these Merlin fragments in Drosophila S2 cultured cells and in the imaginal disc epithelium to identify regions within Merlin that are required for proper subcellular localization. Experiments in cultured cells allowed us to examine both the temporal patterning and the subcellular localization of Merlin. As previously reported (McCartney and Fehon, 1996), wild-type Merlin is initially targeted to the membrane, and within 3 h, much of the protein localizes to punctate cytoplasmic structures (Fig. 3, A–C). A similar pattern (membrane-associated and cytoplasmic staining) was also observed in endogenously expressed Merlin in S2 cells (data not shown) and within the imaginal disc epithelium (McCartney and Fehon, 1996; Fig. 3 D). Removal of the conserved COOH-terminal 35 amino acids resulted in a protein, mycMer1–600, that was localized almost exclusively at the plasma membrane and did not appear to internalize (Fig. 3, E–G). This observation suggests that the COOH-terminal 35 amino acid residues play a role in Merlin internalization. In the imaginal epithelia of transgenic larvae, this truncated Merlin protein was localized to the plasma membrane (similar to the localization of wild-type Merlin) but did not display the punctate cytoplasmic localization characteristic of the wild-type protein (Fig. 3 H). More severe COOH-terminal truncations of Merlin, mycMer1–350 (Fig. 3, I–L) and mycMer1–375 (data not shown) appeared to associate less strongly with the plasma membrane, suggesting that within the COOH terminus there are sites required for proper localization. Consistent with this notion, mycMer351–635 (Fig. 3, M–P) and mycMer351–600 (data not shown), which retain most of the COOH-terminal half of Merlin but none of the CNTR, were highly membrane associated in both cultured cells and imaginal tissue. Of the other Merlin constructs, mycMer1–330 localized diffusely throughout the cytoplasm with no distinct subcellular location, while mycMer1–169 and mycMer601–635 had little or no expression (data not shown), owing possibly to their inherent instability. We conclude from these results that components of both the NH2- and COOH-terminal halves of Merlin are required for its correct targeting to the plasma membrane.
Figure 3.
Subcellular localization of myc-tagged Merlin truncations in Drosophila S2 cells and wing imaginal disc epithelia: mycMer+ (A–D), mycMer1–600 (E–H), mycMer1–350 (I–L), and mycMer351–635 (M–O). All samples were stained with mouse anti-myc antibody (9c10) and a CY3-conjugated secondary antibody. In the cultured cell experiments, samples were collected from three time points AHS induction: 1 h AHS (A, E, I, and M); 3 h AHS (B, F, J, and N); and 6 h AHS (C, G, K, and O). Localization of the Merlin truncations was also examined in larval wing imaginal discs (D, H, L, and P). All images of the imaginal epithelium are tangential optical sections of the apical-most focal plane. In S2 cells, mycMer+ (A–C) has a temporal and spatial pattern similar to that described previously for expressed wild type protein (McCartney and Fehon, 1996). Initially, Merlin is targeted to the plasma membrane (A); after 3 h, much of the protein is found in the cytoplasm in punctate cytoplasmic structures (B and C). The localization of mycMer+ in the imaginal epithelium showed characteristic localization to the apical plasma membrane and in the cytoplasm (D). The COOH-terminal truncation, mycMer1–600, localized to the plasma membrane but had no punctate cytoplasmic staining (E–H). Initially in S2 cells, much of the 350– amino acid NH2-terminal Merlin fragment was distributed throughout the cytosol (I and J); however, 6 h AHS, much of the protein is found at the plasma membrane (K). In an imaginal epithelia, this protein is found in the apical plasma membrane and throughout the cytoplasm (L). Deletion of the NH2-terminal domain in mycMer351–635 results in a protein that localizes to the plasma membrane in both cultured cells and imaginal epithelium (M–P). Bar, 5 μm.
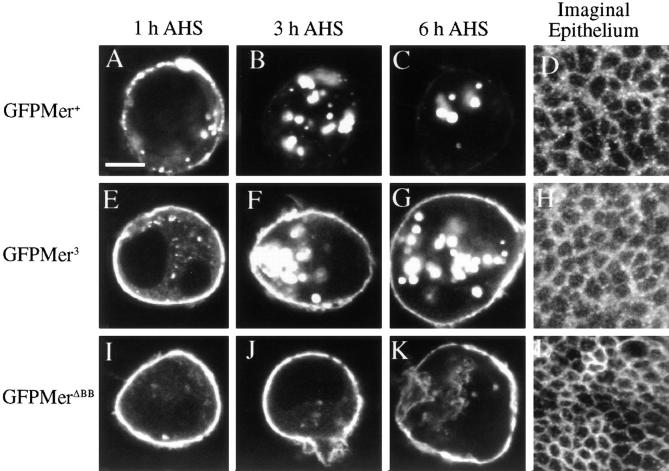
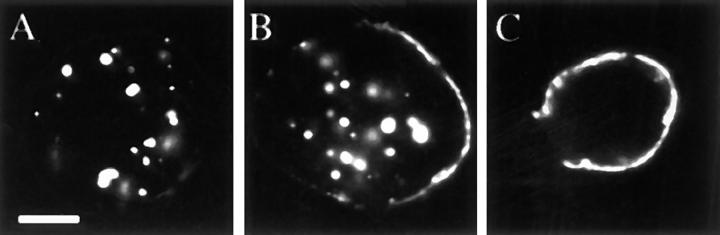
To assess the role of the Blue Box (BB) region in Merlin's subcellular localization, we examined the subcellular distribution of wild-type and mutant proteins fused to green fluorescent protein (GFP). Wild-type Merlin-GFP (GFPMer+) retains full wild-type rescue function; thus, the GFP moiety does not interfere with Merlin function (data not shown). In S2 cells, GFPMer3 remained associated with the membrane 3–5 h after heat shock (Fig. 4, E–G), unlike GFPMer+, which had all internalized by this time (Fig. 4, A–C). Removal or replacement of the entire BB resulted in proteins that were properly targeted to the plasma membrane but were not internalized (Fig. 4, I–K). Similarly, in imaginal epithelia, MerΔBB was exclusively plasma membrane associated (Fig. 4 L).
