The Dynamic Organization of the Perinucleolar Compartment in the Cell Nucleus (original) (raw)
Abstract
The perinucleolar compartment (PNC) is a unique nuclear structure preferentially localized at the periphery of the nucleolus. Several small RNAs transcribed by RNA polymerase III (e.g., the Y RNAs, MRP RNA, and RNase P H1 RNA) and the polypyrimidine tract binding protein (PTB; hnRNP I) have thus far been identified in the PNC (Ghetti, A., S. PinolRoma, W.M. Michael, C. Morandi, and G. Dreyfuss. 1992. Nucleic Acids Res. 20:3671–3678; Matera, A.G., M.R. Frey, K. Margelot, and S.L. Wolin. 1995. J. Cell Biol. 129:1181–1193; Lee, B., A.G. Matera, D.C. Ward, and J. Craft. 1996. Proc. Natl. Acad. Sci. USA. 93: 11471–11476). In this report, we have further characterized this structure in both fixed and living cells. Detection of the PNC in a large number of human cancer and normal cells showed that PNCs are much more prevalent in cancer cells. Analysis through the cell cycle using immunolabeling with a monoclonal antibody, SH54, specifically recognizing PTB, demonstrated that the PNC dissociates at the beginning of mitosis and reforms at late telophase in the daughter nuclei. To visualize the PNC in living cells, a fusion protein between PTB and green fluorescent protein (GFP) was generated. Time lapse studies revealed that the size and shape of the PNC is dynamic over time. In addition, electron microscopic examination in optimally fixed cells revealed that the PNC is composed of multiple strands, each measuring ∼80–180 nm diam. Some of the strands are in direct contact with the surface of the nucleolus. Furthermore, analysis of the sequence requirement for targeting PTB to the PNC using a series of deletion mutants of the GFP–PTB fusion protein showed that at least three RRMs at either the COOH or NH2 terminus are required for the fusion protein to be targeted to the PNC. This finding suggests that RNA binding may be necessary for PTB to be localized in the PNC.
Many nuclear functions including DNA replication, RNA transcription, processing, and transport have been extensively investigated at the biochemical and molecular levels. However, much less is understood regarding the spatial organization of these events within the three-dimensional context of the mammalian cell nucleus. Light and electron microscopic examination of cell nuclei has revealed many readily identifiable nuclear structures including the nucleolus, electron dense heterochromatin, and a variety of granular and fibrillar structures including interchromatin granules, perichromatin granules, and perichromatin fibrils (for review see Spector, 1993). The use of increasingly sophisticated molecular techniques and the availability of a large number of antibodies and also nucleic acid probes has advanced our understanding of the temporal and spatial organization of nuclear functions, as well as revealed the complex nature of the mammalian cell nucleus.
In addition to the ubiquitous features of the nucleus, nuclear bodies have been described in specific cell types or cells at different physiological states (Bouteille et al., 1967). One of the more extensively studied examples is the coiled body. Coiled bodies were first described by Ramon and Cajal (1903) as nucleolar accessory bodies. These generally round structures, 0.5–1.0 μm diam, consist of coiled fibrillar strands (Monneron and Bernhard, 1969). In addition to small nuclear RNPs, several nucleolar components and a coiled body–specific protein, coilin, have been found in these structures (for reviews see Brasch and Ochs, 1992; Lamond and Carmo-Fonseca, 1993; Gall et al., 1995). However, [3H]uridine incorporation studies showed little to no labeling of these bodies after a short pulse (Moreno Diaz de la Espina et al., 1980), suggesting that coiled bodies are unlikely to be the sites of active RNA synthesis. In addition, the absence of essential pre-mRNA splicing factors such as SC35 and SF2/ASF (Raska et al., 1991; Huang and Spector, 1992; Lamond and Carmo-Fonseca, 1993; Krainer, A., and D. Spector, unpublished observations) in these structures suggests that they are probably not the sites of active splicing. The number of coiled bodies per nucleus and the percentage of cells that contain coiled bodies increase dramatically in immortalized cells or cancer cells as compared to primary cells (Spector et al., 1992). In addition, coiled bodies have been found to be within the nucleoli in some cell lines derived from breast cancer tissues (Ochs et al., 1994). However, the function of the coiled body remains elusive. More recently, a novel nuclear structure, gems, was identified in close proximity to coiled bodies (Liu and Dreyfuss, 1996). Gems are similar to coiled bodies in number and size, response to metabolic conditions, and their dynamics through the cell cycle. However, gems and coiled bodies contain different macromolecular components. Survival of motor neuron protein, involved in the genetic disease spinal muscular atrophy, is localized in gems. Components that are localized in coiled bodies such as snRNPs, coilin, and fibrillarin are not present in gems (Liu and Dreyfuss, 1996). The function of gems is currently unknown.
Another intensively studied nuclear body is the promyelocyte (PML)1 oncogenic domain (POD), also named PML or ND10, which consists of a dense fibrillar ring flanking a central core (Ascoli and Maul, 1991; Dyck et al., 1994; Koken et al., 1994, 1995; Weis et al., 1994; Terris et al., 1995). The POD was defined by immunolabeling with an antibody specifically recognizing the PML protein in hematopoietic cells. Several other autoimmune antibodies also react with components in the POD (Ascoli and Maul, 1991). The POD becomes fragmented into a large number of microparticulates in acute promyelocytic leukemia (Dyck et al., 1994; Koken et al., 1994; Weis et al., 1994). The break up of the POD into microparticulates is related to the formation of the PML–retinoic acid receptor α fusion protein resulting from a t(15;17) translocation (Dyck et al., 1994; Koken et al., 1994; Weis et al., 1994). When these cells are treated with retinoic acid, the fragmented PML particulates fuse and reassemble into PODs along with the initiation of cellular differentiation and the loss of the PML–retinoic acid receptor α fusion protein. This observation provided evidence that alterations of nuclear structure may play a role in this form of carcinogenesis (Dyck et al., 1994; Koken et al., 1994; Weis et al., 1994). In addition to being present in hematopoietic cells, the POD has recently been detected in a variety of tissues including hepatocytes, endothelium, epithelium, and connective stroma (Koken et al., 1995; Terris et al., 1995). The expression of PML and its immunolabeling pattern, in some of the samples examined, appeared to change depending upon the proliferating or cancer state. However, the function of the POD is presently unknown.
More recently, a unique structure localized at the periphery of the nucleolus, the perinucleolar compartment (PNC), was identified (Ghetti et al., 1992; Matera et al., 1995). The PNC has been shown to contain several small RNAs transcribed by RNA polymerase III, including RNase P, MRP RNAs, and multiple Y RNAs (Matera et al., 1995; Lee et al., 1996), as well as the polypyrimidine tract binding protein (PTB; Ghetti et al., 1992; Matera et al., 1995). However, the structural and functional characteristics of the PNC have not yet been extensively studied. In this paper, we examined the PNC in human cancer and normal cell lines. We also characterized the PNC in fixed cells using light and electron microscopy and in living cells through time lapse observations. In addition, we analyzed the sequence requirements to target a green fluorescent protein (GFP)–PTB fusion protein to the PNC.
Materials and Methods
Cell Culture
Several human cell lines were grown to subconfluence on 22 × 22 mm glass coverslips in 35-mm Petri dishes in the appropriate culture medium. HeLa (epithelial carcinoma, cervix), T84 (colon carcinoma), SW620 (colon adenocarcinoma), WI-38 VA13 (WI-38 cells transformed with SV40 T), Wacar (normal skin fibroblasts), Homa (normal skin fibroblasts), and MRC5 (normal lung fibroblasts) were grown in DME supplemented with 10% fetal bovine serum (GIBCO BRL, Gaithersburg, MD). The breast cancer cell lines including MDA-MB-468 (adenocarcinoma) and Hs 578T (ductal carcinoma) cells were grown in DME supplemented with 15% FBS. Normal human mammary epithelium (NHME) cells were grown in mammary epithelium basal medium (MEBM) supplemented with 5 mg/liter transferrin, 1 mg/liter hydrocortisone, 5 μM isoproterenol, 10 mg/liter insulin, 5 μg/liter epithelium growth factor (EGF), and 0.2% bovine pituitary extract. MG63 (osteosarcoma), Detroit 551 (normal skin fibroblasts), and WI-38 (normal lung fibroblasts) cells were grown in minimal essential medium (MEM) supplemented with 10% FBS. Cells were maintained at 37°C with 10% CO2.
