EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer (original) (raw)
Abstract
Recent experiments have demonstrated that the Polycomb group (PcG) gene EZH2 is highly expressed in metastatic prostate cancer and in lymphomas. EZH2 is a component of the PRC2 histone methyltransferase complex, which also contains EED and SUZ12 and is required for the silencing of HOX gene expression during embryonic development. Here we demonstrate that both EZH2 and EED are essential for the proliferation of both transformed and non-transformed human cells. In addition, the pRB-E2F pathway tightly regulates their expression and, consistent with this, we find that EZH2 is highly expressed in a large set of human tumors. These results raise the question whether EZH2 is a marker of proliferation or if it is actually contributing to tumor formation. Significantly, we propose that EZH2 is a bona fide oncogene, since we find that ectopic expression of EZH2 is capable of providing a proliferative advantage to primary cells and, in addition, its gene locus is specifically amplified in several primary tumors.
Keywords: E2F/EED/EZH2/Polycomb/pRB
Introduction
The retinoblastoma protein (pRB) pathway plays a critical role in regulating progression through the mammalian cell cycle. The high frequency with which alterations have been found in the core members of this pathway in human tumors has led to the suggestion that its inactivation is an obligatory event for the development of all human cancers (Hanahan and Weinberg, 2000). The most studied and best understood targets for pRB are members of the E2F transcription factor family (Dyson, 1998). The E2Fs are essential for the transcriptional regulation of a number of genes whose products control cell cycle progression. These include both genes essential for the entry into the S phase of the cell cycle, such as CCNE1 (Cyclin E1) and CCNA2 (Cyclin A2), and genes that are involved in the regulation of DNA replication, such as CDC6, DHFR and TK1 (Helin, 1998; Trimarchi and Lees, 2002).
Recently, a number of laboratories, including ours, have used gene expression profiling and ChIP assays to identify novel E2F-regulated genes (Müller et al., 2001; Weinmann et al., 2001, 2002; Ren et al., 2002). Due to the critical role of the E2Fs in controlling the expression of key regulators of cell proliferation, it is very likely that several novel key regulators of cell proliferation are amongst these many identified genes. The characterization of these will be critical for both the understanding of normal proliferation control and its deregulation in cancer.
The identification of a number of Polycomb group (PcG) genes, Enhancer of Zeste Homolog 2 (EZH2), Embryonic Ectoderm Development (EED) and Suppressor of Zeste 12 (SUZ12) as potential E2F targets was one of the most interesting findings in our screens (Müller et al., 2001). PcG genes are best known for their role in maintaining the repression of HOX genes during development (for reviews, see Brock and van Lohuizen, 2001; Jacobs and van Lohuizen, 2002). The PcG proteins are believed to act by forming multiprotein complexes that, through modification of chromatin structure, repress target gene expression. Two distinct PcG protein complexes, Polycomb repressive complex (PRC)1 and PRC2 have been genetically and biochemically defined (Jacobs and van Lohuizen, 2002). Interestingly, the three potential E2F regulated PcG genes, EZH2, EED and SUZ12 constitute the PRC2 complex (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Müller et al., 2002). PRC1 contains several other PcG proteins including the oncogene BMI1, HPC, HPH2 and SCML (Jacobs and van Lohuizen, 2002). Functionally, EZH2 is the catalytically active component of PRC2 and is capable of methylating lysine 9 (H3K9) and lysine 27 (H3K27) of histone H3 (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Müller et al., 2002). Interestingly, HPC2 of the PRC1 complex can specifically bind to methylated K27 (Cao et al., 2002; Kuzmichev et al., 2002). When taken together with the previous demonstrations that PRC1 cannot associate with chromatin without a functional PRC2, this suggests a model in which K27 methylation by EZH2 results in the recruitment of PRC1 and subsequent transcriptional repression.
Here, we show that the expression of the two PRC2 members EZH2 and EED is controlled by the E2F transcription factors. We demonstrate that EZH2 and EED are required for cell proliferation acting as essential downstream mediators of E2F function. Furthermore, we show that EZH2 is highly expressed and amplified in primary human tumors, and that ectopic expression of EZH2 confers a growth advantage in primary mouse embryo fibroblasts. Thus, in addition to identifying EZH2 as being a strong candidate oncogene, our data provide a direct link between the pRB-E2F growth control pathway and chromatin modifications regulated by an essential PcG complex.
Results
The pRB-E2F pathway regulates the expression of the EED and EZH2
Previously, we identified three PRC2 genes EZH2, EED and SUZ12 as being induced upon overexpression of E2F in the tumor cell line U2OS (Müller et al., 2001). To establish if these genes are relevant physiological targets of E2F function, we have performed a number of experiments as described below.
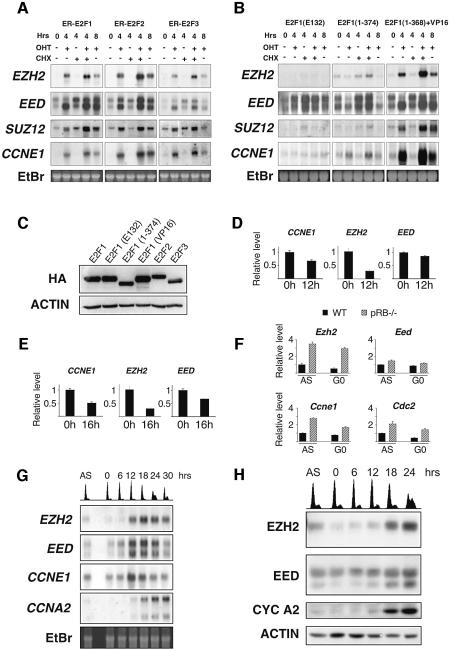
First, to establish whether E2Fs were also capable of inducing the PRC2 genes in a non-transformed cell line, northern blot analysis was performed in WI38 human diploid fibroblasts. This analysis showed that the activation of E2F1–3 dramatically increased the levels of the three PRC2 genes (Figure 1A). This up-regulation occurred in the absence of de novo protein synthesis. To test the mechanism by which the PRC2 genes were induced we used specific mutants of E2F1 (Figure 1B). The expression levels of all fusion proteins were determined by western blotting (Figure 1C). A DNA-binding mutant E2F1(E132) and a transactivation domain (TA) mutant E2F1(1–374) were not capable of inducing the PRC2 genes. However, a mutant in which the TA and the pRB-binding domain of E2F1 are substituted with the TA of another transcription factor, VP16, was capable of inducing the PcG genes. These results demonstrate that E2F-mediated transactivation of the PRC2 genes requires functional transactivation and DNA-binding domains.