Figure 4.
Blue Box mutations are retained at the plasma membrane. Subcellular localization of Merlin Blue Box mutant proteins in S2 cells and wing imaginal epithelium: GFPMer+ (A–C), mycMer+ (D), GFPMer3 (E–G), mycMer3 (H), GFPMerΔBB (I– K), and mycMerΔBB (L). All GFP Merlin fusion proteins were visualized using the inherent fluorescence of the GFP moiety; Mer3 and MerΔBB were visualized in the imaginal disc epithelium using guinea pig anti-Merlin and a CY3-conjugated secondary antibody. For experiments in S2 cells, samples were collected from three time points: 1 h AHS (A, E, and I), 3 h AHS (B, F, and J), and 6 h AHS (C, G, and K). The subcellular localization of the Merlin BB mutant proteins was examined in larval wing imaginal epithelia (D, H, and L). GFPMer+ has a wild-type Merlin distribution in S2 cells and in tissue (A–D). Upon induction, GFPMer3 is targeted to the plasma membrane of S2 cells (E). Unlike GFPMer+, much of the GFPMer3 protein remains at the plasma membrane at later time points: 3 h AHS (F) and 6 h AHS (G). In tissue, mycMer3 displays both punctate and cytoplasmic localization with no discernible difference from wild type (H). MerΔBB is exclusively associated with the plasma membrane in both S2 cells (I–K) and imaginal disc tissue (L). Bar, 5 μm.
Characterization of In Vivo Function
To assay the in vivo function of the mutant Merlin proteins, we used the Gal4/UAS system (Brand and Perrimon, 1993) to express Merlin truncations in mutant and wild-type genetic backgrounds. In these studies, we addressed two questions: First, what is the minimal protein region required for Merlin function, and second, do any mutant Merlin proteins produce antimorphic (dominant-negative) phenotypes when expressed in vivo? In the genetic rescue experiments, UASMerlin constructs were driven under a ubiquitously expressing Gal4 enhancer trap (T80Gal4) in a Mer4 mutant background and tested for their ability to rescue Mer4 lethality. Complementation of the other lethal Merlin alleles (Mer1 and Mer2) was also performed with similar results (data not shown). To test for possible dominant activity of a Merlin truncation, we examined the nonmutant class flies from these same experiments for phenotypes similar to those expressed by Merlin mutants.
In the control experiments, we observed almost complete genetic rescue with the mycMer + transgene (Fig. 2 A). Surprisingly, even stronger genetic rescue was observed with a truncated Merlin protein missing the COOH-terminal 35 amino acid residues (mycMer1–600). This improved genetic rescue over mycMer + was consistently observed with independent transgenic lines, suggesting that the truncated form has increased in vivo function. We also observed partial rescue of Mer4 lethality with two even shorter NH2-terminal Merlin transgenes, mycMer1–350 and mycMer1–375 (Fig. 2 A). All of the rescued flies were phenotypically wild-type and did not possess any of the characteristic Mer3 phenotypes. Expression of mycMerΔBB and mycMerBBA failed to rescue Mer4 lethality (Fig. 2 A), indicating that this region has essential functions. In contrast, expression of mycMer3 partially rescued Mer4 lethality. However, unlike rescued flies from other experiments, all of the mycMer3 rescued flies expressed wing phenotypes similar to those expressed by flies carrying the original Mer3 allele. No rescue was observed with expression of any other Merlin truncation. Taken together, the results indicate that all essential Merlin functions reside in the NH2-terminal 350 amino acids of the protein.
In the same genetic experiments, nonmutant class siblings that carried ubiquitously expressing Mer transgenes were examined for dominant phenotypes. None of the NH2- or COOH-terminal deletions displayed any dominant phenotype in a Mer + background. In contrast, we observed dominant phenotypes resulting from expression of the Merlin BB mutant proteins. This dominant phenotype included a broadening of the wing blade, variably penetrant ectopic wing vein material (primarily around the second wing vein), anterior and posterior cross vein defects, and ectopic bristles or sense organs (Fig. 5, C and D and data not shown). The most striking quality of this dominant phenotype was the enlargement of the wing blade, resulting in a curvature of the wing surface. Furthermore, high levels of expression of the Merlin BB mutant proteins in the wing blade under two different Gal4 drivers (engrailedciBeGal4, apterousmd5 Gal4) resulted in an outheld wing phenotype associated with alterations in the wing hinge morphology (data not shown).