Construction of GFP Fusion Proteins
Human PTB cDNA (Gil et al., 1991) was amplified by PCR using Vent DNA polymerase (New England Biolabs Inc., Beverly, MA). The amplified fragment was inserted in frame into a GFP expression construct, pEGFP-C1 (Clontech Laboratories, Inc., Palo Alto, CA), at the HindIII and BamH1 sites. The fusion protein contained GFP at the NH2 terminus of PTB. Subsequently, deletion fragments of PTB were generated by PCR using specific primers and were inserted into pEGFP-C1, which gave rise to a series of mutant fusion proteins.
Transfection
Expression constructs were transiently transfected into HeLa cells by electroporation (Sambrook et al., 1989). Briefly, subconfluent cells in a 100-mm culture dish were collected by trypsinization and mixed with 20 μg of DNA including 7 μg target DNA and 13 μg sheared salmon sperm DNA. A 280 μl mixture of cells in DME with 10% FCS and DNA was electroporated in a BioRad (Richmond, CA) electroporator at 270 V and 960 μFaraday. Cells were subsequently seeded onto glass coverslips in 35mm petri dishes and were grown for either 7 or 24 h.
Immunolabeling
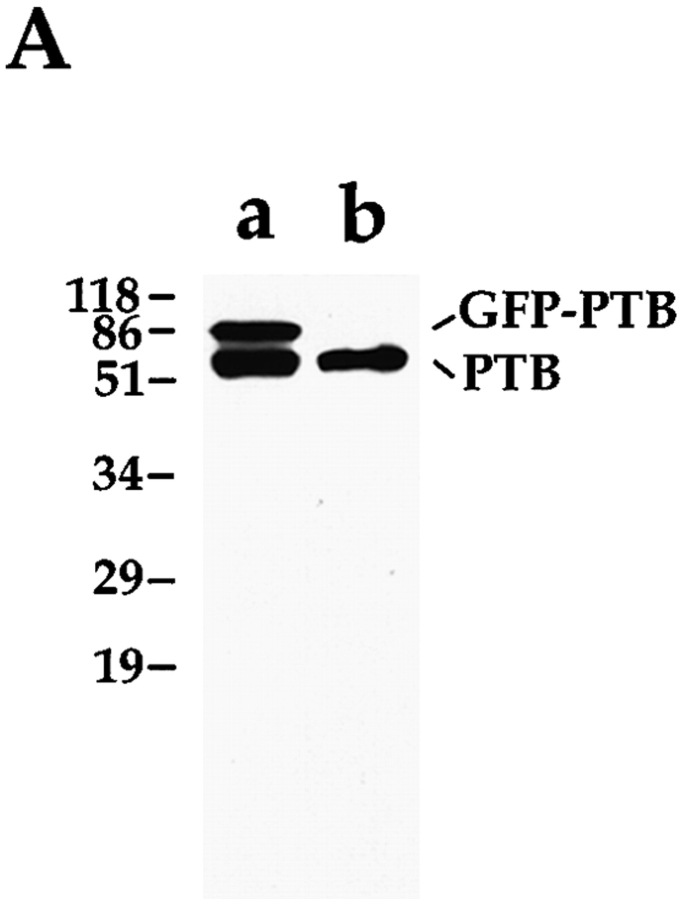
SH54 monoclonal antibody was raised against HeLa nuclear extracts. The antibody was selected by its specific labeling of the PNC in the nucleus. SH54 recognizes a 57-kD protein on a Western blot (see Fig. 4 A) and was confirmed to bind PTB specifically.
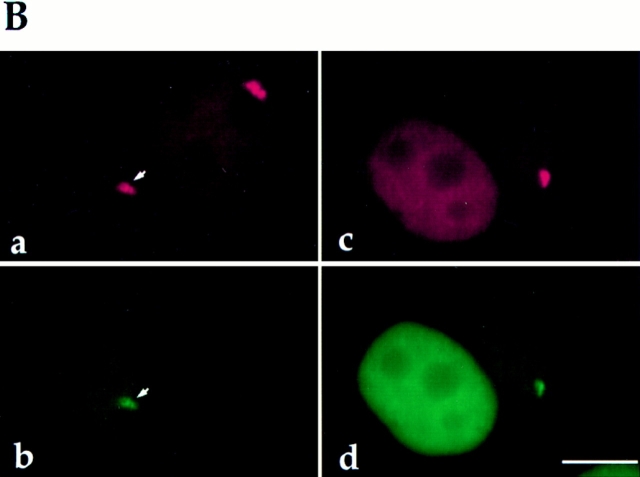
Figure 4.
(A) Both the GFP– PTB fusion protein and endogenous PTB are expressed in transfected cells and are detected at the expected size by Western blot using antibody SH54 (lane a). Only endogenous PTB is detected in cells transfected with the GFP construct alone (lane b). (B) The subcellular localization of GFP–PTB is indistinguishable from endogenous PTB. The endogenous PTB (a, red) and the GFP–PTB fusion protein (b, green) are colocalized to both the PNC (arrows) and the nucleoplasm. The PNC observed in transfected cells is not the result of the overexpression of the protein, as some cells do not contain a PNC in spite of expressing large amounts of the fusion protein (c and d, left cell). c shows both the immunolabeling of endogenous PTB and transiently expressed GFP–PTB; d shows GFP–PTB. Bar, 10 μm.
Subconfluent cells grown on the glass coverslips were fixed in freshly made 2% formaldehyde in PBS for 15 min, and cells were washed 3 × 10 min each in PBS and incubated with anti-PTB primary antibody SH54 at a dilution of 1:300, anti-Sm antibody (Lerner and Steitz, 1979) at 1:1,000, anti-SC35 (Fu and Maniatis, 1990) at 1:1,000, or anti-fibrillarin (Sigma Chemical Co., St. Louis, MO) at 1:5 for 1 h at room temperature. Cells were rinsed in PBS and then incubated with Texas red–conjugated goat anti–human or FITC-conjugated goat anti–mouse antibody at a dilution of 1:40 for 1 h at room temperature followed by three washes in PBS. The coverslips were mounted onto glass slides with mounting medium containing 90% glycerol in PBS with 1 mg/ml paraphenylenediamine as an antifade agent. The mounting medium was adjusted to pH 8.0 with 0.2 M bicarbonate buffer. Cells were examined with a microscope (FXA; Nikon Inc., Melville, NY) equipped with epifluorescence and differential interference contrast optics. Images were captured by a SenSys cooled CCD camera (Photometrics, Tucson, AZ) using Oncor Image software.
Photooxidation
Immunoelectron microscopic localization was performed according to the preembedding photooxidation procedure (Deerinck et al., 1994). After incubation of the primary antibody (as described in the immunolabeling procedure), cells were washed twice for 2 min in 0.1 M PBS and incubated in 0.1 M PBS with 1% normal goat serum (NGS), 1% cold water fish gelatin, and 1% BSA (fraction V) for 20 min to block nonspecific staining. Cells were then incubated with goat anti–mouse eosin-5-isothiocyanate (conjugated as previously described, Deerinck et al., 1994) diluted in 0.1 M PBS with 1% BSA and 1% NGS for 1 h. Unbound conjugate was removed by washing six times for 5 min in 0.1 M PBS with 1% BSA and 1% NGS followed by two times in 5 min washes with 0.1 M sodium cacodylate, pH 7.4. All washes and incubations were at 4°C.
