Molecular Bases for the Recognition of Tyrosine-based Sorting Signals (original) (raw)
Targeting of transmembrane proteins to different compartments of the endocytic and late (post-Golgi) secretory pathways is largely dependent upon sorting signals contained within the cytosolic domains of the proteins (reviewed in reference 19, 24). The signals are thought to interact with specific recognition molecules, which are components of the machinery involved in the formation of membrane-bound transport intermediates (e.g., coated vesicles; reference 2). The interaction of signals with their recognition molecules is thus considered to be the key event leading to selective recruitment of cargo transmembrane proteins into the nascent transport intermediates. Studies over the past 30 years have provided extensive evidence for the occurrence of this basic mechanism of protein sorting at multiple sites within the cell. However, the molecular details of the signal-recognition event have only recently begun to be unraveled. This mini-review will focus on recent progress in the elucidation of the molecular bases for the recognition of a subset of sorting signals, referred to as tyrosine-based signals, by a family of adaptor protein (AP)1 complexes.
Tyrosine-based Signals: A Degenerate Family
Tyrosine-based signals constitute a family of degenerate motifs minimally defined by the presence of a critical tyrosine residue (see reference 22 and references therein). Most tyrosine-based signals conform to the consensus motifs YXXØ (Y is tyrosine, X is any amino acid, and Ø is an amino acid with a bulky hydrophobic side chain; reference 5) or NPXY (N is asparagine and P is proline; reference 6). YXXØ signals are currently the best understood from a structural standpoint and thus will be the primary subject of our discussion. YXXØ signals can be found within the cytosolic domains of all types of transmembrane proteins, including type I (e.g., lamp-1), type II (e.g., the transferrin receptor), and multi-spanning (e.g., CD63). They can be most easily identified within short cytosolic tails (i.e., <35 amino acid residues), although they have also been shown to exist within the large cytosolic domains of some signaling receptors (e.g., the epidermal growth factor receptor) and retroviral envelope glycoproteins (e.g., HIV-1 gp41). The presence of a sequence conforming to the YXXØ motif within a large cytosolic domain, however, is not necessarily predictive of sorting information since signals must be presented in an appropriate context to be active. In mammalian cells, virtually all YXXØ signals mediate rapid internalization from the cell surface. Some YXXØ signals can additionally mediate lysosomal targeting, localization to specialized endosomal-lysosomal organelles such as antigen-processing compartments, delivery to the basolateral plasma membrane of polarized epithelial cells or localization to the TGN (reviewed in reference 19, 22, 24). The multiple functions of YXXØ signals raise the question of how the same type of signal can mediate sorting to different cellular compartments. A hypothesis that has been put forth to explain the various roles of YXXØ signals is that they must interact selectively with a family of recognition molecules associated with different sites of protein sorting. Recent findings that YXXØ signals are capable of interacting with several AP complexes provide a framework for testing the validity of this hypothesis.
Recognition of YXXØ Signals by the μ2 Subunit of AP-2
Glickman et al. (14) pioneered the use of in vitro affinity-binding methods to study the interactions of the cytosolic tails of membrane receptors with AP complexes. In the course of these studies, they demonstrated a tyrosine-dependent interaction of the cytosolic tail of the cation-independent mannose 6-phosphate receptor with AP-2, a plasma membrane, clathrin-associated complex composed of two large subunits (α and β2), one medium subunit (μ2), and one small subunit (σ2) (Fig. 1 A). Generalization of this biochemical approach to other transmembrane proteins, however, was hampered by the low affinity of the interactions in vitro. Further progress required the development of more sensitive protein interaction assays based on techniques such as the yeast two-hybrid system and surface plasmon resonance spectroscopy. The use of the yeast two-hybrid system, for instance, was instrumental in the identification of μ2 as a recognition molecule for YXXØ signals (29). Mutational and combinatorial analyses demonstrated that the Y residue is essential for binding to μ2 and cannot be effectively substituted even by the structurally related phenylalanine or phosphotyrosine residues (4, 28, 36). Leucine is the preferred residue at the Ø position, although isoleucine, phenylalanine, methionine, and, to a lesser extent, valine, are tolerated (4, 27, 28). Many residues are permitted at the X positions, although arginine and proline are favored at the second X position (4, 27, 28). All of these preferences are consistent with the requirements for optimal function of YXXØ signals in rapid internalization, and thus provide strong correlative evidence for the physiological role of YXXØ-μ2 interactions.