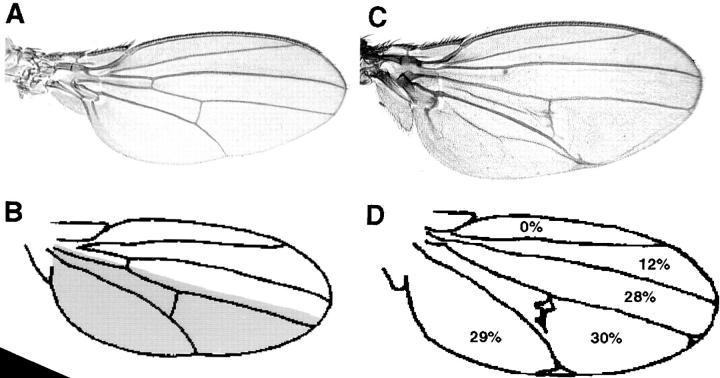
Figure 5.
Expression of MerΔBB results in an increase in the size of the wing blade. (A) Expression of UASMer + under the engrailed GAL4 driver. (C) Expression of two copies of UASMerΔBB under the engrailed GAL4 driver results in the expansion of the wing blade. Camera lucida drawings of mounted wings expressing UASMer + (B) and UASMerΔBB (D) under the engrailed GAL4 driver were made to compare the overall wing area (E) and to determine and compare the surface area of individual regions of the wing blade. The shaded area in B delineates the approximate domain of expression under this driver. Values shown in D represent the percent change in area in these regions of the wing blade when compared with wings expressing Mer +. The overgrowth phenotype observed in wings expressing UASMerΔBB is limited to the region of the wing where the construct is expressed (D). Loss of area in regions of the wing blade where the construct is not expressed may be due to a compensation mechanism. The drawing in D also displays several of the venation defects associated with expression of MerΔBB. (E) The overall wing area is increased in wings expressing Merlin Blue Box mutant proteins. The severity of this effect is dependent on the severity and the dose of the mutation. When the t test was applied to these values the difference in wing area between wings expressing wild-type Merlin and those expressing Merlin Blue Box mutants was found to be significant (P < 0.001).
The broadening of the wing blade we observed with expression of Merlin BB mutations is characteristic of mutations that cause increased cell proliferation in the wing (Mahoney et al., 1991; Boedigheimer and Laughon, 1993; Bryant et al., 1993). To examine this phenotype in more detail, we compared the surface area of wings expressing Merlin BB proteins or wild-type Merlin under the engrailed GAL4 driver. As expected of an overproliferation phenotype, we observed a significant increase in the total surface area of wings expressing BB mutant proteins (Fig. 5 E). The area of overgrowth was localized to the posterior half of the wing where the engrailed Gal4 specifically drives expression (Fig. 5 B, shaded area), although a decrease in surface area was observed between wing veins 2 and 3, a region anterior to the expression of the transgene. To determine whether the increase in area we observed was due to increased cell number or increased cell size, we measured the densities of wing hairs at several positions within the wing blade. (Each wing blade cell secretes a single hair.) The overall wing hair density in wings expressing BB Merlin proteins was indistinguishable from that in wings expressing a wild-type protein (data not shown), indicating that the broadening of the wing results from an increase in cell number rather than in cell size. As shown in the somatic mosaic analysis, loss of Mer function resulted in overproliferation of mutant cells. Similarly, ectopic expression of BB mutant proteins resulted in increased proliferation. These results suggest that the BB mutant proteins act in a dominant-negative manner and therefore interfere with the activity of wild-type protein.
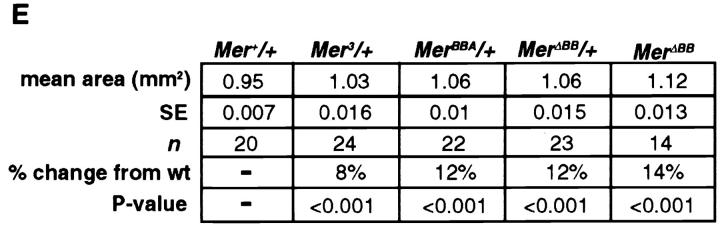
To confirm that the BB mutations have dominant-negative activity, we examined the modification of the dominant MerΔBB wing phenotype in response to alteration of endogenous gene dose. As would be expected for an antimorphic allele, dominant MerΔBB wing phenotypes were enhanced by a reduction of endogenous gene dose (Mer4/+) and were suppressed in response to an increase in the level of endogenous Merlin (+/+;P{cos Mer + }; Table I). These results confirm that Merlin BB proteins act as dominant-negative proteins.
Table I.