Cells were examined using a 35-mm microscope (Axiovert; Zeiss Inc.) equipped with an MRC-1024 laser scanning confocal system (BioRad) using the 488 nm excitation line from an argon/krypton laser. During imaging the cells were kept at 10°C using a cold stage and were in 0.1 M sodium cacodylate that had been deoxygenated by bubbling with argon to retard photobleaching.
Once an area had been selected using a 40× 1.3 NA objective (Planapo; Zeiss Inc.), a cold oxygenated solution of 1 mg/ml diaminobenzidine tetrahydrochloride (DAB; Sigma Chemical Co.) in 0.1 M sodium cacodylate, pH 7.2, was added to the cells. Photooxidation of the DAB by eosin was accomplished by illuminating the region of interest with 515 nm light from 100 W mercury lamp. The progress of the reaction was monitored by transmitted light, and illumination was halted when a brownish reaction product became visible (10–20 min). Both experimental and control preparations were reacted under identical conditions.
After photooxidation, the cells were rinsed five times for 2 min in 0.1 M sodium cacodylate and then postfixed in 1% osmium tetroxide for 1 h. Cells were then rinsed in double distilled water, dehydrated in an ethanol series, and infiltrated with Durcupan ACM resin (Electron Microscopy Sciences, Ft. Washington, PA). After polymerization of the resin for 24 h at 60°C, the bottom glass coverslip was removed from the dish, and the region of interest was cut out and mounted for ultramicrotomy with an ultramicrotome (Ultracut E; Leica Inc., Deerfield, IL) using a diamond knife (Diatome U.S., Ft. Washington, PA). Electron micrographs were recorded from sections 80 nm in thickness at 60–80 kV with a transmission electron microscope (100CX; Jeol Ltd., Tokyo, Japan) and sections 1 μm thick at 300 kV with an intermediate voltage transmission electron microscope (4000EX; Jeol Ltd.). Stereo-pair images were recorded by tilting the specimen stage to ± 8°. Sections of photooxidized cells were not poststained.
Structural Characterization by Correlative Electron Microscopy
To examine the structure of the PNC in optimally fixed cells at high resolution, transiently expressed GFP–PTB was used as a marker to indicate the localization of the PNC. The corresponding nuclear region identified by fluorescence of the GFP–PTB fusion protein was examined by electron microscopy. Specifically, transfected cells were seeded on gridded coverslips. 12 h after transfection, cells were fixed in 4% paraformaldehyde with 0.05% glutaraldehyde in PBS, and cells that showed the PNC were quickly examined and photographed using an epifluorescence microscope (FXA; Nikon Inc.) equipped with a SenSys cooled CCD camera (Photometrics Inc., Tucson, AZ) using Oncor Image software. Subsequently, cells were fixed in 2% glutaraldehyde for 20 min and washed in PBS containing 0.3 M glycine and 0.1 M cacodylate buffer. Cells were then postfixed in 1% osmium tetroxide for 1 h and dehydrated by incubation in a series of ascending concentrations of ethanol and embedded in epon/araldite at 60°C for 48 h. 80-nm sections were poststained with uranyl acetate/lead citrate and examined at 75 kV with a transmission electron microscope (H-7000; Hitachi Scientific Instrs., Mountain View, CA). The same cell photographed at the fluorescence microscopic level was located, and the nuclear region that corresponded to the PNC was identified and photographed.
Observation of the PNC in Living Cells
Living cell studies were performed using an FCS2 live cell chamber (Bioptechs, Inc., Butler, PA). Transfected cells were grown on the customized coverslips that were assembled into the FCS2 chamber, where temperature was maintained at 37°C, and cells were supplemented with a controlled flow of fresh medium. The chamber was directly mounted onto the specimen stage of an inverted epifluorescence microscope (Axiovert 405M; Zeiss, Inc.) equipped with a cooled CCD (NU200; Photometrics Inc.) camera. Images were captured using Oncor Image software.
Results
The Presence of the PNC Correlates with the Transformed Phenotype
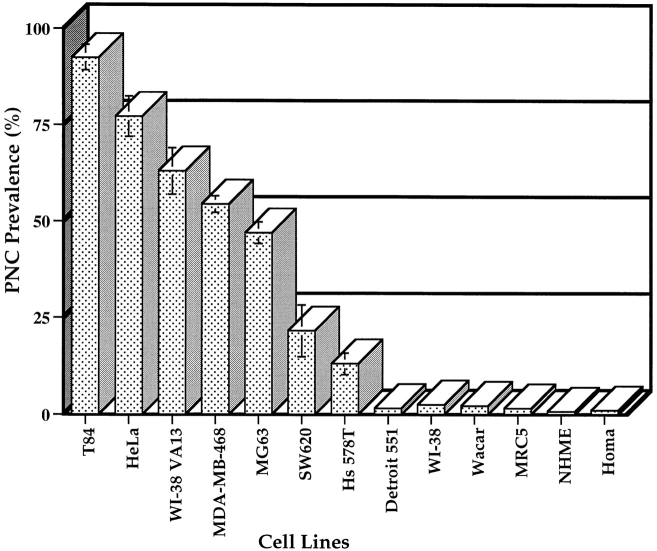
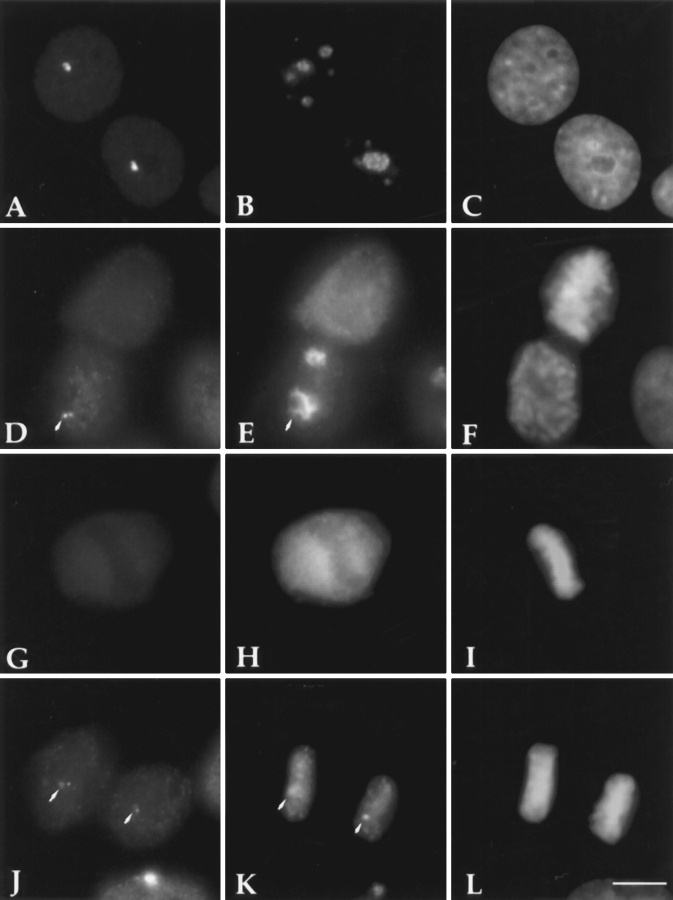
Since PTB is the only protein component of the PNC thus far identified, we used a monoclonal antibody, SH54, that specifically recognizes PTB (see Materials and Methods and Fig. 4 a) to characterize the PNC in cell nuclei. A large number of human cancer cell lines and normal human diploid cell lines were examined for the presence of the PNC. The results showed that the PNC is predominantly present in cancer cells and is rarely observed in normal primary cells (Figs. 1 and 2). PNC prevalence, defined as the percentage of cells that contains one or more PNCs, showed a large diversity among the cancer cell lines examined. Some of the cell lines such as HeLa (endothelial carcinoma, cervix) and T84 (colon carcinoma) showed a PNC prevalence of >80%. In contrast, the PNC prevalence of other cell lines such as SW620 and MG63 ranged from 25 to 50%. The large variability of PNC prevalence among cancer, transformed, and primary diploid cells suggests that PNC prevalence may be correlated with the degree of malignancy. To examine this possibility, we compared PNC prevalence in two human breast cell lines. We found that PNC prevalence appeared to be fivefold higher in MDAMB-468 cells (American Type Culture Collection, Rockville, MD) isolated from a metastatic mammary adenocarcinoma that induces tumors in nude mice, than in Hs 578T cells (American Type Culture Collection) isolated from a mammary ductal carcinoma that does not induce tumors in nude mice (Fig. 2). In addition, when primary human diploid fibroblasts (WI-38 cells) were transformed by SV40 large T antigen (WI-38 VA13), the PNC prevalence rose to >60% compared to 2% in the nontransformed parental cells (Fig. 2). It is also noticeable that the size of PNCs varies from cell to cell (Fig. 1). In some cancer cells, the structure tended to be larger (Fig. 1, HeLa and _WI_-38 VA13 cells) and occasionally extended into the nucleolus, whereas, in other cancer cells, the structure was mostly observed as small dots (Fig. 1, MG63 cells). However, in the very small percentage of normal cells that contains the PNC, the PNC invariably appeared to be a small dot (data not shown).