Fig. 1. The expression of EZH2 and EED is regulated by E2F and pRB. (A) Northern blot analysis of EZH2, EED, SUZ12 and CCNE1. WI38 fibroblasts expressing ER-E2F1, ER-E2F2 or ER-E2F3 were grown as indicated in the presence (+) or absence (–) of OHT and cycloheximide (CHX). mRNA was extracted at the indicated hours after addition of 4-hydroxy tamoxifen (OHT) and/or CHX. (B) Northern blot analysis as described in panel A using WI38 expressing ER-E2F1 mutants. (C) Western blot analysis of WI38 expressing ER-E2F fusion proteins. (D) EZH2 and EED expression is repressed by pRB. Real-time quantitative PCR (qPCR) was used to determine relative mRNA expression levels upon induction of pRbΔcdk in U2OS cells. (E) EZH2 and EED expression is repressed by p16. qPCR was used to determine the effects of p16 overexpression. (F) Ezh2 expression is derepressed in _Rb_–/– MEFs. qPCR was used to determine Ezh2, Eed, Ccne1 and Cdc2 mRNA levels in pRb-deficient and wild-type (WT) asynchronously growing (AS) and serum-starved (G0) MEFs. (G) The expression of EZH2 and EED is cell growth regulated. WI38 cells were serum starved for 72 h and re-stimulated to enter the cell cycle by the addition of serum. mRNA was prepared for northern blot analysis at the indicated time points. Propidium iodide (PI) FACS analysis is presented at the top of the panel. (H) EZH2 and EED proteins are cell growth regulated. Western blot analysis of cell lysates prepared from an IMR90 cell growth experiment similar to that described in (C). PI FACS analysis is presented at the top of the panel.
Next, we performed experiments to determine if the expression of two upstream regulators of E2F, namely pRB and p16 affected EZH2 and EED transcription. The expression of both pRbΔcdk and p16 results in repression of E2F activity and subsequent cell cycle arrest (Jiang et al., 1998; Lukas et al., 1999). To avoid secondary effects on transcription as a result of this cell cycle arrest, cells were harvested before measurable changes in cell cycle progression were observed (data not shown). In U2OS cells, the expression of pRbΔcdk and p16 led to significant repression of EZH2 and to a lesser extent, EED mRNA levels (Figure 1D and E). In addition, the mRNA levels of Ezh2 and in part Eed were increased in Rb–/– mouse embryonic fibroblasts (MEFs) compared with wild-type MEFs (Figure 1F). Taken together, these data suggest that both EZH2 and EED are physiological targets of the pRB-E2F pathway, and that deregulation of the pathway, as is frequently observed in human cancer, would result in higher levels of EZH2 and EED.
EZH2 and EED expression is restricted to growing cells
In differentiated cells, the mRNA levels of both EZH2 and EED are down-regulated. However, their expression is not cell cycle regulated (see Supplementary figure 1 available at The EMBO Journal Online). Therefore, to determine if the expression of EZH2 and EED is cell growth regulated, WI38 cells were serum starved for 3 days and subsequently re-stimulated to enter the cell cycle by the addition of serum. As shown in Figure 1G, both EZH2 and EED mRNA transcripts are strongly cell growth regulated and accumulate at the G1/S transition consistent with being E2F cell growth regulated genes. Western blot analysis demonstrated that the protein levels of both EZH2 and EED are also strongly cell growth regulated (Figure 1H). At least two specific forms of EED were recognized using a monoclonal antibody generated against the full-length mouse Eed protein (Figure 1H). The slower migrating form could be a modified form of the EED protein since ectopic expression of EED also generated this slower migrating band (Figure 4C). In addition both bands are reduced in intensity upon specific EED siRNA (Figure 3B) confirming the specificity of the antibody. Further studies will be required to understand the nature of this slower migrating modified EED protein. Significantly, only the faster migrating form of EED is strongly cell growth regulated.
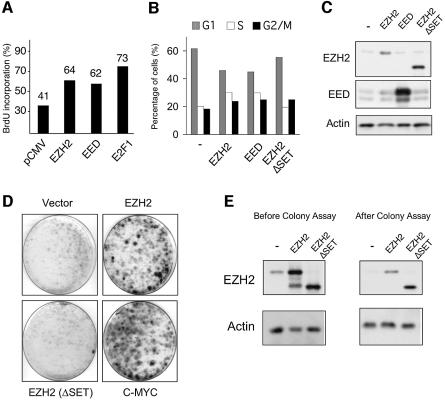
Fig. 4. Ectopic expression of EZH2 results in shorter G1 and prolongs the life span of primary MEFs. (A) Ectopic expression of EZH2 and EED increases the number of cells in S-phase. Serum-starved, quiescent Rat1 cells were injected with pCMV plasmids expressing either EZH2 or EED as indicated. Cells injected with E2F1 were used as a positive control. Serum was added to the serum-starved injected cells for 14 h prior to a 45 min pulse of BrdU. Relative DNA synthesis was determined by assessing BrdU incorporation by immunofluorescence. One representative experiment of five independent experiments is shown. (B) EZH2 ectopic expression results in S phase accumulation. Cell cycle phases of HeLa cells, transfected with the indicated plasmids, were determined 48 h after transfection by PI-FACS. (C) Western blot analysis of HeLa cells show similar expression of the wild type and mutant EZH2 proteins. (D) Expression of wild type EZH2, but not a SET-domain mutant, allows primary MEFs to form colonies, when plated at low density. Primary MEFs were infected with retroviral vectors expressing the indicated proteins. The experiment shown is representative of five independent experiments using different MEF preparations. (E) Western blot analysis of MEFs infected with the indicated retroviral vectors before and after the colony assay.
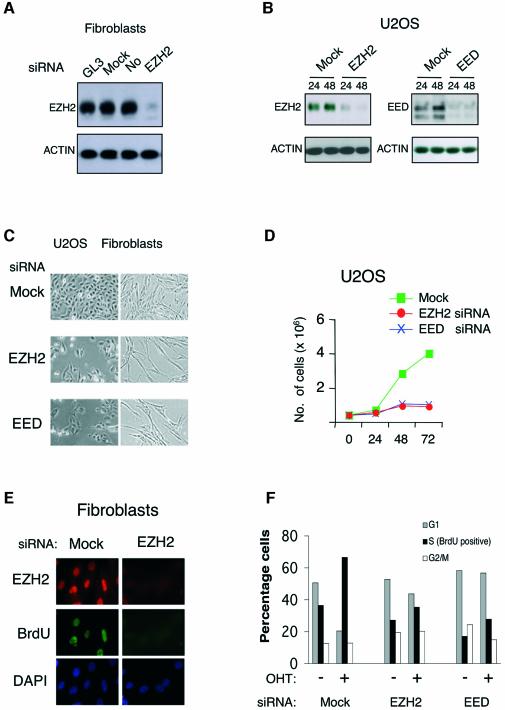
Fig. 3. EZH2 and EED are essential for proliferation of both normal and cancer cells. (A) Specific inhibition of EZH2 expression by siRNA in TIG3 fibroblasts. TIG3 cells were transfected with H2O (Mock), luciferase siRNA oligos (GL3) and oligos specific for EZH2 mRNA. Cell lysates were prepared 44 h after transfection and the EZH2 protein levels were detected by western blot. (B) Specific inhibition of EZH2 and EED expression by siRNA in U2OS cells. Cells were transfected with the indicated siRNA oligos and cell lysates were prepared for western blot analysis 24 and 48 h after transfection. (C) Inhibition of EZH2 and EED expression results in changes in cellular morphology and slower growth. Representative photos of TIG3 (fibroblast) and U2OS cells taken 48 h after siRNA transfection. (D) Inhibition of EZH2 and EED expression results in slower growth. Growth curves of EZH2, EED and mock siRNA treated U2OS are presented. The growth curves were determined in triplicate, and they are representative of three independent experiments. (E) siRNA to EZH2 results in specific decrease in EZH2 levels and inhibition of DNA synthesis. The levels EZH2 protein and the amount of BrdU incorporation were determined by immunofluorescence 44 h after transfection of BJ1 cells. (F) EZH2 and EED are required for E2F induced proliferation. ER-E2F3 expressing U2OS cells were treated with mock, EZH2 and EED siRNA for 36 h in the absence of serum. OHT was added for 14 h and the biological effect of E2F3 overexpression was determined by BrdU FACS after a 10 min pulse of BrdU.