Genetic Characterization of MerlinΔBB
| Genotype* | n | Ectopic wing material‡ | Defects in cross veins§ |
|---|---|---|---|
| +/+; +/+ | 100 | 0 | 0/0 |
| +/+; Mer +/+ | 100 | 0 | 0/0 |
| +/+; MerΔBB/+ | 350 | 53 | 4/1 |
| Mer 4/+; MerΔBB/+ | 74 | 78 | 12/4 |
| +/+; MerΔBB/+;P[cosMer + ]/+ | 74 | 28 | 1/0 |
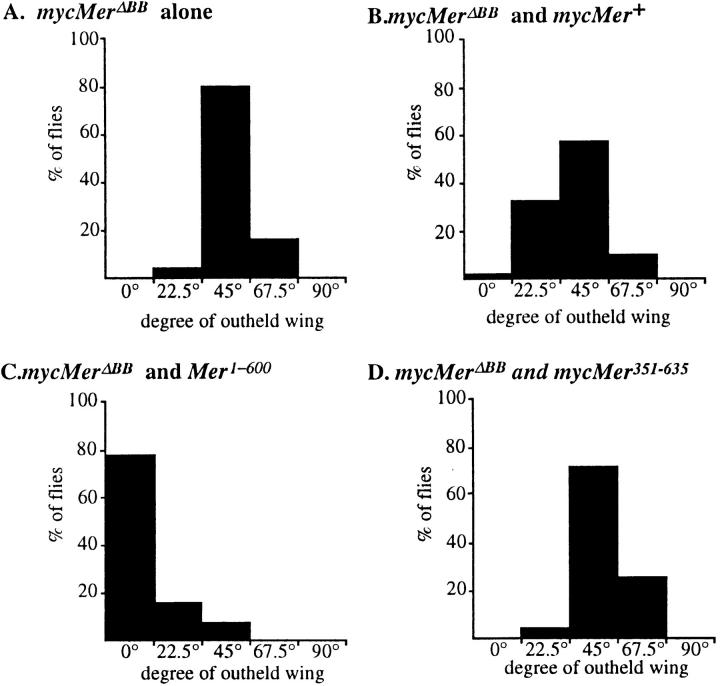
Based on the in vivo activity assay, mycMer1–600 appears to behave as an activated protein. An additional way to test this proposal is to determine whether mycMer1–600 is able to rescue the dominant-negative activity of the BB mutant proteins. We examined the BB phenotype of wings coexpressing MerΔBB and either wild-type Merlin (mycMer+) or the putative activated form (mycMer1–600). Flies expressing mycMerΔBB under the apterous Gal4 driver display wings held at an average angle of 45° from the body axis (Fig. 6 A). This phenotype appears to be caused by increased proliferation in the wing hinge region (data not shown). Coexpression of mycMer+ with mycMerΔBB resulted in a slight suppression in the degree of outheld wings (Fig. 6 B). However, when mycMer1–600 was coexpressed with the mycMerΔBB, we observed a dramatic suppression of the dominant BB wing phenotype (Fig. 6 C). In a control experiment, the COOH-terminal half of Merlin was coexpressed with mycMerΔBB, and no suppression of the phenotype was observed (Fig. 6 D). The ability of mycMer1–600 to suppress the dominant BB phenotype supports the idea that this is an activated form of the Merlin protein.
Figure 6.
Constitutively activated COOH-terminal truncation of Merlin suppresses the dominant MerΔBB phenotype. Flies coexpressing either wild-type or truncated forms of Merlin under the apterousmd52 GAL4 (apGal4) enhancer trap were examined for suppression of the MerΔBB outheld wing phenotype. Flies were placed into one of five classes of wings, each corresponds to the degree of which the wings were outheld from the central body axis: 0°, 22.5°, 45°, 67.5°, and 90°. The y-axis shows the percent of flies within a class. When outcrossed, flies with the apGal4 UASMerΔBB genotype hold their wings out at an average angle of 45° (A). Flies expressing both UASMer + and UASMerΔBB show only a slight difference from the control (B). However, expression of UASMer1–600 greatly suppresses the outheld wing phenotype (C); with two independent insertion lines of mycMer1–600, we observed almost complete suppression. As a negative control, the expression of the nonrescuing UASMer351–635 has a profile similar to the outcrossed stock (D).
Competition Experiments
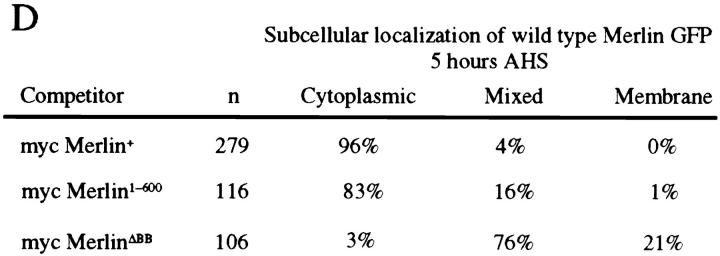
To directly observe the dominant-negative behavior of mycMerΔBB, we examined the localization of wild-type Merlin protein in the presence of mycMerΔBB. We cotransfected Drosophila S2 cells with GFPMer+ and either mycMer+ or mycMerΔBB and examined the localization of both proteins. In S2 cells cotransfected with either the mycMer+ or mycMer1–600 competitor proteins, the majority of GFPMer+ in these cells was associated with characteristic punctate cytoplasmic structures 5 h after induction (Fig. 7, A and D) as was shown earlier (Figs. 3 C and 4 C). However, in almost all S2 cells cotransfected with mycMerΔBB competitor, wild-type GFPMerlin remained at the plasma membrane (Fig. 7, B–D). This result suggests that localization to the plasma membrane is not sufficient for Merlin activity and that mycMerΔBB interferes with wild-type Merlin function by causing it to accumulate at the plasma membrane in a nonfunctional state.