Figure 1.
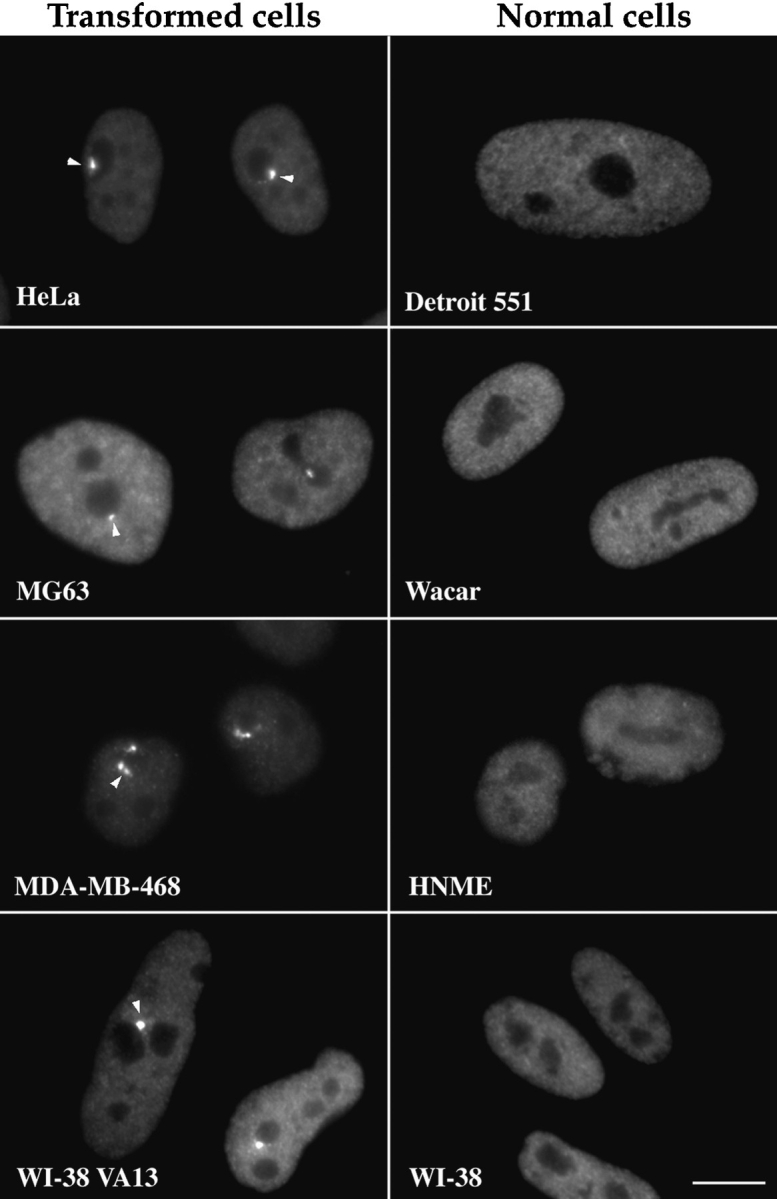
The PNC is predominantly present in human cancer cells. PNCs were detected by immunolabeling using antibody SH54. The left column shows the presence of the PNC in human cancer cells (arrowheads), including HeLa (epithelia carcinoma), MG63 (osteosarcoma), MDA-MB-468 (mammary adenocarcinoma), and WI-38 VA13 (human lung fibroblast cells WI-38 transformed by SV40 T). The right column shows that the PNC is generally not present in normal human diploid cells including Detroit 551 (skin fibroblast), Wacar (skin fibroblast), HNME (human normal mammary epithelium), and WI-38 (lung fibroblast). Bar, 10 μm.
Figure 2.
The percentage of cells that contain the PNC (PNC prevalence) is correlated with the transformed phenotype. The histogram indicates the statistical evaluation of PNC prevalence among human cancer cells and normal diploid cells. 500 cells from each cell line were examined.
Characterization of the PNC through the Cell Cycle
We have further characterized the PNC in HeLa cell nuclei throughout the cell cycle by immunolabeling using SH54. The PNC is clearly associated with nucleoli in interphase cells. The shape and size of the PNC varies from cell to cell within the same cell line. As cells progress through mitosis, the PNC dissociates in prophase cells (Fig. 3 D). The dissociation appears to be a gradual event. When double labeled with the anti-fibrillarin antibody, a more concentrated PTB labeling is still visibly associated with the partially disassembled nucleolus at early prophase (Fig. 3, D and E, arrows). Both PTB and fibrillarin are diffusely localized in metaphase cells (Fig. 3, G and H). At late telophase, the PNC begins to form in the new daughter cell nuclei before the re-entry of the entire nucleoplasmic population of PTB (Fig. 3 J, arrows). Double labeling experiments using the anti-fibrillarin antibody showed that the initial detectable PNCs are spatially associated with nucleolar regions (Fig. 3, J and K, arrows). Double labeling experiments using the anti-Sm antibody showed that the re- establishment of splicing factors in the new daughter cell nuclei takes place before the formation of the PNC (data not shown). In addition, the formation of the PNC did not appear to be affected by the presence of cycloheximide (data not shown), suggesting that the newly formed PNC is composed of cellular components from the previous cell cycle.
Figure 3.
The PNC dissociates at prophase and reforms at late telophase. The horizontal rows show different stages of mitosis. The left column shows the immunolabeling of the PNC with antibody SH54, the center column the immunolabeling of fibrillarin with human anti-fibrillarin antibody, and the right column DNA staining by Dapi. The dissociation of the PNC at prophase (D) appears to be a gradual event. A concentrated PTB labeling is still spatially linked with the partially dissociated nucleolus (D and E, arrows). Both PTB (G) and fibrillarin (H) are diffusely distributed in metaphase cells. The earliest detectable PNCs (J, arrows) in the daughter cell nuclei are associated with nucleolar regions (K, arrows). Bar, 10 μm.