Figure 1.
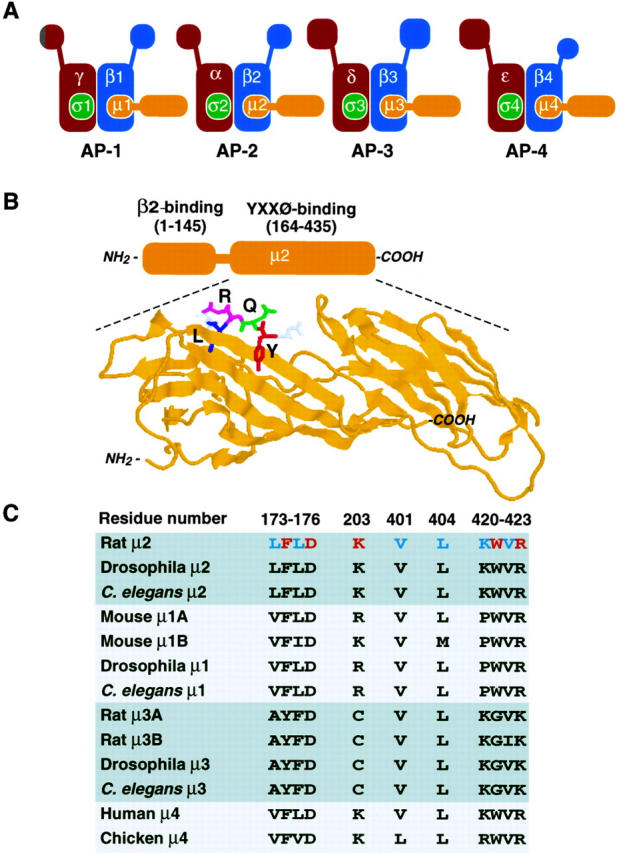
(A) Schematic representation of AP complexes. Each AP complex consists of two large subunits (γ/α/δ/ε and β1-4), one medium subunit (μ1-4), and one small subunit (σ1-4). Some subunits exist in more than one isoform. (B) Bipartite structure of μ2 (approximate residue numbers indicated in parentheses) and ribbon representation of its YXXØ-binding domain complexed to a DYQRLN peptide (adapted from reference 32; PDB accession code 1BXX). (C) Residues of rat μ2 involved in interactions with YXXØ signals and corresponding residues in other members of the AP μ family. Y- and Ø-binding residues are indicated in red and blue, respectively.
Structure-function analyses of μ2 have established that this polypeptide has a bipartite structure with the NH2-terminal third of the molecule (amino acid residues ∼1–145) being involved in assembly with β2, and the remaining two-thirds (amino acid residues ∼164–435) in interactions with YXXØ signals (1) (Fig. 1 B). In a landmark study, David Owen and Philip Evans (32) have recently solved the crystal structure of the YXXØ-binding domain of μ2 complexed to peptides containing either the YQRL signal from the protein TGN38 or the YRAL signal from the epidermal growth factor receptor. The YXXØ-binding domain of μ2 has a banana-shaped structure consisting of 16 β-sheet strands arranged into two subdomains (Fig. 1 B). YXXØ signals bind in an extended conformation (rather than as a tight turn, as was previously believed) to a region of the molecule having pockets for both the Y and Ø residues. This mode of interaction, resembling a two-pronged plug fitting into a two-holed socket, is reminiscent of that of phosphotyrosine-containing motifs with SH2 domains (40), although the topographic features of the binding sites and the details of the interactions differ considerably. The aromatic ring of the critical Y residue is involved in hydrophobic interactions with μ2 residues F174 and W421, as well as stacking on the guanidinium group of R423. In addition, the phenolic hydroxyl group of the Y residue is engaged in a network of hydrogen bonds with D176, K203, and R423 of μ2 (Y-binding residues are indicated in red in Fig. 1 C; reference 32). These characteristics of the Y-binding pocket explain why phenylalanine and phosphotyrosine residues substitute poorly or not at all for tyrosine residues in the signals: phenylalanine residues would be unable to establish hydrogen bonds with residues at the bottom of the pocket, while phosphotyrosine residues would be too bulky to fit into the pocket and would elicit electrostatic repulsion by D176. Residues lining the Ø pocket include L173, L175, V401, L404, V422, and the aliphatic portion of K420 (Ø-binding residues are indicated in blue in Fig. 1 C; reference 32). The hydrophobicity and flexibility of the side chains of these residues allow accommodation of different bulky hydrophobic side chains at the Ø position, with leucine providing the best fit. Although interactions through the Y and Ø residues provide the main means of attachment of signals to μ2, specific X residues at positions between the Y and Ø residues may contribute additional contact points. For example, the R residue at the second X position of the YQRL signal is engaged in hydrophobic interactions with W421 and I419 and hydrogen bonding with K420 thus explaining