E2Fs transactivate and bind to the EED and EZH2 promoters
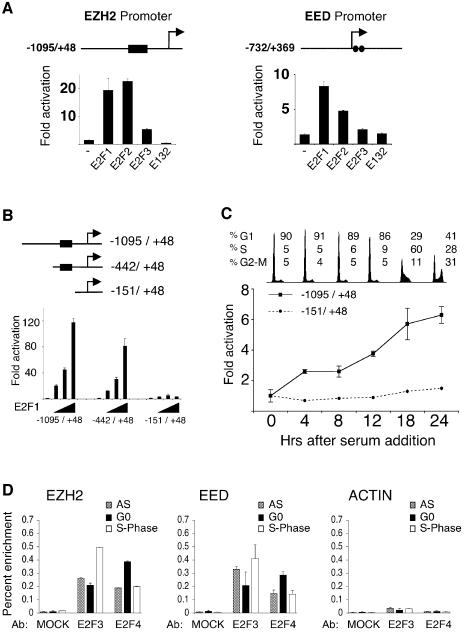
To investigate the mechanism by which the E2Fs can regulate the expression of EZH2 and EED, we identified their promoters in the NCBI database. Inspection of the sequences of the two promoters revealed that they each contain at least two potential E2F DNA binding sites (Figure 2A). The indicated promoters cloned into a luciferase reporter construct, responded to ectopic expression of E2F1, E2F2 and E2F3, but not to the E132 DNA binding mutant of E2F1 (Figure 2A). Generation of truncated versions of the EZH2 promoter demonstrated that removal of four potential E2F DNA binding sites rendered the promoter unresponsive to ectopic expression of E2F1 (Figure 2B). To determine if these E2F responsive elements in the EZH2 promoter are required for its cell growth regulation, Rat1 cells were transfected with the EZH2 promoter constructs, serum-starved and subsequently induced to enter the cell cycle by the addition of serum. As shown in Figure 2C, the longest EZH2 promoter construct, which is efficiently transactivated by the E2Fs, is induced upon entry into the cell cycle, consistent kinetically with the cell growth-regulated expression of the EZH2 mRNA. In contrast, the EZH2 promoter construct lacking the putative E2F binding sites, does not respond to serum addition, indicating that _EZH2_’s cell growth regulation is dependent on E2F. Interestingly, the shorter construct is approximately four times as active as the longer promoter construct in quiescent cells, suggesting that the E2F DNA binding sites are also required for the efficient repression of transcription in non-growing quiescent cells (data not shown). As a final test for a functional role of E2F in the regulation of the EZH2 and EED expression, quantitative chromosomal immunoprecipitation (ChIP) assays using antibodies specific for E2F3 and E2F4 were performed. As shown in Figure 2D, E2F3 and E2F4 are bound to the EZH2 and EED promoters, demonstrating that the EZH2 and EED promoters are direct targets of the E2F transcription factors in vivo. Significantly, the enrichment of E2F3 is greatest in S phase while the enrichment of E2F4 is strongest in G0.
Fig. 2. E2F binds to the EZH2 and EED promoters to regulate their expression. (A) E2F transactivates the EZH2 and EED promoters. Schematic presentation of the genomic regions corresponding to the promoters of both EZH2 and EED. The black box in the EZH2 promoter represents a region of four putative E2F sites, while the two circles in the EED promoter each represent a putative E2F DNA binding site. The +1 represents the most 5′ nucleotide in the longest identified cDNA published in the NCBI database. The numbers on the promoter constructs are relative to +1. Luciferase reporter analysis of EZH2 and EED promoter constructs co-transfected with pCMVE2F1, pCMVE2F2, pCMVE2F3 or pCMVE2F1(E132) DNA binding mutant in 293T cells. (B) Delineation of E2F DNA binding sites in the EZH2 promoter. U2OS cells were transfected with the indicated luciferase reporter constructs and different concentrations of pCMVE2F1 and pCMVE2F1(E132). (C) E2F is required for the cell growth regulated expression of EZH2. Rat1 cells were transfected either with the indicated EZH2 promoters,serum starved for 48 h and then released into the cell cycle by the addition of serum. A plasmid expressing β-galactosidase was co-transfected to normalize luciferase activity for transfection efficiency. Fold activations refer to the empty pCMV transfection. Standard deviation of the mean is shown in each experiment. (D) E2F3 and E2F4 chromatin immunoprecipitations of TIG3 fibroblasts serum starved (G0), growing (AS) and serum released cells in S phase (S).
EZH2 and EED are required for proliferation of both normal and cancer cells and confer a proliferative advantage when overexpressed
To test if EZH2 and EED are essential for proliferation, double stranded RNA interfering oligonucleotides (siRNA oligos) were designed for both EZH2 and EED. As shown in Figure 3A, siRNA to EZH2 specifically prevented EZH2 synthesis in TIG3 diploid human fibroblasts. Almost complete depletion of both EZH2 and EED were also observed in U2OS cells (Figure 3B) and in HeLa cells (data not shown).
Cells depleted for EZH2 and EED appeared larger than control transfected cells with a senescent-like phenotype. Inspection of transfected cultures indicated that lack of EZH2 and EED resulted in less proliferation (Figure 3C). This effect was quantified by measuring the proliferation rate of the transfected cells (Figure 3D). A decrease in either EZH2 or EED expression resulted in a strong inhibition of cell proliferation in U2OS cells. Similar reductions in growth rates were also observed in other transformed cells such as HeLa and SAOS2, as well as in the diploid fibroblasts TIG3, WI38 and HEL299 (data not shown).
To understand if EZH2 and EED are required at a specific point during the mammalian cell cycle, human diploid BJ1 cells were transfected with EZH2 siRNA or control siRNA, and BrdU incorporation was measured 48 h after transfection (Figure 3E). Whereas EZH2 was easily detected in mock-transfected cells, no EZH2 expression was detected in cells that had received EZH2 siRNA. Moreover, consistent with the notion that both EZH2 and EED are required for cell proliferation, cells transfected with EZH2 and EED siRNA ceased DNA replication (Figure 3E). Similar results were obtained in U2OS and HeLa cell lines (data not shown). Furthermore FACS analysis showed that all cell lines transfected with siRNA to EED or EZH2 ceased proliferation with increases in the number of cells in G1, G2 or both (data not shown), indicating that EZH2 and EED required for both entry into and progression through the S phase of the cell cycle.