Characterization of the PNC in Living Cells
To directly visualize the PNC in living cells, a mammalian expression vector was constructed that expresses a GFP– PTB fusion protein. Human PTB cDNA (Gil et al., 1991) was amplified by PCR and inserted into the pEGFP-C1 vector (Clontech Laboratories, Inc.) generating a fusion protein in which PTB is at the COOH-terminal end of GFP. The construct was transiently transfected into HeLa cells, and the expression of GFP–PTB was examined. Proteins from cells transfected with either GFP–PTB or GFP alone were analyzed by Western blot using the SH54 antibody (Fig. 4 A). Cells transfected with the GFP–PTB construct expressed the fusion protein of expected size (84 kD) and the endogenous PTB (57 kD; Fig. 4 A, lane a). Cells transfected by GFP alone showed only the endogenous PTB protein (Fig. 4 A, lane b). To examine if the subcellular localization of the fusion protein is similar to that of the endogenous PTB, the intranuclear localization of the fusion protein and endogenous PTB was compared (Fig. 4 B). 24 h after transfection, cells were fixed, immunolabeled with the SH54 antibody, which recognizes both endogenous PTB and GFP–PTB, and subsequently probed with Texas red–conjugated secondary antibody (Fig. 4 B, a, red signal). GFP–PTB alone emitted a green signal (Fig. 4 B, b). Comparisons between a and b reveal that the GFP–PTB fusion protein and the endogenous PTB were colocalized to the same nuclear regions, which included both the PNC (Fig. 4 B, a and b, arrows) and the nucleoplasm. To exclude the possibility that the formation of the PNC may be due to the overexpression of the GFP–PTB fusion protein, we examined cells that do not contain a PNC, as determined by immunolabeling. We found that the expression of GFP–PTB at a high level (roughly evaluated by the intensity of the green fluorescence) did not induce the formation of the PNC (Fig. 4 B, c and d, left cell).
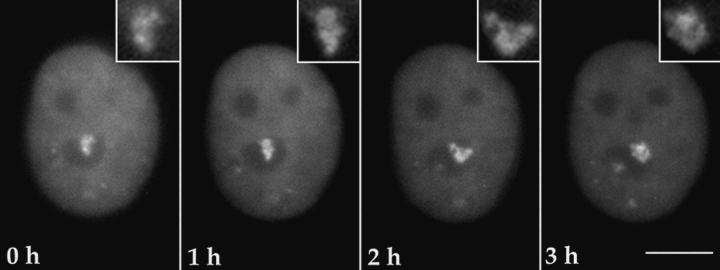
Using the GFP–PTB fusion protein as a probe, we observed the PNC in living cells. Cells were transiently transfected with the GFP–PTB construct. 12 h after transfection, cells grown on glass coverslips were moved to a live cell chamber (see Materials and Methods). The chamber was mounted onto the stage of a fluorescence microscope, and images were captured at various time intervals. The exposure of cells to the excitation light was kept minimal. Time lapse observations demonstrated that the PNC is a dynamic structure that makes small movements at the periphery of and occasionally into the nucleolus over time (Fig. 5). Closer examination revealed that the PNC appears to contain substructures whose organization also changes through time (Fig. 5, insets). These substructures appeared to be strand like in some cases (Fig. 5, inset, 2 h). In many cells, the PNC changed shape and position within 2–3 h. GFP–PTB did not appear to have an immediate toxicity to transfected cells, as cells expressing GFP–PTB progressed through multiple cell cycles and newly divided cells expressed the fusion protein (data not shown).
Figure 5.
The PNC is a dynamic structure with small movements over time when observed in living cells. This figure illustrates the observation of a single cell through time as indicated in all frames. The PNC changes its shape and position over a 3-h period. In the insets, the PNC appears to contain substructures that are reorganized through time. Bar, 10 μm.
High Resolution Electron Microscopic Examination of the PNC
To further analyze the structural characteristics of the PNC at higher resolution, we examined the PNC in HeLa cells by electron microscopy. Photooxidation pre-embedding immunolabeling was used to achieve a high degree of sensitivity and good morphological preservation (Deerinck et al., 1994). The results of these experiments revealed that the PNCs are in direct contact with the surface of the nucleolus (Figs. 6 and 7), and occasionally, they extend into the nucleolus (Fig. 6 C). In addition, the PNCs are irregular in shape. A clear margin of the structure was not observed at this level as compared to what was revealed at the light microscopic level. The immunolabeling of PTB in the PNC appeared to be heterogeneous, with some areas more intensely stained than others. Higher magnification micrographs (Fig. 6, B and C) or stereo images from 1-μm– thick sections viewed at 300 kV (Fig. 7 B) revealed strandlike structures across the PNC (Figs. 6 and 7 B).
Figure 6.
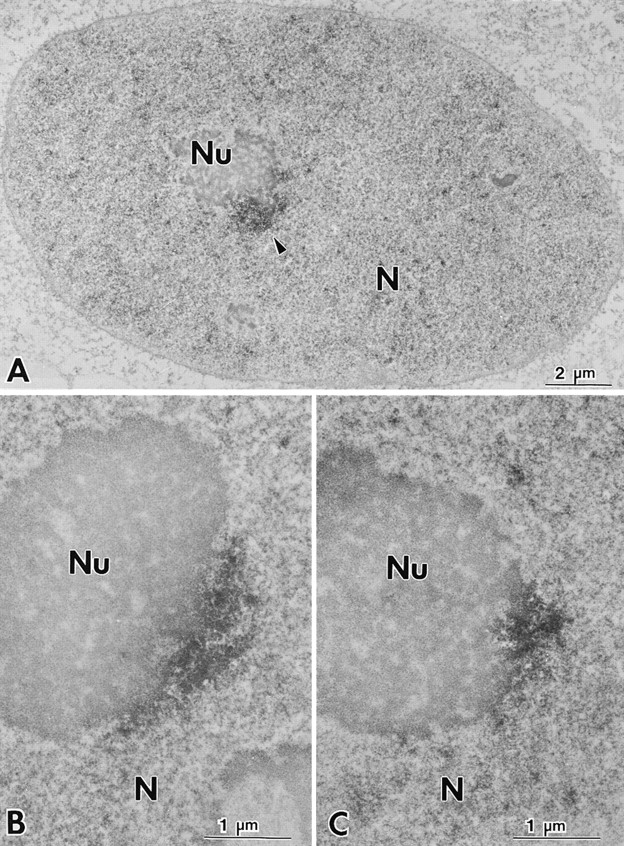
The PNC is spatially associated with the nucleolus when examined at high resolution by electron microscopy. Photooxidation pre-embedding immunocytochemical labeling of the PNC with SH54 demonstrates that the PNC is in direct contact with the nucleolus (A–C), and PTB is heterogenously distributed in the PNC (B and C). The arrowheads indicate the PNC.
Figure 7.
Stereo pair images of 1-μm thick sections observed at 300 kV using intermediate voltage electron microscopy reveal in three dimensions that the PNC is directly linked to the surface of the nucleoli (A). Images at higher magnification show strand-like structures. Bars: (A) 4 μm; (B) 1 μm.