the preference for R at this position (4, 27, 28). Neither NPXY-type signals (6) nor dileucine-based signals (another type of signal having a critical pair of bulky hydrophobic residues; reference 17, 21) can be accommodated in the YXXØ-binding site of μ2 (32), in agreement with the failure to isolate peptides conforming to these motifs in combinatorial screens (4, 27), as well as with the inability of these signals to compete with YXXØ signals for the sorting machinery in vivo (23, 42). In fact, recent studies have shown that NPXY and dileucine-based signals bind to other recognition molecules, namely the terminal domain of clathrin (18) and the β subunits of AP-1 and AP-2 (15, 34), respectively.
Interactions of YXXØ Signals with Other AP μ Subunits
The finding that the μ2 subunit of AP-2 interacts with YXXØ signals raised the possibility that analogous subunits of other AP complexes could similarly function in recognition of YXXØ. To date, three additional complexes structurally related to AP-2 have been described in mammals: AP-1, AP-3, and AP-4 (Fig. 1 A). Each of these AP complexes contains a μ subunit that displays significant homology to μ2 over the entire sequence. μ1A (formerly called μ1; reference 25) is a component of the AP-1 complex in most cell types, whereas a closely related isoform, μ1B, may be a subunit of this complex in polarized epithelial and glandular cells (30). μ3A and μ3B are alternative components of AP-3 (10, 37, 38); μ3A is widely expressed, whereas μ3B expression is mainly restricted to cells of neuronal origin (33). Finally, μ4 (originally known as μ-ARP2; reference 41) is a subunit of the recently described AP-4 complex (9). Sequence alignments indicate that most of the μ2 residues directly involved in interactions with the Y and Ø residues of YXXØ signals are conserved in other AP μ family members (Fig. 1 C). Indeed, μ1A, μ1B, μ3A, and μ3B have all been shown to interact with YXXØ signals, albeit with lower affinity relative to μ2 (10, 27–30, 34, 39). The conservation of Y- and Ø-binding residues also extends to μ4, as well as to AP μ orthologs from nonmammalian organisms (Fig. 1 C). This suggests that these molecules may also be capable of recognizing YXXØ signals.
The identification of a family of proteins that interact with YXXØ signals supports the hypothesis that the functional specificity of these signals may be dictated by their selective interaction with different recognition molecules. As mentioned above, μ2 tolerates many different amino acid side chains surrounding the critical Y and Ø residues, although it prefers arginine at the second X position of the YXXØ signal (4, 27, 28). Similar analyses have revealed that μ1A and μ3A prefer non-polar and acidic residues, respectively, at that position (27). Although the functional significance of the μ1A preferences is unclear, μ3A preferences are suggestive of a role in lysosomal targeting since the signals of several proteins localized to lysosomes and lysosome-related organelles (e.g., CD63, lamp-2a, and GMP-17) contain acidic residues at positions adjacent to the tyrosine residue.
Physiological Roles of YXXØ-μ Subunit Interactions
Having just identified a family of YXXØ-recognition molecules, an important next question that needs to be addressed is: what sorting events are mediated by interaction of YXXØ signals with each of these molecules?