To evaluate if EZH2 and EED are required for E2F induced cell proliferation, we established an assay in which E2F activation in human cells increases the number of cells in S phase. To do this, we deprived U2OS cells expressing ER-E2F3 of serum for 36 h, resulting in a 2-fold decrease in the number of BrdU incorporating cells (Figure 3F; data not shown). In these conditions, activation of E2F3 for 14 h led to a 2-fold increase in the number of BrdU incorporating cells. Significantly, this increase was dependent on EZH2 and EED suggesting that both genes are essential downstream mediators of E2F-induced proliferation.
Since EZH2 and EED siRNA depleted cells cease DNA replication, we wished to determine if the PRC2 proteins could accelerate S phase entry in cells released from growth arrest. To test this, we microinjected EZH2 and EED expression plasmids into quiescent Rat1 cells (Figure 4A). These cells were subsequently released from serum-starvation and monitored for S phase entry. E2F1 was used as a positive control in these experiments. A representative example of three independent experiments is shown in Figure 4A, and demonstrates that overexpression of EZH2 and EED shortens the G0/G1-S transition. Consistent with this result, overexpression of EZH2 and EED in HeLa cells resulted in an increased number of cells in S phase (Figure 4B). Significantly, this effect of EZH2 required a functional SET domain. The overexpression of EZH2, EED and EZH2ΔSET was confirmed by western blot analysis (Figure 4C).
Next, we wished to determine if ectopic expression of EZH2 could confer a growth advantage to primary cells. It is known that primary cells when plated at low density undergo G1 arrest (Sherr and DePinho, 2000). However, loss of all three members of the pRB family, inactivation of p53 or ARF, or overexpression of oncogenes, such as MYC can allow these cells to continue to grow (Beier et al., 2000; Sage et al., 2000; Sherr and DePinho, 2000). To test EZH2 in this assay, we infected primary MEFs with retroviral vectors expressing EZH2, EZH2ΔSET, and MYC as a positive control. The infected cells were plated at low density and grown for 14 days before staining with crystal violet. The expression levels of EZH2 were controlled both prior to and after the experiment (Figure 4E). As demonstrated in Figure 4D, expression of EZH2, like MYC, allows a substantial number of cells to form colonies. Significantly, EZH2 requires a functional SET domain for this biological effect. The colonies formed were picked and passaged further to determine their characteristics. These cells grew very slowly, suggesting they were not immortal and therefore that EZH2 and MYC are not acting as immortalizing oncogenes in this assay. In fact, of the colonies tested, all contained a functional p53 pathway as demonstrated by the accumulation of p53 and p21 following exposure to UV (data not shown). Therefore in summary, overexpression of EZH2, like MYC (Beier et al., 2000) shortens the G1 phase of the cell cycle, results in accumulation of cells in the S phase of the cell cycle, and confers a proliferative advantage when ectopically expressed in primary MEFs.
EZH2 is highly expressed and amplified in human cancer
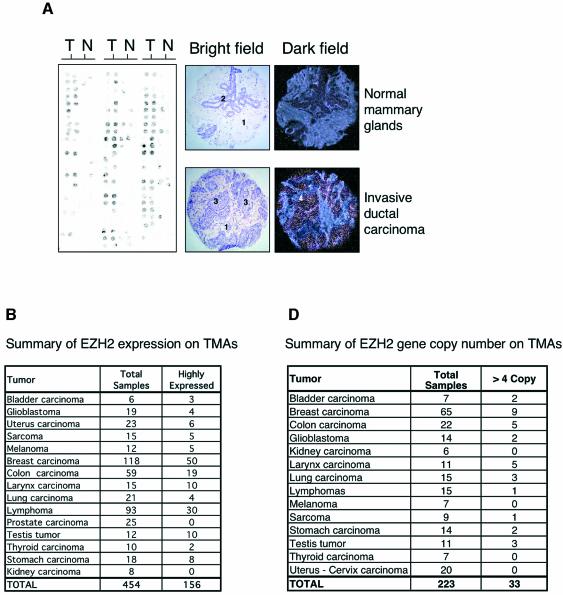
Since our data demonstrate that ectopic expression of EZH2 confers a growth advantage, and previous results have shown that a subset of genes regulated by the E2Fs are overexpressed in cancer, we tested the expression of EZH2 in primary human tumors by mRNA in situ hybridization (ISH). We evaluated the levels of EZH2 expression in the in situ analysis using a scale of 0–3, where 0 = negative staining intensity, 1 = weak staining intensity, 2 = moderate staining intensity and 3 = strong staining intensity. Most normal non-tumor tissue presented negative or weak staining. In contrast, a total of 34% (156/454) of the tumor samples analyzed presented moderate/strong EZH2 expression levels. Therefore we consider EZH2 in these tumors to be ‘highly expressed’. These data suggest that EZH2 is overexpressed in a larger fraction of human tumors than previously thought. Significantly, we did not observe high expression of EZH2 in 25 primary prostate cancers analyzed, which could appear to contradict a recent publication showing that EZH2 overexpression correlates with aggressiveness of metastatic prostate cancers (Varambally et al., 2002). However, we do not believe so, since we did not have any metastatic prostate cancers among the 25 samples analyzed and most of them showed low proliferative index (<10% as assessed by Ki67 IHC).
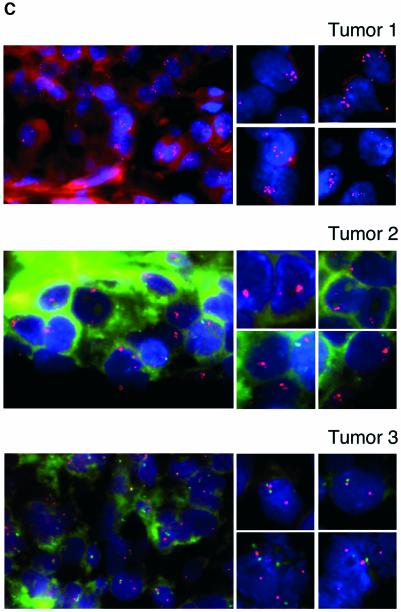
Since the pRB pathway controls the expression of EZH2, and this pathway is deregulated in human tumors, the high expression of EZH2 observed in the tumors could simply be due to specific genetic alterations in core members of the pRB pathway. Alternatively, as has been shown for some E2F-regulated genes such as CCNE1 (Lin et al., 2000), MYBL2 (B-MYB; Kononen et al., 1998) and MYB (Wallrapp et al., 1997), the high expression of EZH2 could be the result of amplification of the gene locus. Therefore to determine if EZH2 is amplified in primary human tumors, we isolated a BAC clone containing EZH2 located on chromomsome 7q35 (Cardoso et al., 2000). The fluorescent-labeled BAC clone was used for FISH analysis on tissue microarrays together with a probe specific for the centromeric region of chromosome 7 (Figure 5C). In all, but two cases, we observed two copies of the centromeric region of chromosome 7 (data not shown). Significantly, in ∼15% of the 225 tumors analyzed (Figure 5D) we observed amplification of EZH2 (defined as more than four copies per tumor) (Figure 5C, top panel) or a tight cluster of signals (Figure 5C, middle panel). Several cases presented with 8–10 copies of the EZH2 gene per cell (Figure 5C, top panel; data not shown). In a recent report, in which the gene expression profile and the gene copy number of several primary breast cancers were determined, the investigators found a strong correlation between DNA copy number and levels of transcription (Pollack et al., 2002). Specifically, these authors found that on average a 2-fold change in DNA copy number is associated with a 1.5-fold change in mRNA levels. Therefore, our FISH results suggest that EZH2 is overexpressed between 3- and 8-fold in the analyzed breast tumors, which is consistent with our mRNA ISH analysis. Very importantly, we found that all the tumors, which had increased DNA copy number of EZH2, also overexpressed the gene at the mRNA level. Taken together, our data show that EZH2 is highly expressed in a large number of human cancers, and that, at least in a subset of these cases, the high level of expression is likely to be due to a specific amplification of the EZH2 gene.