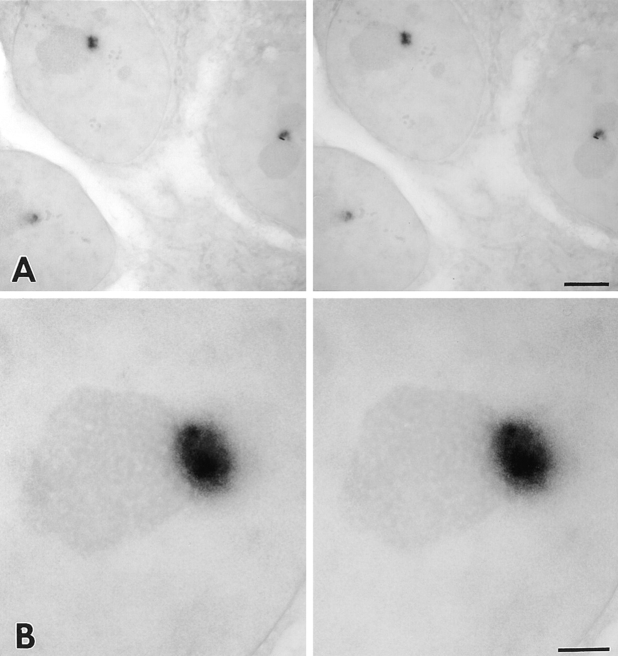
To visualize the basic structural features of the PNC in optimally fixed cells at high resolution, we examined the PNC in the same cell by fluorescence and electron microscopy. To mark the PNC without immunocytochemical manipulations, the expression of GFP–PTB was used as a marker to indicate the localization of the PNC in transiently transfected cells. Transfected cells were photographed, optimally fixed, and processed for the electron microscopic examination (see Materials and Methods). The fluorescence and electron microscopic images of the same cell are shown in Fig. 8. The nuclear region corresponding to the PNC appeared to be an electron-dense structure, which is composed of thick, short strands measuring ∼80– 180 nm diam. Each strand appears to be surrounded by lesser electron-dense areas. The strand-like structure may correspond to the heterogeneous labeling of the PNC observed in the immunoelectron microscopic examination using SH54 (Fig. 6). Some of the strands are directly linked to the surface of the nucleolus (Fig. 8 C, arrows). Similar structures were observed in multiple sections. In some sections, the strands were as long as 1 μm. It is possible that in three dimensions the strands are connected, forming a continuous structure.
Figure 8.
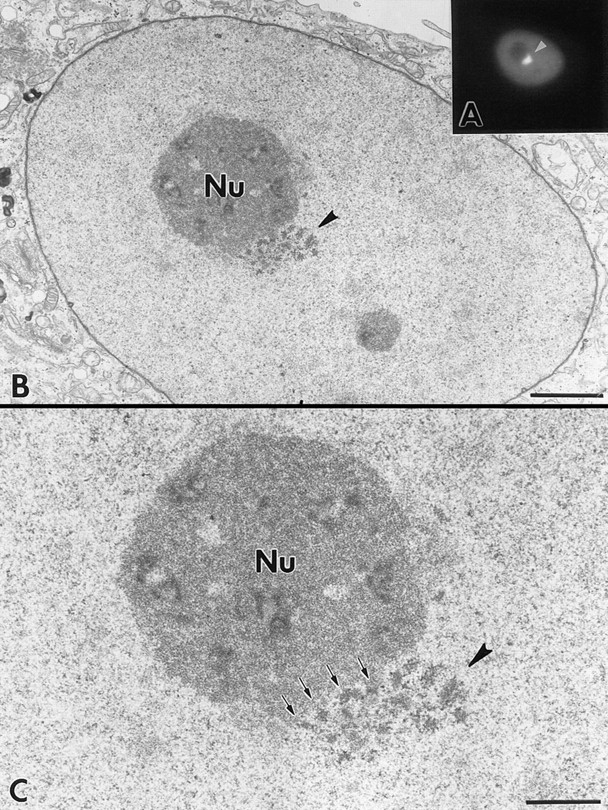
Fluorescence and electron microscopic examination of the PNC in the same cell. The nuclear region corresponding to the PNC in the fluorescence micrograph was examined in poststained, 80-nm–thick sections. Arrowheads indicate the corresponding nuclear region from fluorescence to electron microscopic images. The PNC is composed of electron-dense, thick, short strands measuring 80–180 nm diam. These strands are surrounded by less electron-dense areas. Some of the strands are in direct contact with the surface of the nucleolus. Bars: (A) 10 μm; (B) 2 μm; (C) 1 μm.
Sequence Requirement for Targeting PTB to the PNC
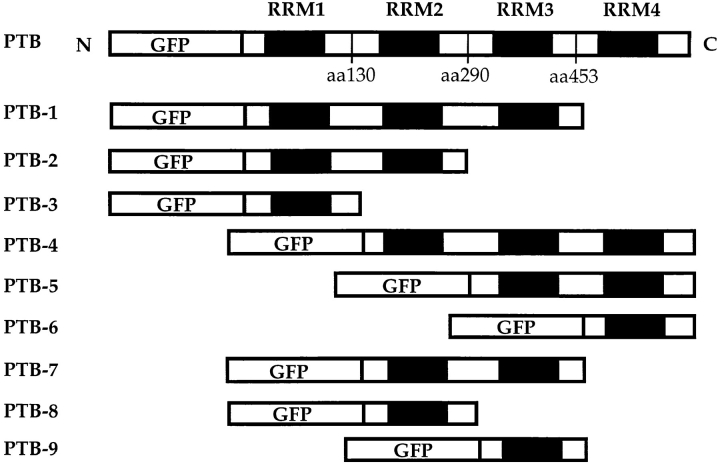
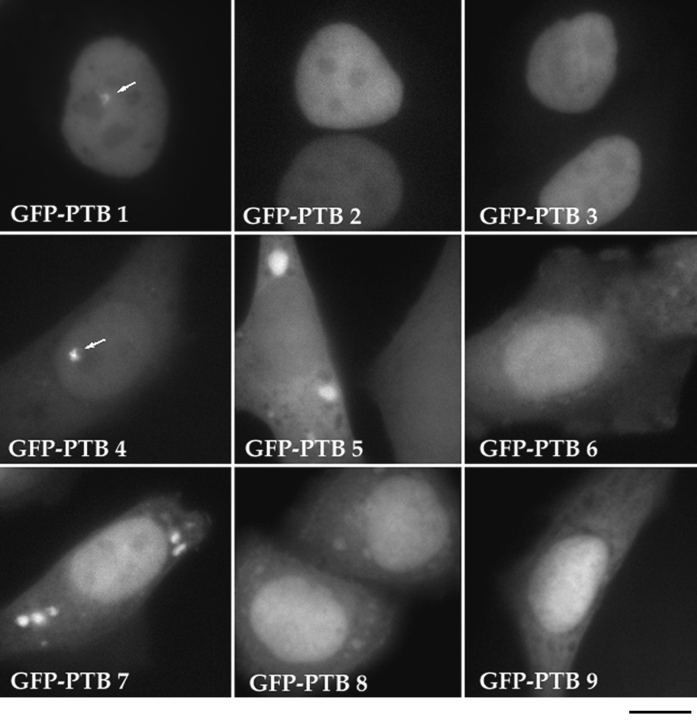
To determine the sequence requirement for targeting GFP–PTB to the PNC, we generated a series of deletion mutants of the GFP–PTB fusion protein and analyzed the subcellular localization of these mutant fusion proteins. The mutant proteins were designed according to the structural characteristics of PTB, which has been previously shown to contain four RNA recognition motifs (RRMs; Patton et al., 1991; Ghetti et al., 1992). Nine mutants (Fig. 9) containing various numbers of RRMs from either the NH2 or COOH terminus of the protein were made. The inserts in all mutant constructs were verified by restriction enzyme analysis (data not shown). The constructs were transiently transfected into HeLa cells, and the expression of the mutant proteins was examined by fluorescence microscopy. All mutant fusion proteins were expressed at a detectable level, as monitored by the expression of GFP. The subcellular localization of these mutants was examined (Fig. 10) and compared to the localization of endogenous PTB (Table I). The result of the analysis demonstrated that only two mutant fusion proteins, PTB-1 and -4, both of which contain three RRMs, were localized to the PNC in transfected cells (Fig. 10 and Table I). Mutants with a combination of two RRMs, including combinations of RRMs 1 and 2, 3 and 4, or 2 and 3, as well as mutants with only one RRM were not able to target the fusion protein to the PNC (Fig. 10 and Table I). These observations suggest that a minimum of three RRMs are required for GFP– PTB to be efficiently targeted to the PNC.
Figure 9.
Deletion mutants were generated to analyze the sequence requirement for PTB to be localized to the PNC. The mutants are designed according to the four RNA recognition motifs of the protein.
Figure 10.