AP-1 has been localized mainly to the TGN at steady state, where it is thought to mediate transport of lamp-1 and mannose 6-phosphate receptors to compartments of the endosomal-lysosomal system (13, 16). Recent studies, however, have raised the possibility that AP-1 may be involved in protein sorting to the basolateral plasma membrane of polarized epithelial cells (12, 31).
As the only AP complex localized to the plasma membrane, AP-2 is an obvious candidate for mediating rapid internalization through recognition of YXXØ signals. Recently, Nesterov et al. have provided compelling evidence for a role of μ2 in this process using a dominant negative genetic approach (26). These investigators constructed a μ2 variant with mutations in D176 and W421, which are critical elements of the YXXØ-binding site (Fig. 1 C). This mutant μ2 was unable to bind YXXØ signals but competed with endogenous μ2 for incorporation into the AP-2 complex. Interestingly, overexpression of mutant μ2 inhibited internalization of the transferrin receptor (26), which is known to be mediated by the YXXØ-type signal YTRF (7).
The intracellular localization of the AP-3 complex is not known with certainty, although published evidence suggests an association with endosomes and/or the TGN (8, 10, 37, 38). Evidence for a role of AP-3 in sorting mediated by YXXØ signals has recently been obtained from the analysis of AP-3–deficient cells. These cells were either generated by using an antisense RNA methodology (20) or derived from two patients with Hermansky-Pudlak syndrome carrying mutations in the AP-3 β3A subunit (11). In both cases, the AP-3 deficiency resulted in increased routing of YXXØ-containing, lysosomal membrane proteins through the plasma membrane, thus suggesting a function for AP-3 in YXXØ-mediated targeting to lysosomes. In contrast, the trafficking of non-lysosomal membrane proteins having YXXØ signals (e.g., the transferrin receptor) was not noticeably altered (11). This differential effect, which is consistent with the preference of the AP-3 μ3A subunit for YXXØ signals found in lysosomal membrane proteins (11, 27, 39), lends support to the notion that selective interaction with AP complexes underlies the functional specificity of YXXØ signals. The fact that a substantial fraction of lysosomal membrane proteins are still targeted to lysosomes in AP-3–deficient cells (11, 20) suggests that other AP complexes may provide alternative means of delivery to lysosomes. Perhaps this is a function of AP-1, or of the recently described AP-4 complex, which appears to be localized to the TGN or a neighboring compartment (9).
In conclusion, the hypothesis advanced to explain the involvement of YXXØ signals in multiple sorting events can now be made more explicit: YXXØ signals are recognized with characteristic preferences by the medium (μ) subunits of several AP complexes. The factors that determine the fidelity of sorting processes in vivo, however, remain poorly understood. First, although each μ subunit displays preferences for certain X and Ø residues, there is nonetheless a significant overlap in sequence specificity (27). Contextual factors such as the position of the signal within the cytosolic domain (35), the oligomeric state of the transmembrane protein (3), and the presence of other signals in the cytosolic domain, may contribute to differential interactions with the AP complexes. Second, there still may be additional YXXØ-binding proteins to be discovered. As discussed above, μ4 is a likely candidate for one such molecule. Finally, transmembrane proteins moving along trafficking pathways may meet the AP complexes sequentially rather than simultaneously. This means that the trajectory followed by a protein, as well as potential biochemical modifications along the way, may determine which interactions actually take place. Further research will be needed to assess the contribution of these factors to the selectivity of sorting by YXXØ signals. With a solid molecular foundation now in place, however, we can anticipate rapid progress toward the decipherment of this protein sorting code.
Acknowledgments
We thank Jennifer Lippincott-Schwartz, Mickey Marks, and Larry Samelson for helpful comments on the manuscript.