Figure 7.
MerΔBB alters subcellular distribution of wild-type Merlin in S2 cells. The localization of GFPMer+ was examined in S2 cells cotransfected with either mycMer+ or mycMerΔBB 5 h AHS. Cells were stained with an anti-myc antibody to confirm expression of the second protein (data not shown). In S2 cells cotransfected with a mycMer+ (A) or mycMer1–600 (data not shown), all of the GFPMer+ protein localizes to cytoplasmic structures 5 h AHS. However, in S2 cells cotransfected with mycMerΔBB, a considerable fraction of the GFPMer+ accumulates at the plasma membrane (B and C). In D, the data from this experiment are summarized. Three classes of GFPMer+ localization were observed 5 h AHS. Bar, 5 μm.
Discussion
Although the protein 4.1 family has typically been characterized as a group of membrane–skeletal proteins, recent studies have revealed a diversity of function, some of which may not be primarily related to the canonical membrane–cytoskeletal linker function (for review see McCartney and Fehon, 1997). In addition, recent genetic studies in particular have implicated protein 4.1 family proteins in mediating the intercellular interactions that regulate cell proliferation. For example, mutations in Drosophila expanded, a divergent member of the protein 4.1 superfamily that localizes to the adherens junction, result in overproliferation of the cells that form the adult wings (Boedigheimer and Laughon, 1993). Previous experiments have shown that proliferation in this tissue is controlled by cell–cell interactions, mediated by at least three known signal transduction pathways, wingless, decapentapelegic, and Epidermal Growth Factor (Edgar and Lehner, 1996). Studies of Drosophila Coracle, a protein 4.1 homologue, indicate that it is associated with the septate junction, a structure that has previously been implicated in the regulation of cell growth (Woods and Bryant, 1993), and that coracle mutations interact genetically with a hypermorphic mutation of the Drosophila Epidermal Growth Factor-receptor homologue (Fehon et al., 1994). In addition, the human NF2 tumor-suppressor gene Merlin clearly has a role in maintaining proper regulation of cell proliferation.
In NF2, as well as in other diseases involving tumor-suppressor genes, a genetically heterozygous individual experiences somatic loss of the second copy of the gene in certain tissues (called loss of heterozygosity or LOH), resulting in the formation of tumors in those tissues. These individuals are then genotypically mosaic with respect to the tumor-suppressor gene. Although NF2 was originally characterized as a disease of the central nervous system (CNS), somatic loss of NF2 is believed to occur in the Schwann cells that form the myelin sheath surrounding CNS axons. The glial cells form a polarized epithelium that isolates the axons from the surrounding environment. Thus, although Merlin is expressed in the Drosophila CNS (McCartney and Fehon, 1996), we have concentrated our analysis of Drosophila Merlin function in epithelial tissues, which also express Merlin and are readily amenable to experimental analysis.
We simulated loss of heterozygosity in Drosophila using somatic mosaic analysis in which the effects of homozygous loss of gene function in a patch of cells (a clone) were examined in an otherwise heterozygous background. Using this system, we demonstrated that loss of Merlin function during larval development results in a two- to threefold increase in proliferation relative to the wild-type sister clone. This hypertrophied tissue is morphologically normal, indicating that loss of Merlin function specifically affects proliferation rather than differentiation or morphogenesis. This conclusion is supported by the proliferation defect we observed in tissues overexpressing Merlin Blue Box mutant proteins. Interestingly, as with human NF2, the overproliferation we observed seems moderate and does not result in the extreme hyperplasia observed in other Drosophila “tumor suppressors” (Mahoney et al., 1991; Bryant et al., 1993; Xu et al., 1995) or in the malignant transformation associated with many human cancers. As we demonstrated in this study, the degree of overproliferation observed was dependent on the mutant allele of Merlin examined. Similarly, individuals with NF2 display a range of tumor growth rates that may reflect differences in their Merlin genotypes. In general, nonsense mutations in human NF2 result in more severe phenotypes than missense mutations. Alternatively, it is possible that some of the observed differences in the severity of NF2 is due to second site modifying loci. Currently, we are using the genetic techniques available in the Drosophila system to identify second site modifiers of Merlin mutant phenotypes.
To understand how Merlin functions in the cell and how these functions relate to the regulation of proliferation, we identified important functional domains within Merlin required for its activity and proper subcellular localization. Genetic rescue experiments using a series of NH2- and COOH-terminal truncations of Merlin indicate that all essential Merlin functions reside in the CNTR. This result, though somewhat surprising, is consistent with recent results from other protein 4.1 superfamily members. Studies of Drosophila Coracle, a protein 4.1 homologue, indicate that the CNTR of this protein performs several essential functions within the septate junction (Ward et al., 1998). In addition, recent studies of the ERM proteins indicate that sequences COOH-terminal to the CNTR may play a primarily regulatory role, a model that also seems to apply to Merlin (see below). While there are clearly family members that have a more complex functional organization, taken together these results indicate that the CNTR of many protein 4.1 superfamily members acts as an independent functional domain. The diversity of identified protein 4.1 superfamily members suggests that during the course of evolution, the essential function of this domain, probably to serve as a targeting sequence to a particular region of the cell membrane, has been adapted repeatedly to different proteins (Fehon et al., 1997).