The subcellular localization of a series of deletion mutants of GFP–PTB fusion proteins. Deletion of either RRM 1 or 4 does not affect the localization of the fusion protein to the PNC (GFP–PTB 1 and 4, arrows). However, deletion mutants that contain fewer than three RRMs result in the inability to target the fusion proteins to the PNC (other frames). Deletion mutants that contain RRM1 show a predominantly nuclear localization (top row), whereas mutants missing RRM1 show a much more cytoplasmic localization (other frames). The mutant GFP–PTB with RRMs 3 and 4 shows that the cytoplasmic localization becomes dominant over the nuclear localization. Bar, 10 μm.
Table I.
Characteristics of the GFP–PTB Deletion Mutant Fusion Proteins
| PTBs | GFP–PTBs in the PNC | Endogenous PTB in the PNC | Fibrillarin | SC35 | Nucleus | Cytoplasm |
|---|---|---|---|---|---|---|
| GFP–PTB | ++++ | + | + | + | ++++ | −−−− |
| GFP–PTB 1 | +++ | + | + | + | ++++ | −−−− |
| GFP–PTB 2 | −−−− | + | + | + | ++++ | −− |
| GFP–PTB 3 | −−−− | + | + | + | ++++ | + |
| GFP–PTB 4 | ++++ | + | + | + | ++++ | ++ |
| GFP–PTB 5 | −−−− | + | + | + | + | +++ |
| GFP–PTB 6 | −−−− | + | + | + | ++++ | ++ |
| GFP–PTB 7 | −−−− | + | + | + | +++ | ++ |
| GFP–PTB 8 | −−−− | + | + | + | ++++ | +++ |
| GFP–PTB 9 | −−−− | + | + | + | ++++ | +++ |
To examine if the expression of the mutant GFP–PTBs interfered with the localization of endogenous PTB in the PNC, the location of the endogenous PTB was examined by immunolabeling with SH54 (summarized in Table I). The result showed that the expression of the wild-type GFP–PTB and GFP–PTB mutants did not affect the localization of the endogenous PTB in the PNC (Table I). In addition, the expression of these fusion proteins did not appear to affect the subcellular distribution of the nucleolar protein, fibrillarin, or splicing factors such as SC35 (Table I).
In addition to the requirement of three RRMs for the localization of GFP–PTB to the PNC, we have also observed some interesting features of these mutant proteins with respect to their subcellular localization. The GFP– PTB fusion proteins with an intact NH2 terminus gave rise to a predominantly nuclear localization. The cytoplasmic localization of these proteins was hardly detectable (Fig. 10, top row and Table I). In contrast, deletion of the NH2terminal RRM resulted in a much more prominent cytoplasmic localization of these mutant proteins (Fig. 10 and Table I). Particularly, the mutant PTB-5, which contained two RRMs at the COOH terminus, showed a predominantly cytoplasmic localization (Fig. 10). However, neither RRM3 nor RRM4 alone is sufficient to direct a strong cytoplasmic localization (Fig. 10). These findings suggest that the NH2 terminus of PTB may be responsible for the nuclear localization of this protein.
Discussion
We have examined a large number of human cancer and normal diploid cells and have found that the presence of the PNC correlates with the transformed phenotype. Our findings agree with and further extend unpublished data described in Matera et al. (1995), stating that the PNC is found predominantly in transformed cells. The correlation between the PNC and cancer cells suggests that the formation of the PNC may be the result of oncogenic transformation. Further support for this suggestion comes from the controlled observation of a pair of cell lines in which PNC prevalence increased >25-fold from WI-38 cells, normal human lung fibroblasts, to their transformed derivative, WI-38 VA13 cells. In addition, we found that PNC prevalence varies tremendously among different cancer cell lines examined, suggesting that PNC prevalence may correspond to the degree of malignancy. This possibility is supported by the examination of breast cancer cells, in which PNC prevalence appears to be 5-fold higher in MDA-MB-468 cells, derived from a breast metastatic carcinoma that induces tumors in nude mice, than in Hs 578T cells, derived from a breast ductal carcinoma that does not induce tumors in nude mice. Investigations are currently underway to examine the correlation between the PNC and human cancer tissues.
The development of cancer is clearly a multi-step process (Knudson, 1971), a chain of events in which the consequence of one change unleashes a cascade of subsequent alterations. Although the function of the PNC is unknown, its preferential presence in cancer cells suggests that the formation of the PNC is part of an evolving process that transforms normal cells into cancer cells. The formation of the PNC may result from initial abnormalities and in turn may promote additional changes during the progression of cancer. However, it is also possible that the formation of these nuclear structures merely represents a cellular response to overall physiological changes that occur during cancer formation.
Observations in living and fixed cells at the light and electron microscopic levels have demonstrated a close association between the PNC and the nucleolus. The association begins during nucleologenesis at late telophase and extends to prophase before nucleolar dissociation. Such a close interaction suggests that the PNC may be linked to nucleolar activities. It has been documented that nucleoli undergo significant changes during carcinogenesis (for review see Busch, 1981), including increases in the expression of certain nucleolar proteins as well as alterations in the number and shape of nucleoli. Immunocytochemical labeling of nucleolar proteins has provided useful markers in cancer diagnosis and prognosis (Busch, 1981, 1990). However, the functional significance of the changes in nucleoli during carcinogenesis remains unclear. In normal cells, the function of the nucleolus has long been shown to be primarily involved in the biogenesis of preribosomal particles (for reviews see Busch and Smetana, 1970; Hadjiolov, 1985; Scheer and Benavente, 1990); more recently, studies in the yeast system revealed that nucleoli may also play a role in the processing and transport of poly(A)+ RNA (Schneiter et al., 1995; Tani et al., 1995). During carcinogenesis, nucleolar activities may change to accommodate the changes in cellular physiology. The spatial association between PNCs and the nucleolus as well as the correlation between PNCs and oncogenic transformation raise the possibility that the PNC may participate in the changes in nucleolar activities that occur during carcinogenesis. However, only when structural components and the function of the PNC are better understood, can its role in carcinogenesis be addressed.
The PNC is an electron-dense structure consisting of multiple strands, some of which are directly linked to the surface of the nucleolus. These thick strands may correspond with strand-like substructures observed in living cells (Fig. 5). As compared to the coiled body, which is densely packed with coiled fibrils (Ramon y Cajal, 1903), the PNC is loosely packed with thick strands that are surrounded by areas of lesser electron density. The PNC seems to bear some resemblance to a structure described by Cohen et al. (1984; Chung et al., 1984). These investigators identified protuberances that develop at the nucleolar periphery in estrogen-stimulated nerve cells (Chung et al., 1984; Cohen et al., 1984). Examination by electron microscopy showed these structures to be electron dense without well defined margins. Similar to the PNC, they are linked to the surface of the nucleolus by strands of electron-dense material (Cohen et al., 1984). Sodium tungstate staining and DNase digestion on resinless sections suggested that they contain DNA (Chung et al., 1984; Cohen et al., 1984). Studies are underway to determine if these protuberances and the PNC are the same structure.