Abbreviation used in this paper
AP
adaptor protein
Footnotes
Address all correspondence to Juan S. Bonifacino, Cell Biology and Metabolism Branch, NICHD, Building 18T, Room 101, National Institutes of Health, Bethesda, Maryland 20892. Tel.: (301) 496-6368. Fax: (301) 402-0078. E-mail: juan@helix.nih.gov
References
- 1.Aguilar RC, Ohno H, Roche KC, Bonifacino JS. Functional domain mapping of the clathrin-associated adaptor medium chains μ1 and μ2. J Biol Chem. 1997;272:2760–2766. doi: 10.1074/jbc.272.43.27160. [DOI] [PubMed] [Google Scholar]
- 2.Anderson RGW, Goldstein JL, Brown MS. A mutation that impairs the ability of lipoprotein receptors to localise in coated pits on the cell surface of human fibroblasts. Nature. 1977;270:695–699. doi: 10.1038/270695a0. [DOI] [PubMed] [Google Scholar]
- 3.Arneson LS, Miller J. Efficient endosomal localization of major histocompatibility complex class II-invariant chain complexes requires multimerization of the invariant chain targeting sequence. J Cell Biol. 1995;129:1217–1228. doi: 10.1083/jcb.129.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boll W, Ohno H, Songyang Z, Rapoport I, Cantley LC, Bonifacino JS, Kirchhausen T. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO (Eur Mol Biol Organ) J. 1996;15:5789–5795. [PMC free article] [PubMed] [Google Scholar]
- 5.Canfield WM, Johnson KF, Ye RD, Gregory W, Kornfeld S. Localization of the signal for rapid internalization of the bovine cation-independent mannose 6-phosphate/insulin-like growth factor-II receptor to amino acids 24-29 of the cytoplasmic tail. J Biol Chem. 1991;266:5682–5688. [PubMed] [Google Scholar]
- 6.Chen J-J, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- 7.Collawn JF, Stangel M, Kuhn LA, Esekogwu V, Jing SQ, Trowbridge IS, Tainer JA. Transferrin receptor internalization sequence YXRF implicates a tight turn as the structural recognition motif for endocytosis. Cell. 1990;63:1061–1072. doi: 10.1016/0092-8674(90)90509-d. [DOI] [PubMed] [Google Scholar]
- 8.Dell'Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 9.Dell'Angelica EC, Mullins C, Bonifacino JS. AP-4, a novel protein complex related to clathrin adaptors. J Biol Chem. 1999;274:7278–7285. doi: 10.1074/jbc.274.11.7278. [DOI] [PubMed] [Google Scholar]
- 10.Dell'Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO (Eur Mol Biol Organ) J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal membrane proteins in Hermansky-Pudlak syndrome due to mutations in the β3A subunit of the AP-3 adaptor complex. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- 12.Futter CE, Gibson A, Allchin EH, Maxwell S, Ruddock LJ, Odorizzi G, Domingo D, Trowbridge IS, Hopkins CR. In polarized MDCK cells basolateral vesicles arise from clathrin-gamma-adaptin-coated domains on endosomal tubules. J Cell Biol. 1998;141:611–623. doi: 10.1083/jcb.141.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geuze HJ, Slot JW, Strous GJ, Hasilik A, von Figura K. Possible pathways for lysosomal enzyme delivery. J Cell Biol. 1985;101:2253–2262. doi: 10.1083/jcb.101.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glickman JN, Conibear E, Pearse BM. Specificity of binding of clathrin adaptors to signals on the mannose-6-phosphate/insulin-like growth factor II receptor. EMBO (Eur Mol Biol Organ) J. 1989;8:1041–1047. doi: 10.1002/j.1460-2075.1989.tb03471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 16.Höning S, Hunziker W. Cytoplasmic determinants involved in direct lysosomal sorting, endocytosis, and basolateral targeting of rat lgp120 (lamp-I) in MDCK cells. J Cell Biol. 1995;128:321–332. doi: 10.1083/jcb.128.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson KF, Kornfeld S. A His-Leu-Leu sequence near the carboxyl terminus of the cytoplasmic domain of the cation-dependent mannose 6-phosphate receptor is necessary for the lysosomal enzyme sorting function. J Biol Chem. 1992;267:17110–17115. [PubMed] [Google Scholar]