Fig. 5. EZH2 is highly expressed and amplified in primary human tumors. (A) EZH2 is highly expressed in primary breast tumors. The expression of EZH2 mRNA levels was determined by ISH on TMA. The left panel shows a representative example of ISH for EZH2 of a breast cancer TMA. Two samples from each tumor (T) and normal (N) counterparts were taken for the TMA. The bright field panels show the morphology of the tissue sample as revealed by hematoxylin and eosin counterstaining. The dark field panels show the ISH analysis of EZH2 expression levels in a breast invasive ductal carcinoma and in a normal mammary gland. In the panels the numbers indicate: 1, stroma; 2, mammary gland epithelial cells; 3, tumor tissue; 4, EZH2 expressing cells. (B) EZH2 is highly expressed in a large number of primary human tumors. Summary table of EZH2 expression on the TMAs tested. (C) Subsets of primary tumors show an amplification of the EZH2 gene (top and middle panel). FISH analysis of tumor TMAs of EZH2 gene copy number. The two panels show two representative breast carcinoma tissues with significant amplifications of EZH2. The EZH2 specific probe was labeled with Cy5 and a centromeric probe for chromosome 7 with FITC. Due to the fast fading of the FITC probe and the relative long microscopic analysis of each TMA we were able to count, but not to take pictures of the specific staining of the centromere for several of the analyzed samples. (C, lower panel) Example of a breast tumor tissue containing a normal copy number of EZH2. FISH analysis of an unrelated gene (HECTH9) on the same TMAs revealed no amplification in all the samples tested (data not shown). (D) Summary table of EZH2 copy number of the tested TMAs.
PRC2 is required for the expression of proliferative genes
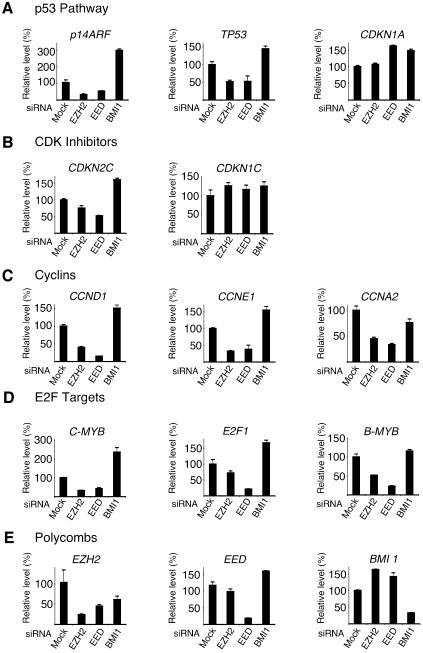
The identification of target genes for the PRC2 members will be essential for the determination of the mechanism by which they promote proliferation in both normal and cancer cells. Previously, Bmi1, a component of PRC1, has been shown to regulate cell proliferation by repressing p19ARF (p14ARF in humans) (Jacobs et al., 1999). Since the PRC2 complex is believed to be required for the repressive capacity of the PRC1 complex, we reasoned that the PRC2 complex could also repress p14ARF. Therefore, we tested if depletion of either EED or EZH2 resulted in elevated levels of p14ARF, which would explain the block in proliferation observed in PRC2 depleted cells. Relative mRNA levels were quantified by real time quantitative PCR in EZH2, EED, BMI1 and control GL3 siRNA treated TIG3 cells (Figure 6A). As expected, in BMI1 depleted cells, p14ARF levels increased 3-fold compared to control GL3 siRNA transfected cells. However, to our surprise, we found that p14ARF mRNA levels were significantly reduced in both EED and EZH2 siRNA treated cells (Figure 6A). Next, we extended our analysis to other known inhibitors of cell cycle progression and found the mRNA levels of p53 (TP53) were also reduced in PRC2 depleted cells (Figure 6A). In addition, we did not find significant increases in the expression of any of the cyclin-dependent kinase inhibitors: p16 (CDKN2A), p18 (CDKN2C), p21 (CDKN1A), p27 (CDKN1B) and p57 (CDKN1C) (Figure 6B; data not shown). Therefore, we also tested the expression of a number of ‘positive regulators’ of cell proliferation. In fact, in EED and EZH2 siRNA-treated cells the mRNA levels of all cyclins tested, namely cyclin D1 (CCND1), cyclin E1 (CCNE1), cyclin A2 (CCNA2) and cyclin B1 (CCNB1), were significantly reduced (Figure 6C; data not shown). The mRNA levels of all E2F-regulated genes tested, namely MYB, B-MYB, CDC6, E2F1 and CDC2, were also significantly reduced (Figure 6D; data not shown). In contrast, in BMI1-depleted cells, the mRNA levels of CCND1, CCNE1, C-MYB, E2F1 and B-MYB increased, similar to the effect observed for p14ARF mRNA.
Fig. 6. EZH2 and EED are required for the active transcription of several genes. Expression level analysis by qPCR of TIG3 cells tranfected for 44 h with siRNA specific for EZH2, EED and BMI1. (A) mRNA levels of members of the p53 pathway. (B) mRNA levels of CDK inhibitors. (C) mRNA levels of cyclins. (D) mRNA levels of E2F regulated genes. (E) mRNA levels of PcG genes.
Discussion
In this manuscript we demonstrate that the pRB-E2F pathway controls the expression of the PRC2 genes EZH2 and EED. This is consistent with the high expression of PRC2 genes in proliferating cells and their down-regulation in non-growing or differentiated cells. Furthermore, we demonstrate that the cell growth regulated expression of EZH2 and EED is dependent on the E2F transcription factors. Both PRC2 members are required for cellular proliferation in all cell types tested and their overexpression results in induction of S phase. In addition, EZH2 is capable of extending the growth capacity of primary cells, when they are subjected to growth inhibitory conditions. This suggests that EZH2 could contribute to neoplastic growth. Finally, we demonstrate that EZH2 is highly expressed in multiple primary tumors, which is consistent with the fact that the pRB pathway is deregulated in most cancers. However, our demonstration that the EZH2 locus is amplified in 10–15% of the tested tumors shows a specific genetic selection for tumors overexpressing EZH2 and suggests that the increased expression of EZH2 can also occur independently of a deregulated pRB pathway.