In humans, many of the mutations associated with NF2 are predicted to generate truncated forms of Merlin as a result of nonsense mutations (Jacoby et al., 1996; Welling et al., 1996). Given the results presented here, some of these truncated products should retain partial function, just as the mycMer1–350 allele appears to be partially functional. Although it is possible that this apparent discrepancy results from differences in human and Drosophila Merlin function, expression of human NF2 is sufficient for genetic rescue of lethal Drosophila Merlin alleles, indicating that their functions are conserved (McCartney, B., V. Ramesh, and R. Fehon, unpublished observations). Unlike other protein 4.1 family members (Algrain et al., 1993; Ward et al., 1998), the CNTR of Merlin is poorly targeted to the plasma membrane, thereby decreasing functional protein levels. Thus, even when overexpressed, the CNTR provides only partial genetic rescue. It is therefore not surprising that human NF2 alleles that truncate COOH-terminally to the CNTR produce severe phenotypes even though the proteins they encode may retain all essential Merlin functions. Further tests need to be performed to confirm this hypothesis. If correct, stabilization of mutant NF2 products containing an intact CNTR (but lacking COOH-terminal targeting sequences) could be an effective therapeutic strategy for some patients afflicted with NF2 because it would increase the levels of a partially functional Merlin protein.
We have shown that mycMer1–600 acts as an activated protein that displays greater rescuing activity and suppresses the phenotypes produced by a dominant-negative form of Merlin. Pulsed expression of Mer1–600 in S2 cultured cells under an inducible promoter indicates that the levels of expression and the stability of this protein are not significantly different from wild-type Merlin (data not shown). Thus, the simplest explanation for this phenomenon is that the COOH-terminal region contains a domain important for reducing the activity of Merlin. Recent studies demonstrate that ERM protein activity is regulated by a Rho-based signaling pathway (Takaishi et al., 1995; Hirao et al., 1996; Mackay et al., 1997). Rho-Kinase has been shown to phosphorylate a conserved threonine within the COOH-terminal 35 amino acids of all ERM proteins and thereby regulate the conformational change that occurs during a putative transition from an inactive to an active state (Matsui et al., 1998). Although the COOH-terminal 35 amino acids of Merlin are divergent from those of the ERM proteins, the threonine residue phosphorylated by Rho-Kinase is conserved (McCartney and Fehon, 1996), suggesting that Merlin could use a similar mechanism for switching between an inactive and an active state. Creation of an activated Merlin protein by the removal of the COOH-terminal regulatory region is consistent with this notion, but further studies are required to confirm this regulatory role. It is interesting to note in this regard that an isoform of human and mouse NF2 that alters the COOH terminus of Merlin has been identified (isoform II; Haase et al., 1994; Pykett et al., 1994). Recent studies indicate that isoform II, which retains the conserved threonine residue, is less functional than isoform I in suppressing growth (Sherman et al., 1997), consistent with the idea that the COOH terminus of Merlin has regulatory functions.
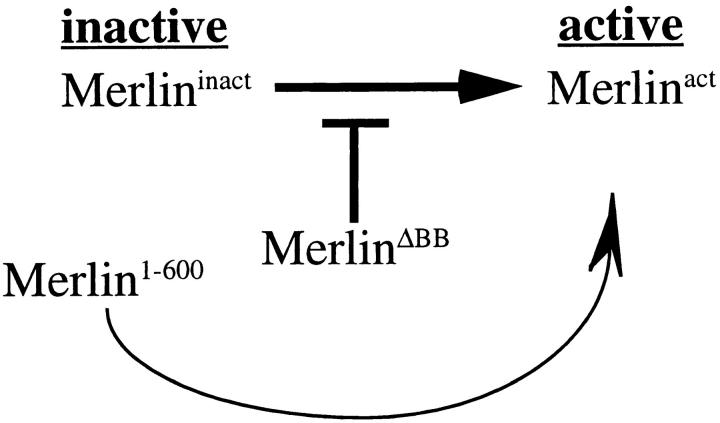
Two Merlin mutations described here, the dominant-negative MerΔBB and the activated Mer1–600, are found primarily at the plasma membrane. This apparently contradictory result, that two mutant forms of Merlin with opposite functions both localize to the plasma membrane, indicates that localization to the plasma membrane is not sufficient for Merlin function. We propose that Merlin normally undergoes an activation process that occurs at the cytoplasmic face of the plasma membrane (Fig. 8). In our model, not only is MerΔBB refractory to activation, but it also inhibits activation of endogenous wild-type Merlin, thus causing wild-type Merlin to accumulate at the plasma membrane in a nonfunctional state and producing a dominant phenotype. In contrast, Mer1–600 exists in a constitutively activated state, thereby evading the block presented by MerΔBB and suppressing the dominant BB mutant phenotype. The two functional states of wild-type Merlin may represent two distinct conformations of the protein, as has been shown for ERM proteins (Berryman et al., 1995; Bretscher et al., 1995). Alternatively, Merlin may associate with two distinct binding partners at the membrane. In either case, one state may serve to associate Merlin with the membrane, and the second may be required for Merlin activation.