- 18.Kibbey RG, Rizo J, Gierasch LM, Anderson RG. The LDL receptor clustering motif interacts with the clathrin terminal domain in a reverse turn conformation. J Cell Biol. 1998;142:59–67. doi: 10.1083/jcb.142.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchhausen T, Bonifacino JS, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Op Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 20.Le Borgne R, Alconada A, Bauer U, Hoflack B. The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J Biol Chem. 1998;273:29451–29461. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- 21.Letourneur F, Klausner RD. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- 22.Marks MS, Ohno H, Kirchhausen T, Bonifacino JS. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- 23.Marks MS, Woodruff L, Ohno H, Bonifacino JS. Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J Cell Biol. 1996;135:341–354. doi: 10.1083/jcb.135.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellman I. Endocytosis and molecular sorting. Annual Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama Y, Goebl M, O'Brine B, Greco, Lemmon S, Pingchang E, Chow, Kirchhausen T. The medium chains of the mammalian clathrin-associated proteins have a homolog in yeast. Eur J Biochem. 1991;202:569–574. doi: 10.1111/j.1432-1033.1991.tb16409.x. [DOI] [PubMed] [Google Scholar]
- 26.Nesterov A, Carter RE, Sorkina T, Gill GN, Sorkin A. Inhibition of the receptor-binding function of clathrin adaptor protein AP-2 by dominant-negative mutant μ2 subunit and its effects on endocytosis. EMBO (Eur Mol Biol Organ) J. 1999;18:2489–2499. doi: 10.1093/emboj/18.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno H, Aguilar RC, Yeh D, Taura D, Saito T, Bonifacino JS. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J Biol Chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- 28.Ohno H, Fournier MC, Poy G, Bonifacino JS. Structural determinants of interaction of tyrosine-based sorting signals with the adaptor medium chains. J Biol Chem. 1996;271:29009–29015. doi: 10.1074/jbc.271.46.29009. [DOI] [PubMed] [Google Scholar]
- 29.Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 30.Ohno H, Tomemori T, Nakatsu F, Okazaki Y, Aguilar RC, Fölsch H, Mellman I, Saito T, Shirasawa T, Bonifacino JS. μ1B, a novel member of the adaptor medium chain family expressed in polarized epithelial cells. FEBS Lett. 1999;449:215–220. doi: 10.1016/s0014-5793(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 31.Orzech E, Schlessinger K, Weiss A, Okamoto CT, Aroeti B. Interactions of the AP-1 Golgi adaptor with the polymeric immunoglobulin receptor and their possible role in mediating brefeldin A-sensitive basolateral targeting from the trans-Golgi network. J Biol Chem. 1999;274:2201–2215. doi: 10.1074/jbc.274.4.2201. [DOI] [PubMed] [Google Scholar]
- 32.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pevsner J, Volknandt W, Wong BR, Scheller RH. Two rat homologs of clathrin-associated adaptor proteins. Gene. 1994;146:279–283. doi: 10.1016/0378-1119(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 34.Rapoport I, Chen YC, Cupers P, Shoelson SE, Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO (Eur Mol Biol Organ) J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohrer J, Schweizer A, Russell D, Kornfeld S. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol. 1996;132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiratori T, Miyatake S, Ohno H, Nakaseko C, Isono K, Bonifacino JS, Saito T. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity. 1997;6:583–589. doi: 10.1016/s1074-7613(00)80346-5. [DOI] [PubMed] [Google Scholar]
- 37.Simpson F, Bright NA, West MA, Newman LS, Darnell RB, Robinson MS. A novel adaptor-related protein complex. J Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens DJ, Banting G. Specificity of interaction between adaptor-complex medium chains and the tyrosine-based sorting motifs of TGN38 and lgp120. Biochem J. 1998;335:567–572. doi: 10.1042/bj3350567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Kiliman MW. Identification of two new μ-adaptin-related proteins, μ-ARP1 and μ-ARP-2. FEBS Lett. 1997;402:57–61. doi: 10.1016/s0014-5793(96)01500-1. [DOI] [PubMed] [Google Scholar]
- 42.Warren RA, Green FA, Stenberg PE, Enns CA. Distinct saturable pathways for the endocytosis of different tyrosine motifs. J Biol Chem. 1998;273:17056–17063. doi: 10.1074/jbc.273.27.17056. [DOI] [PubMed] [Google Scholar]