A functional link between the pRB-E2F pathway and PRC2 expression
A major finding presented here is the fact that the E2F transcription factors are required for the transcriptional regulation of EZH2 and EED. Taken together with the demonstration that ectopic expression of pRB and p16 represses their expression and that their expression is higher in pRb-deficient MEFs, this finding provides a functional link between the pRB pathway and the presence of the PRC2 complex. Whilst the pRB-E2F pathway is best known for its role in the regulation of cell proliferation and tumorigenesis, several results obtained in mouse, Drosophila and Xenopus have shown that the integrity of this pathway is also essential for differentiation and proper embryonic development (Lipinski and Jacks, 1999; Myster et al., 2000; Suzuki and Hemmati-Brivanlou, 2000). Our demonstration that the pRB-E2F pathway regulates the PRC2 genes could provide an explanation for the molecular mechanisms by which pRB and E2F are involved in the regulation of development.
The PRC2 complex is required for cell proliferation
The tight control of the PRC2 genes by the pRB-E2F pathway is further supported by our results showing that both EZH2 and EED mRNAs and proteins are strongly increased in proliferating cells as compared to non-proliferating and differentiated cells. These results are consistent with previous data showing that EZH2 and EED proteins are highly expressed in proliferating, but not in differentiated, B- and T-cells (Raaphorst et al., 2000, 2001). The expression of EZH2 and EED in actively cycling cells strongly suggests that the PRC2 complex exerts a biological role in proliferating cells. Indeed, we demonstrate that the complex is required for cellular proliferation, since siRNAs specific for EED and EZH2 block DNA replication and stop cell growth. Work from other laboratories further support this concept. Fukuyama et al. (2000) demonstrated that an antisense oligonucleotide to EZH2 suppressed DNA synthesis in HL-60 cells, while O’Caroll et al. (2001) speculated that Ezh2 has a more global role in the control of proliferation. This suggestion was based on the observation that mice deficient for EZH2 died during gastrulation; a period of embryogenesis characterized by vigorous growth and short cell cycles. Moreover, attempts to derive ES cells from blastocysts resulted in non-ES-like cells that failed to proliferate. We suggest that the PRC2 complex is required for the proliferation of most, if not all, cell types.
The PRC2 complex and tumorigenesis
One of our working hypotheses is that amongst the E2F target genes there exists as yet unidentified oncogenes. This concept is based on the fact that the deregulation of the pRB-E2F pathway is a frequent event in human cancer, and that some E2F target genes like CCNE1, MYB and MYBL2 are bona fide oncogenes (Wallrapp et al., 1997; Kononen et al., 1998; Lin et al., 2000). Here, we have extended previous observations and shown that EZH2 is highly expressed in a wide range of human cancers and not only in prostate and lymphatic cancer types (Visser et al., 2001; Varambally et al., 2002). In addition, we have demonstrated that EZH2 is amplified in 10–15% of the tested primary tumors strongly suggesting that it contributes to tumor formation, and it is not just a marker of proliferation. This suggestion was further supported by our findings that ectopic expression of EZH2 shortens the G1 phase of the cell cycle and confers a proliferative advantage in primary MEFs. Based on these findings, we suggest that EZH2 should be considered a very strong candidate oncogene. To further test this we are currently generating transgenic mice overexpressing EZH2 in specific tissues, thereby establishing a more relevant model for the study of tumorigenesis than fibroblasts.
Supporting the concept that deregulation of the PRC2 complex can contribute to cancer, is the recent finding that SUZ12 is frequently found as a fusion protein in endometrial sarcomas of the cervix (Koontz et al., 2001). Although the authors did not demonstrate that the fusion protein is causative for the development of the sarcomas, the high frequency with which it was found, taken together with the relatively low frequency with which chromosomal translocations occur in tumorigenesis, strongly suggest that the fusion protein contributes significantly to sarcoma development. These findings together with the results presented in this paper suggest that specific genetic alterations resulting in the deregulation of PRC2 activity is a frequent event in cancer.
The PRC2 complex—molecular mechanisms of action and target genes
To understand the potential mechanism by which the PRC2 complex regulates proliferation, we tested a number of potential target genes. Interestingly, the abrigation of EED or EZH2 expression did not result in increased levels of a number of negative regulators of the cell cycle, including p14ARF. However, we found that the expression of several E2F regulated genes, including the G1/S expressed cyclins, was significantly decreased in cells lacking both EZH2 and EED.
We speculate that the PRC2 complex may be required for either the activation or the maintainance of the activated state of a number of these genes. Contrary to this argument are several studies from Drosophila showing that mutations in PcG genes lead to ectopic or increased expression of homeotic (Hox) genes in developing embryos (Jacobs and van Lohuizen, 2002). The inference from this has been that the function of PcGs is to either repress or maintain the repression of target genes. However, a strong temperature-sensitive E(z) mutant in Drosophila demonstrated that lack of E(z) resulted in both an increase in the levels of certain Hox genes but significantly, a loss of expression of others (LaJeunesse and Shearn, 1996). Thus, the authors speculated that E(z) may also have a role in the maintenance of the activated transcriptional state of certain genes. These observations have been extended by the same group showing that several PcG genes, including E(z), can enhance the phenotype of TrxG mutants (Gildea et al., 2000). Based on these results, the authors proposed to call this novel group of genes for ‘Enhancer of Trithorax and Polycomb’ or ETP (reviewed in Brock and van Lohuizen, 2001). Thus, these intriguing results raise the possibility that EZH2 and EED may also have a role in the maintenance of the activated state of certain target genes. Mechanistically, we propose that the PRC2 complex is required for the activation or the maintainance of the activated state of certain genes in proliferating cells, via its capacity to methylate K27 on histone H3. The continued presence of the PRC2 complex would be required to maintain this activated state since depletion of both EZH2 and EED results in the down regulation of potential target genes such as E2F regulated genes. The PRC2 target genes that are active in growing cells could be subsequently repressed in differentiated cells via the binding of PRC1 to K27 methylated histone H3. Consistent with this, Dahiya et al. (2001) have shown that the PRC1 member, HPC2 binds to certain E2F regulated genes in association with pRB in cells induced to exit the cell cycle. However, to further investigate this novel concept it will be necessary to determine whether H3K27 methylation is associated with actively transcribing genes. In addition, further studies will be required to demonstrate the presence of PRC2 members on the promoters of actively transcribing genes or alternatively to demonstrate that the methylation of histones by PRC2 proteins plays a role in either transcriptional activation or in the maintenance of activation.