Using deletion mutagenesis we have analyzed the sequence requirement for PTB targeting to the PNC. We have found that at least three RRMs at either the COOH or NH2 terminus are necessary and sufficient for the truncated fusion proteins to be localized to the PNC. It is unlikely that a specific signal involving only a small stretch of conserved sequences is responsible for the localization of the protein to the PNC. Analysis of translational regulation by PTB in virus-infected human cells has previously shown that three RRMs are essential and sufficient for an efficient binding of PTB to the internal ribosomal entry sites of viral RNA transcripts (Kaminski et al., 1995). Our finding that three RRMs are needed for PTB to be localized to the PNC suggests that the localization of PTB in the PNC may be due to its binding to polypyrimidine tract containing RNA(s). In addition, we have recently found that the presence of PTB in the PNC is sensitive to RNase A treatment (data not shown), further supporting the idea that the presence of PTB in the PNC is dependent upon its RNA binding. Our observations agree with and augment the finding and suggestion by Matera et al. (1995) that several small RNA polymerase III transcripts containing pyrimidine-rich sequences, including multiple Y RNAs, RNase P, and MRP RNAs, are present in the PNC and that PTB may therefore bind these RNAs. PTB is an RNA-binding protein preferentially recognizing pyrimidine-rich sequences and has been reported to be involved in multiple cellular functions including pre-mRNA splicing (Patton et al., 1993; Gozani et al., 1994; Singh et al., 1995), splice site selection in alternative pre-mRNA splicing (Lin and Patton, 1995), RNA polyadenylation (Lou et al., 1996), and translational regulation of certain viral RNA transcripts (Hellen et al., 1994; Kaminski et al., 1995; Witherell et al., 1995). Its capability of shuttling between the nucleus and cytoplasm suggests that PTB may also be involved in RNA transport (Pinol-Roma and Dreyfuss, 1992; Michael et al., 1995). PTB appears to participate in these functions through the binding to pyrimidine-rich RNA sequences. Thus, it is possible that PTB serves as a bridge between the pyrimidine tract RNAs and different macromolecules in fulfilling different cellular functions. However, the association and function of PTB in the PNC remains to be determined.
In summary, we have extensively characterized the PNC both in fixed and living cells using PTB as a probe. We have found that the PNC is a dynamic structure that is in direct contact with the nucleolus. The association initiates at the beginning of the cell cycle and ends at prophase. The PNC appears to be an electron-dense structure. It is composed of multiple strands, each measuring 80–180 nm diam. Some of the strands are in direct contact with the surface of the nucleolus. Deletion mutagenesis of GFP– PTB indicates that at least three RRMs, either NH2 or COOH terminal are required for PTB to be localized to the PNC, suggesting that the role of PTB in the PNC may involve its binding to polypyrimidine tract-containing RNAs. Furthermore, the presence of the PNC is closely associated with oncogenic transformation. Studies are underway to further analyze the structural components and functional characteristics of the PNC.
Footnotes
1. Abbreviations used in this paper: GFP, green fluorescent protein; PML, promyelocyte; PNC, perinucleolar complex; POD, PML oncogenic domain; PTB, polypyrimidine tract binding protein; RRM, RNA recognition motif.
We would like to express our special gratitude to Tamara Howard for her excellent technical assistance. We would also like to thank Dr. G. Dreyfuss (University of Pennsylvania, Philadelphia, PA) and his colleagues for confirming the specificity of the antibody SH54 to PTB, and Dr. E. Wimmer (S.U.N.Y., Stony Brook, NY) for the gift of the human PTB cDNA clone. We are very grateful to Drs. D. Helfman (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), G. Matera (Case Western Reserve University, Cleveland, OH), P. Mintz (Cold Spring Harbor Laboratory), and T. Misteli (Cold Spring Harbor Laboratory) for their helpful comments on the manuscript.
D.L. Spector is supported by a grant from the National Institutes of Health (GM42694).
Please address all correspondence to Dr. Sui Huang, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724. Tel.: (516) 367-8478; Fax: (516) 367-8876; E-mail: huang@cshl.org
References
- Ascoli AC, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouteille M, Kalifat SR, Delarue J. Ultrastructural variations of nuclear bodies in human diseases. J Ultrastruct Res. 1967;19:474–486. doi: 10.1016/s0022-5320(67)80074-1. [DOI] [PubMed] [Google Scholar]
- Brasch K, Ochs RL. Nuclear bodies (NBs): a newly “rediscovered” organelle. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- Busch, H. 1981. Molecular biology of the nucleolus and its relation to cancer. In Medicine in Transition. E.P. Cohen, editor. University of Illinois Press, Urbana, IL. 259–295.
- Busch H. The final common pathway of cancer: presidential address. Cancer Res. 1990;50:4830–4838. [PubMed] [Google Scholar]
- Busch, H., and K. Smetana. 1970. The Nucleolus. Academic Press, New York. 1–626.
- Chung SK, Cohen RS, Pfaff DW. Ultrastructure and enzyme digestion of nucleoli and associated structures in hypothalamic nerve cells viewed in resinless sections. Biol Cell. 1984;51:23–33. doi: 10.1111/j.1768-322x.1984.tb00280.x. [DOI] [PubMed] [Google Scholar]
- Cohen RS, Chung SK, Pfaff DW. Alteration by estrogen of the nucleoli in nerve cells of the rat hypothalamus. Cell Tissue Res. 1984;235:485–489. doi: 10.1007/BF00226943. [DOI] [PubMed] [Google Scholar]
- Deerinck TJ, Martone ME, Lev-Ram V, Green DP, Tsien RY, Spector DL, Huang S, Ellisman MH. Fluorescence photooxidation with eosin: a method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol. 1994;126:901–910. doi: 10.1083/jcb.126.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WH, Jr, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature (Lond) 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/ coiled body a universal nuclear component? . Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Sharp PA, Jamison SF, Garcia-Blanco MA. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO (Eur Mol Biol Organ) J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov, A.A. 1985. The nucleolus and ribosome biogenesis. In Cell Biology Monographs. Springer-Verlag New York, Inc., New York. 1–263.
- Hellen CU, Pestova TV, Litterst M, Wimmer E. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci USA. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Hunt SL, Patton JG, Jackson RJ. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- Knudson AGJ. Mutation and cancer. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken MH, Puvion-Dutilleul F, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Caivo F, Chomienne C. The t(15:17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO (Eur Mol Biol Organ) J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken MH, Linares CG, Quignon F, Viron A, Chelbi AM, SobczakThepot J, Juhlin L, Degos L, Calvo F, de-The H. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Lee B, Matera AG, Ward DC, Craft J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: a possible coordinate role in ribosome biogenesis. Proc Natl Acad Sci USA. 1996;93:11471–11476. doi: 10.1073/pnas.93.21.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Patton JG. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO (Eur Mol Biol Organ) J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Lou H, Gagel RF, Berget SM. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruc Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Moreno Diaz de la Espina, S., A. Sanchez Pina, and M.C. Risueño. The role of plant coiled bodies in the nuclear RNA metabolism. Electron Micros. 1980;2:240–241. [Google Scholar]
- Ochs RL, Stein TWJ, Tan EM. Coiled bodies in the nucleolus of breast cancer cells. J Cell Sci. 1994;107:385–399. doi: 10.1242/jcs.107.2.385. [DOI] [PubMed] [Google Scholar]
- Patton JG, Mayer SA, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature (Lond) 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S. Un sencillo método de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Raska I, Andrande LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with antoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scheer U, Benavente R. Functional and dynamic aspects of the mammalian nucleolus. Bioessays. 1990;12:14–21. doi: 10.1002/bies.950120104. [DOI] [PubMed] [Google Scholar]
- Schneiter R, Kadowaki T, Tartakoff AM. mRNA transport in yeast: time to reinvestigate the function of the nucleolus. Mol Biol Cell. 1995;6:357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and function of higher eukaryotic polypyrimidine tract-binding proteins. Science (Wash DC) 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell. 1992;3:555–569. doi: 10.1091/mbc.3.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T, Derby RJ, Hiraoka Y, Spector DL. Nucleolar accumulation of poly (A) RNA in heat-shocked yeast cells: implication of nucleolar involvement in mRNA transport. Mol Biol Cell. 1995;6:1515–1534. doi: 10.1091/mbc.6.11.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590–1597. [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Witherell GW, Schultz-Witherell CS, Wimmer E. Cis-acting elements of the encephalomyocarditis virus internal ribosomal entry site. Virology. 1995;214:660–663. doi: 10.1006/viro.1995.0081. [DOI] [PubMed] [Google Scholar]