Figure 8.
Model of Merlin activation at the cytoplasmic face of the plasma membrane. Wild-type Merlin exists in two functional states at the plasma membrane, inactive and active. While MerlinΔBB cannot itself be activated, it inhibits the transition of wild-type Merlin from an inactive to an active state. However, Merlin1–600, being constitutively active, bypasses the requirement for activation and therefore the block presented by MerlinΔBB, thus effectively suppressing the dominant Blue Box mutant phenotypes.
In this report, we have shown that Merlin is required for the regulation of proliferation in Drosophila epithelial cells and that it functions at the plasma membrane. The mechanism by which Merlin functions to regulate cellular proliferation is still unclear. Recent evidence suggests that other proteins of the protein 4.1 superfamily operate by organizing functional regions within the plasma membrane (Helander et al., 1996; Ward et al., 1998). Merlin may function in a similar fashion by coordinating interactions of transmembrane signaling molecules and cytoplasmic factors that regulate cellular proliferation. We now plan to elucidate the signaling pathways that Merlin regulates as well as identify genes involved in its regulation.
Acknowledgments
We would like to thank Cathy Laurie for use of the camera lucida, Dan Kiehart and Kevin Edwards for providing the GFP plasmid, and the Bloomington stock center for fly stocks. We thank Team Injection for help with the generation of transgenic Drosophila lines used in this study, Stephen White for technical assistance, and our colleagues in the Fehon lab for suggestions and discussions.
This work was supported by National Institutes of Health grant (R01-NS34783) to R.G. Fehon, National Neurofibromatosis Foundation Young Investigator Awards to D.R. LaJeunesse and B.M. McCartney, and a National Research Service Award postdoctoral fellowship (F32-NS10224) to D.R. LaJeunesse.
Abbreviations used in this paper
AEL
after egg laying
AHS
after heat shock
BB
Blue Box region
BBA
Merlin with the seven Blue Box residues changed to alanine
CNS
central nervous system
CNTR
conserved NH2-terminal region
ΔBB
Merlin with Blue Box region removed
ERM
ezrin-radixin-moesin
FLP
yeast 2 micron Flipase enzyme
FRT
Flipase Recognition Target site
GFP
green fluorescent protein
Mer
Merlin
NF2
Neurofibromatosis type 2
S2 cells
Schneider line 2 cells
UAS
upstream activation sequences of yeast Gal4 transcription factor
Footnotes
Address all correspondence to Richard G. Fehon, B361 LSRC, Research Drive, Duke University, Durham, NC 27708-1000. Tel.: (919) 613-8192. Fax: (919) 613-8177. E-mail: rfehon@acpub.duke.edu
References
- Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane–cytoskeletal linker. J Cell Biol. 1993;120:129–139. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman M, Gary R, Bretscher A. Ezrin oligomers are major cytoskeletal components of placental microvilli: a proposal for their involvement in cortical morphogenesis. J Cell Biol. 1995;131:1231–1242. doi: 10.1083/jcb.131.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedigheimer M, Laughon A. expanded: a gene involved in the control of cell proliferation in imaginal discs. Development (Camb) 1993;118:1291–1301. doi: 10.1242/dev.118.4.1291. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Camb) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Gary R, Berryman M. Soluble ezrin purified from placenta exists as stable monomers and elongated dimers with masked COOH-terminal ezrin-radixin-moesin association domains. Biochemistry. 1995;34:16830–16837. doi: 10.1021/bi00051a034. [DOI] [PubMed] [Google Scholar]
- Brown MA. Tumor suppressor genes and human cancer. Adv Genet. 1997;36:45–135. doi: 10.1016/s0065-2660(08)60307-7. [DOI] [PubMed] [Google Scholar]
- Bryant, P.J., K.L. Watson, R.W. Justice, and D.F. Woods. 1993. Tumor suppressor genes encoding proteins required for cell interactions and signal transduction in Drosophila. Dev. Suppl. 239–249. [PubMed]
- Edgar BA, Lehner CF. Developmental control of cell cycle regulators—a fly's perspective. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MAT, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. . Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Dawson IA, Artavanis-Tsakonas S. A Drosophila homologue of membrane-skeleton protein 4.1 is associated with septate junctions and is encoded by the coraclegene. Development (Camb) 1994;120:545–557. doi: 10.1242/dev.120.3.545. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Oren T, LaJeunesse DR, Melby TE, McCartney BM. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42genes using a simple and efficient reverse-genetic method. Genetics. 1997;146:245–252. doi: 10.1093/genetics/146.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck Z, Gary R, Bretscher A. Moesin, like ezrin, colocalizes with actin in the cortical cytoskeleton in cultured cells, but its expression is more variable. J Cell Sci. 1993;105:219–231. doi: 10.1242/jcs.105.1.219. [DOI] [PubMed] [Google Scholar]
- Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Agosti C, Xu L, Pinney D, Beauchamp R, Hobbs W, Gusella J, Ramesh V. The merlin tumor suppressor localizes preferentially in membrane ruffles. Oncogene. 1996;13:1239–1247. [PubMed] [Google Scholar]
- Haase VH, Trofatter JA, MacCollin M, Tarttelin E, Gusella JF, Ramesh V. The murine NF2 homologue encodes a highly conserved merlin protein with alternative forms. Hum Mol Genet. 1994;3:407–411. doi: 10.1093/hmg/3.3.407. [DOI] [PubMed] [Google Scholar]
- Helander T, Carpen O, Turunen O, Kovanen PE, Vaheri A, Timonen T. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 1996;382:265–268. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Tsukita S. Regulation mechanism of ERM (ezrin/radixin/ moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby LB, MacCollin M, Barone R, Ramesh V, Gusella JF. Frequency and distribution of NF2mutations in schwannomas. Genes Chromosomes Cancer. 1996;17:45–55. doi: 10.1002/(SICI)1098-2264(199609)17:1<45::AID-GCC7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Esch F, Furthmayr H, Hall A. Rho- and rac-dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: an essential role for ezrin/radixin/moesin proteins. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney PA, Weber U, Onofrechuk P, Biessmann H, Bryant PJ, Goodman CS. The fattumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67:853–868. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, Fehon RG. Distinct cellular and subcellular patterns of expression imply distinct functions for the Drosophilahomologues of moesin and the neurofibromatosis 2 tumor suppressor, merlin. J Cell Biol. 1996;133:843–852. doi: 10.1083/jcb.133.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, B.M., and R.G. Fehon. 1997. The ERM family of proteins and their roles in cell-cell interactions. In Cytoskeletal-Membrane Interactions and Signal Transduction. P. Cowin and M.W. Klymkowsky, editors. R.G. Landes Bioscience, Austin, TX. 200–210.
- McClatchey AI, Saotome I, Ramesh V, Gusella JF, Jacks T. The Nf2tumor suppressor gene product is essential for extraembryonic development immediately prior to gastrulation. Genes Dev. 1997;11:1253–1265. doi: 10.1101/gad.11.10.1253. [DOI] [PubMed] [Google Scholar]
- Pykett MJ, Murphy M, Harnish PR, George DL. The neurofibromatosis 2 (NF2) tumor suppressor gene encodes multiple alternatively spliced transcripts. Hum Mol Genet. 1994;3:559–564. doi: 10.1093/hmg/3.4.559. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Suan K, Demczuk S, Desmaze C, Plougastel B, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neurofibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Sainio M, Zhao F, Heiska L, Turunen O, den Bakker M, Zwarthoff E, Lutchman M, Rouleau MA, Jaaskelainen J, Vaheri A, Carpen O. Neurofibromatosis 2 tumor suppressor protein colocalizes with ezrin and CD44 and associates with actin-containing cytoskeleton. J Cell Sci. 1997;110:2249–2260. doi: 10.1242/jcs.110.18.2249. [DOI] [PubMed] [Google Scholar]
- Sherman L, Xu HM, Geist RT, Saporito-Irwin S, Howells N, Ponta H, Herrlich P, Gutmann DH. Interdomain binding mediates tumor growth suppression by the NF2 gene product. Oncogene. 1997;15:2505–2509. doi: 10.1038/sj.onc.1201418. [DOI] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kameyama T, Tsukita S, Tsukita S, Takai Y. Translocation of activated Rho from the cytoplasm to membrane ruffling area, cell-cell adhesion sites and cleavage furrows. Oncogene. 1995;11:39–48. [PubMed] [Google Scholar]
- Takeuchi K, Sato N, Kasahara H, Funayama N, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Perturbation of cell adhesion and microvilli formation by antisense oligonucleotides to ERM family members. J Cell Biol. 1994;125:1371–1384. doi: 10.1083/jcb.125.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev Biol. 1987;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Eldridge R, Kley N, Menon AG, Pulaski K, Haase H, Ambrose CM, Munroe D, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- Turunen O, Wahlström T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward REI, Lamb RS, Fehon RG. A conserved functional domain of DrosophilaCoracle is required for localization at the septate junction and has membrane organizing activity. J Cell Biol. 1998;140:1463–1474. doi: 10.1083/jcb.140.6.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welling DB, Guida M, Goll F, Pearl DK, Glasscock ME, Pappas DG, Linthicum FH, Rogers D, Prior TW. Mutational spectrum in the neurofibromatosis type 2 gene in sporadic and familial schwannomas. Hum Genet. 1996;98:189–193. doi: 10.1007/s004390050188. [DOI] [PubMed] [Google Scholar]
- Woods DF, Bryant PJ. Apical junctions and cell signalling in epithelia. J Cell Sci. 1993;17(Suppl.):171–181. doi: 10.1242/jcs.1993.supplement_17.25. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin G. Analysis of genetic mosaics in developing and adult Drosophilatissues. Development (Camb) 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila latsgene encodes a putative protein kinase. Development (Camb) 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]