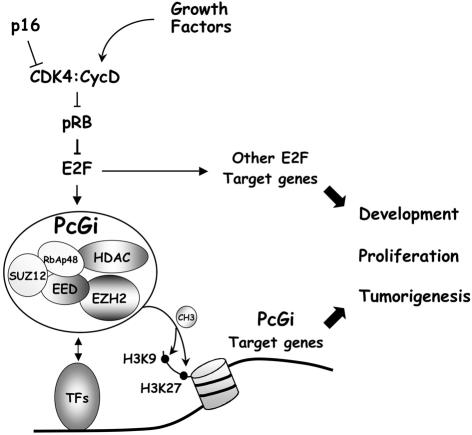
In summary, we propose a model (Figure 7) in which the transcription of the PRC2 genes is controlled by growth factors through the pRB-E2F pathway. The pRB-E2F pathway is best characterized for its role in controlling cell proliferation and its deregulation commonly contributes to tumorigenesis. The PRC2 complex in contrast has until recently been primarily functionally associated with development control. Therefore the implications of this work are several. First, we propose that the PRC2 complex is also central to proliferation control acting downstream of the pRB-E2F pathway. Significantly, its deregulation seems likely to contribute to cancer. Secondly, the role of E2F and pRB in the control of embryonic development is likely related to their ability to regulate the abundance of the PRC2 complex.
Fig. 7. Model for the cooperative role of the pRB pathway and the PRC2 complex. See Discussion for details.
Materials and methods
Quantitative PCR
cDNA was generated by RT–PCR using the PE Applied Biosystems TaqMan® Reverse Transcription Reagents. Reactions were determined using the SYBR Green I detection chemistry system (Applied Biosystems Foster City, CA), using an ABI Prism 7700 Sequence Detection System. GAPDH was used as a control gene for normalization. The sequences of the primers used are available upon request.
Microinjections
Rat1 cells were incubated in DMEM without serum for 48 h. Cell nuclei were then microinjected directly with 50 ng/µl of the indicated expression plasmids and 2 ng/µl rabbit IgG (Jackson Laboratories) using a Zeiss automatic injection system. After injection, cells were stimulated to re-enter the cell cycle by the addition of DMEM plus 10% serum. BrdU (100 µM) was added after 14 h to the culture medium for 40 min before fixation in 4% paraformaldehyde/PBS and subsequent immunofluorescence staining.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitations were performed and analyzed essentially as described previously (Frank et al., 2001). The antibodies used were anti-E2F3 (sc-878; Santa Cruz), anti-E2F4 (sc-866) and anti-Flag (F3165; Sigma). The imunoprecipitated DNA was quantified by real-time qPCR. The sequences of the PCR primers are available upon request.
Luciferase reporter assays
293T and U2OS cells were transfected in 6-well dishes with 50 ng of pCMV β-Gal plasmid and 100 ng of reporter constructs per well. Different E2Fs constructs were cotransfected at the concentrations indicated in the figure legends. RAT1 cells were cotransfected in 10 cm dishes with 0.4 µg of pCMV β-Gal and 1.6 µg of reporter constructs using the FuGene (Roche) tranfection reagent, and simultaneously serum starved and released as described above.
siRNA interference
Specific siRNA oligos targeting EZH2, EED and BMI-1 mRNAs were designed as indicated by Dharmacon Research. Several oligo sequences were synthesized by Dharmacon Research and the most efficient oligos were: EZH2, AAGACTCTGAATGCAGTTGCT; EED, AAGCACTATGTTGGCCATGGA; and BMI1, AAGGAATGGTCCACTTCCATT. A non-specific oligo (GL3) targeting the luciferase gene was also synthesized. Cells were transfected using Oligofectamine (Invitrogen) and samples for qPCR and western blot analysis were taken at the indicated time points.
MEF colony assay
Primary MEFs were prepared from 129, C57/Bl6 or mixed background of 129-C57/Bl6 embryos taken at day e13.5 during embryogenesis. The obtained results were independent of the genetic background of the MEFs. Early passage MEFs were infected with the indicated retroviral vectors. Infected cells were passaged and plated at low density (10 000 cells/plate) in 100 mm dishes and the media changed every 3–4 days. After 2 weeks, the ability to form colonies was assayed by crystal violet staining.
Tumor material and tissue microarray (TMA) construction
460 formalin fixed and paraffin embedded human tumor samples were used to construct five different TMAs (see Figure 6 for details) as previously described (Kononen et al., 1998). For each tumor sample, two 0.6 mm cylinders from both tumor and normal counterpart tissue were taken.
ISH and immunohistochemistry
EZH2 mRNA expression was assessed by ISH using [35S]UTP-labeled sense and antisense riboprobes. The ISH was performed as previously described (Rugarli et al., 1993). The slides were lightly H&E counterstained and analyzed using a darkfield condenser for the silver grains. The sequence of the probe is available upon request. For Ki67 immunostaining TMA sections were placed for 50 min in 0.25 mM EDTA at 95°C for antigen retrieval and incubated for 3 h with anti-Ki67 monoclonal antibody clone MIB1 (1:200, Immunotech, Westbrook, ME). Bound antibody was revealed using the EnVision Plus/HRP detection system (Dako) and Diaminobenzidine as chromogenic substrate. The tissues were counterstained with Hematoxylin.
Fluorescence ISH
The BAC clone RP11-731B4 localized on 7q35-q36 containing the EZH2 gene locus together with an alphoid probe, pZ75, specific for the human chromosome 7 centromeric region, were labeled with Cy3-dUTP and FluorX-dCTP (Amersham Pharmacia Biotech) respectively. Hybridiz ation specificity was first controlled on normal human buffy coat metaphase chromosome spreads. The BAC clone and the Cr.7 centromeric probe were cohybridized on TMAs as described previously (Andersen et al., 2002).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Daniele Piccini for the generation of anti-EED antibody, Emma Hickman for help in performing MEF assays, Emmanuela Frittoli for help with microinjection experiments, Manuela Nebuloni and Renzo Boldorini for providing tumor material, Micaela Quarto and Marco Bianchi for generation of TMAs and for ISH analysis, Eros Lazzerini-Denchi for his expertise in mouse myoblast differentiation and critical reading of the manuscript and Patrizia Gasparini for technical advice regarding FISH analysis. We thank Bruno Amati, Jiri Bartek, Thomas Jenuwein, Arie Otte and Liang Zhu for providing valuable reagents. We thank Maarten van Lohuizen, Pier Giuseppe Pelicci, Chris Marine and members of the Helin laboratory for discussions. We thank Luciano di Croce and Claire Attwooll for critical reading of the manuscript. This work was supported by grants from AIRC, FIRC, The European Union Fifth Framework Program, The Italian Health Ministry and the Human Science Frontiers Science Program.
References
- Andersen C.L. et al. (2002) High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am. J. Pathol., 161, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier R. et al. (2000) Induction of cyclin E-cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. EMBO J., 19, 5813–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock H.W. and van Lohuizen,M. (2001) The Polycomb group—no longer an exclusive club? Curr. Opin. Genet. Dev., 11, 175–181. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang,L., Wang,H., Xia,L., Erjument-Bromage,H., Tempst,P., Jones,R.S. and Zhang,Y. (2002) Role of Histone H3 Lysine 27 methylation in Polycomb-Group silencing. Science, 298, 1039–1043. [DOI] [PubMed] [Google Scholar]
- Cardoso C., Mignon,C., Hetet,G., Grandchamps,B., Fontes M. and Colleaux,I. (2000) The human EZH2 gene: genomic organisation and revised mapping in 7q35 within a critical region of malignant disorders. Eur. J. Hum. Genet., 8, 174–180. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi,R., McCabe,D., Seitz,V., Imhof,A. and Pirotta,V. (2002) Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell, 111,, 185–196. [DOI] [PubMed] [Google Scholar]
- Dahiya A, Wong,S., Gonzalo,S., Gavin,M. and Dean,D.C. (2001). Linking the Rb and polycomb pathways. Mol. Cell, 8, 557–569. [DOI] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Frank S.R., Schroeder,M., Fernandez,P., Taubert,S. and Amati,B. (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev., 15, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama T., Otsuka,T., Shigematsu,H., Uchida,N., Arima,F., Ohno,Y., Iwasaki,H., Fukuda,T. and Niho,Y. (2000) Proliferative involvement of ENX-1, a putative human Polycomb gene, in haematopoietic cells. Br. J. Haematol., 108, 842–847. [DOI] [PubMed] [Google Scholar]
- Gildea J.J., Lopez,R. and Shearn,A. (2000) A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics, 156, 645–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg,R.A. (2000) The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Helin K. (1998) Regulation of cell proliferation by the E2F transcription factors. Curr. Opin. Genet. Dev., 8, 28–35. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J.L. and van Lohuizen,M. (2002) Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim. Biophys. Acta, 1602, 151–161. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J.L., Kieboom,K., Marino,S., DePinho,R.A. and van Lohuizen,M. (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature, 397, 164–168. [DOI] [PubMed] [Google Scholar]
- Jiang H., Chou,H.S. and Zhu,L. (1998) Requirement of Cyclin E-Cdk2 in p16INK4a mediated growth suppression. Mol. Cell. Biol., 18, 5284–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononen J. et al. (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med., 4, 844–847. [DOI] [PubMed] [Google Scholar]
- Koontz J.I., Soreng,A.L., Nucci,M., Kuo,F.C., Pauwels,P., van den Berghe,H., Cin,P.D., Rechter J.A. and Sklar,J. (2001) Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc. Natl Acad. Sci. USA, 98, 6348–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka,K., Erdjument-Bromage,H., Tempst,P. and Reinberg,D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev., 16, 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse D. and Shearn,A. (1996) E(z): a polycomb group gene or a trithorax group gene? Development, 122, 2189–2197. [DOI] [PubMed] [Google Scholar]
- Lin L. et al. (2000) Identification and characterization of a 19q12 amplicon in esophageal adenocarcinomas reveals cyclin E as the best candidate gene for this amplicon. Cancer Res., 60, 7021–7027. [PubMed] [Google Scholar]
- Lipinski M.M. and Jacks,T. (1999) The retinoblastoma gene family in differentiation and development. Oncogene, 18, 7873–7882. [DOI] [PubMed] [Google Scholar]
- Lukas J., Sørensen,C.S., Lukas,C., Santoni-Rugiu,E. and Bartek,J. (1999) p16INK4a, but not constitutively active pRb, can impose a sustained G1 arrest: molecular mechanisms and implications for oncogenesis. Oncogene, 18, 3930–3935. [DOI] [PubMed] [Google Scholar]
- Müller H. et al. (2001) E2Fs regulate the expression of genes involved in differentiation, development, proliferation and apoptosis. Genes Dev., 15, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J. et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell, 111, 197–208. [DOI] [PubMed] [Google Scholar]
- Myster D.L., Bonnette,P.C. and Duronio,R.J. (2000) A role for the DP subunit of the E2F transcription factor in axis determination during Drosophila oogenesis. Development, 127, 3249–3261. [DOI] [PubMed] [Google Scholar]
- O’Caroll D., Erhardt,S., Pagani,M., Barton,S.C., Surani,M.A. and Jenuwein,T. (2001) The polycomb group gene Ezh2 is required for early mouse development. Mol. Cell. Biol., 21, 4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack J.R. et al. (2002) Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl Acad. Sci. USA, 99, 12963–12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst F.M., van Kememade,F.J., Fieret,E., Hamer,K.M., Satijn,D.P.E., Otte,A.P. and Meijer,C.J.L.M. (2000) Cutting edge: Polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J. Immunol., 164, 1–4. [DOI] [PubMed] [Google Scholar]
- Raaphorst F.M., Otte,A.P., van Kemekade,F.J., Blokzijl,T., Fieret,E., Hamer,K.M., Satijn,D.P.E. and Meijer,C.L.J.M. (2001) Distinct BMI-1 and EZH2 expression patterns in thymocytes and mature T cells suggest a role for Polycomb genes in human T cell differentiation. J. Immunol., 166, 5925–5934. [DOI] [PubMed] [Google Scholar]
- Ren B., Cam,H., Takahashi,Y., Volkert,T., Terragni,J., Young,R.A. and Dynlacht,B.D. (2002) E2F integrates cell cycle progression with DNA repair, replication and G2/M checkpoints. Genes Dev., 16, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugarli E.I., Lutz,B., Kuratani,S.C., Wawersik,S., Borsani,G., Ballabio,A. and Eichele,G. (1993) Expression pattern of the Kallmann syndrome gene in the olfactory system suggests a role in neuronal targeting. Nat. Genet., 4, 19–26. [DOI] [PubMed] [Google Scholar]
- Sage J., Mulligan,G.J., Attardi,L.D., Miller,A., Chen,S., Williams,B., Theodorou,E. and Jacks,T. (2000) Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev., 14, 3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. and DePinho,R.A. (2000) Mitotic clock or culture shock? Cell, 102, 407–410. [DOI] [PubMed] [Google Scholar]
- Suzuki A. and Hemmati-Brivanlou,A. (2000) Xenopus embryonic E2F is required for the formation of ventral and posterior cell fates during early embryogenesis. Mol. Cell, 5, 217–229. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees,J.A. (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol., 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Varambally S. et al. (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature, 419, 624–629. [DOI] [PubMed] [Google Scholar]
- Visser H.P.J., Gunster,M.J., Kluin-Nelemans,H.C., Manders,E.M.M., Raaphorst,R.M., Meijer,C.J.L.M., Willemze,R. and Otte,A.P. (2001) The Polycomb group protein EZH2 is upregulated in proliferating, culture mante cell lymphoma. Br. J. Haematol., 112, 950–958. [DOI] [PubMed] [Google Scholar]
- Wallrapp C., Muller-Pillasch,F., Solinas-Toldo,S., Lichter,P., Friess,H., Buchler,M., Fink,T. and Gress,T.M. (1997) Characterization of a high copy number amplification at 6q24 in pancreatic cancer identifies c-myb as a candidate oncogene. Cancer Res., 57, 3135–3139. [PubMed] [Google Scholar]
- Weinmann A.S., Bartley,S.M., Zhang,T., Zhang,M.Q. and Farnham,P.J. (2001) Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol., 21, 6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Yan,P.S., Oberley,M.J., Huang,T.H.-M. and Farnham,P.J. (2002) Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev., 16, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]